Abstract
Cytomegalovirus is a widespread opportunistic pathogen affecting immunocompromised individuals in whom neutrophils may mediate virus dissemination and contribute to progression of disease. Recent sequence analysis suggests that genes absent or altered in attenuated strains may influence pathogenesis. We have found two genes, UL146 and UL147, whose products have sequence similarity to α (CXC) chemokines. UL146 encodes a protein, designated vCXC-1, that is a 117-aa glycoprotein secreted into the culture medium as a late gene product, where its presence correlates with the ability to attract human neutrophils. Recombinant vCXC-1 is a fully functional chemokine, inducing calcium mobilization, chemotaxis, and degranulation of neutrophils. High-affinity vCXC-1 binding is shown to be mediated via CXCR2, but not CXCR1. vCXC-1 exhibits a potency approaching that of human IL-8. As the first example of a virus-encoded α chemokine, vCXC-1 may ensure the active recruitment of neutrophils during cytomegalovirus infection, thereby providing for efficient dissemination during acute infection and accounting for the prominence of this leukocyte subset in cytomegalovirus disease.
Cytomegalovirus (CMV) is an important human pathogen that interacts with different leukocyte subsets during infection (1–9). Little is known, however, about virulence determinants that enable this virus to serve as a primary pathogen causing congenital disease or as an opportunist causing disease in immunocompromised individuals (reviewed in ref. 10). Virus infection and disease are controlled by leukocytes as mediators of innate and adaptive immunity (11), but, paradoxically, leukocytes themselves may also serve as vehicles of viral dissemination (4, 12, 13). After resolution of acute infection, CMV establishes lifelong latency, remaining resident in leukocyte progenitors (6, 14) with virus recoverable from cultured peripheral blood (PB) monocytes after stimulation in culture (9).
Acute human CMV infection is associated with neutrophils in several important and well documented ways. Virus is carried in up to 0.1% of PB neutrophils (PBN) during infection of immunocompromised individuals (2, 15–20), and neutrophil-containing infiltrates are a characteristic of CMV diseases such as retinitis (21, 22), pneumonitis (23), and central nervous system complications (24) affecting AIDS patients. Although the interaction of CMV with neutrophils does not result in productive infection (16, 25, 26), recent cell culture experiments suggest that neutrophils may contribute directly to inflammation and to hematogenous dissemination in immunocompromised hosts (12, 13). This recognition raised our interest in virus-encoded functions that modulate neutrophil behavior.
CMV is species-specific and encodes more than 200 genes (27). The viral genome contains homologs of cellular genes that may modulate the host immune response, including US28, a β (CC) chemokine receptor (28), and an MHC class I homolog (29, 30). The virus also encodes functions that down-modulate host cell MHC class I antigen presentation and help evade cytotoxic T lymphocyte immunity (31, 32). Comparative studies in humans (33, 34) and severe combined immunodeficient mice implanted with human fetal thymus tissue (35) have shown that virulence characteristics of clinical isolates are lost upon laboratory passage. While analyzing sequence differences between lab strains and the virulent Toledo strain (33, 34), a 15-kb pair region with 19 additional ORFs, denoted UL133–UL151, was identified in clinical strains (36). Two of these genes, UL146 and UL147, exhibit size and limited sequence similarity to α (CXC) chemokines, suggesting that they influence the behavior of neutrophils during infection.
Chemokines are small, chemoattractant cytokines that serve to coordinate trafficking of PB leukocytes by stimulating adhesion, chemotaxis, extravasation, and other immune effector functions via G protein-coupled receptors (37, 38). Most known chemokines are categorized as either α (CXC) or β (CC) based on the spacing of cysteines near the N terminus that dictates the spectrum of biological activities. In addition α chemokines generally exert their influence on neutrophils whereas β chemokines affect other cells including monocytes, eosinophils, and basophils. Two functional β chemokine homologs are encoded by Kaposi’s sarcoma herpesvirus, vMIP-I and vMIP-II (39, 40), and one is encoded by the poxvirus molluscum contagiosum, the MC148 gene product (41, 42). Sequence predictions suggest a β chemokine may be encoded by murine CMV (43). vMIP-II and MC148 proteins bind chemokine receptors and can function as antagonists of normal PB leukocyte response to host chemokines (39, 41, 42). An N-terminally processed form of vMIP-II has been reported to have direct stimulatory activity (40). In addition to modulating local inflammatory responses and antiviral immunity (8, 44), stimulatory chemokines may impact on leukocyte recruitment to sites of viral infection. Here we have completed a detailed analysis of the first viral chemokine in the α class, vCXC-1, demonstrating expression and function, that strongly implicates this chemokine as a determinant of host neutrophil behavior in response to CMV infection.
MATERIALS AND METHODS
Viruses, Cells, and Recombinant Protein.
Low-passage (<15 passages) human embryonic lung fibroblasts (HEL) were cultured in DMEM (JRH Biosciences, Lenexa, KS) supplemented with 10% FBS (HyClone) (45). CMV strain Toledo (passage 8) (34, 35) and other viruses were prepared as described previously (45). The Y18 and T23 forms of recombinant vCXC-1 were custom-synthesized by R&D Systems as E. coli-derived recombinant proteins of >98% purity.
DNA Cloning.
Toledo cosmids were derived according to published techniques (46). The termini of cosmid Tol122, derived from the internal junction region of a Toledo genome with an inverted S component, correspond to Toledo nucleotide 2,804 (accession no. U33331) and AD169 nucleotide 206,850 (accession no. 17403). Tol122ΔSca (Toledo nucleotides 2804–15706) was derived from Tol122 by ScaI digestion and ligation to ScaI/PacI adapters (TAAGCGAGT; ACTCGCTTAAT). Tol122ΔEco was derived from Tol122 by partial EcoRI digestion and ligation to EcoRI/PacI adapters (AAACGCCCGGGCGG; TTTGCGGGCCCGCCTTAA). Tol122Δ138–148 deleted Tol122ΔSca from the NsiI site (nucleotide 10,836) to the BamHI site at 3,274. Tol122Δ146–148 deleted Tol122ΔSca from the NsiI site (nucleotide 10,836) to the BamHI site at 9,328 (Fig. 1A). pMP109 was generated by ligating the 4,040-bp ApaI fragment (Toledo nucleotides 9,603–13,644) from Tol122ΔSca into ApaI-digested pGEM7 (Promega). pMP109ΔHpaI was derived by ligation of a double-stranded oligonucleotide (GACTACAAGGACGACGACGACAAG; CTTGTCGTCGTCGTCCTTGTAGTC) encoding the FLAG epitope (47), into HpaI-digested pMP109. Tol146ΔHpaI was constructed by ligation of the 4,064- and 13,043-bp ApaI fragments from pMP109ΔHpaI and Tol122ΔScaI, respectively.
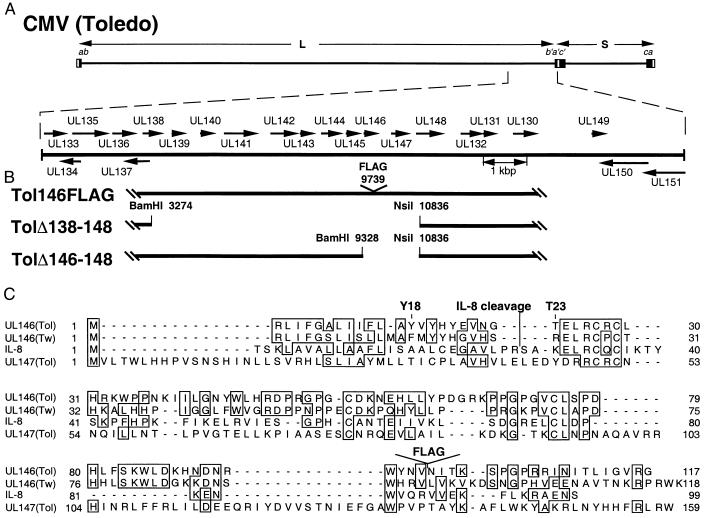
Figure 1.
Schematic representation of strain Toledo and Toledo-derived mutant virus genomes. (A, upper line) CMV strain Toledo genome structure with the invertable L and S genome segments indicated by arrows and inverted repeats (ab-b′a′c′-ca) indicated by boxes. Expanded region is the ULb′ region, with ORFs depicted as arrows. (B) Structures of the recombinant viruses. Nucleotide numbers correspond to Toledo sequences (accession no. U33331). (C) Sequence comparison between UL146 (Toledo), UL146 (Towne), UL147, and human IL-8. Identities are boxed, and the two most likely signal peptide cleavage sites predicted by computer algorithm are marked (Y18 and T23). The cleavage site of mature IL-8 and FLAG insertion site in UL146 (Toledo) also are indicated.
Construction of Recombinant Viruses.
A set of nine overlapping cosmids representing the entire Toledo viral genome was transfected into HEL (46). Tol146ΔHpaI was combined with eight other cosmids to generate Tol146FLAG. Tol146FLAG recombinant viruses from two separate transfection pools were analyzed and evaluated by DNA blot hybridization. Tol122Δ138–148 and Tol122Δ146–148 were used to generate recombinant viruses TolΔ138–148 and TolΔ146–148, respectively. TolΔ138–148 is deleted from the C terminus of UL138 (amino acid 153) through the N terminus of UL148 (amino acids 64), and TolΔ146–148 is deleted from 119 nucleotides upstream of UL146 through the N terminus of UL148 (amino acid 64). These viruses were plaque-purified three times before production of working stocks. Tol122ΔSca was used to generate a cosmid-derived Toledo virus, which was used as a control. For DNA blot analysis, probe DNA was labeled with fluoresceinated uracil according to the manufacturer’s instructions (Tropix, Bedford, MA), hybridized to the immobilized DNA as described previously (45), and developed by using enhanced chemiluminescence (ECL; Amersham).
Protein Preparations and Detection.
HEL were infected at a multiplicity of infection (moi) of 3.0. At various times cells were lysed in lysis buffer [0.5% Triton X-100 (Baker)/50 mM Tris⋅Cl, pH 7.4/150 mM NaCl/protease inhibitors (Boehringer Mannheim)] on ice for 10 min. Antibodies were detected by using the ECL system (Amersham). FLAG-tagged vCXC-1 was immunoprecipitated from cleared supernatants by using Sepharose-bound anti-FLAG M2 Ab (Kodak) according to the manufacturer’s instructions. Peptide N-glycosidase F (PNGase F) treatment was done according to the manufacturer’s instructions (NEB, Beverly, MA). Virions from infected cell supernatants were purified over three, consecutive, 20–70% sucrose gradients by standard techniques (48). For immunofluorescence, HEL on glass coverslips were infected at an moi of 3.0 or mock-infected, fixed, and stained by using published methods (49) and examined on a confocal microscope (Molecular Dynamics).
Neutrophil Chemotaxis Assays.
To assay vCXC-1, HEL were infected at an moi of 3 and incubated in DMEM + 2% dialyzed FBS, and supernatants were cleared of virions and cell debris by centrifugation at 12,000 × g for 30 min. Chemotaxis assays were performed in 24-well chemotaxis chambers (Costar). Migration of neutrophils through a 3-μm polycarbonate filter was measured by determination of β-glucuronidase activity in the lower chamber (50). For the recombinant protein, neutrophil chemotaxis assays were performed in a 96-well format. Migrated cells were stained on the filter using Leukostat (Fisher) and read at OD 540 nm on a plate reader (Bio-Rad) (51). Assays were performed in triplicate on human PBN, isolated as described (52), from 14 different donors.
Binding Analyses.
Fresh PBN or NSO cell transfectants carrying CXCR1 or CXCR2 (38) were incubated at room temperature with 0.1 nM (125I) IL-8 (Amersham Pharmacia) and various unlabeled chemokines (R & D Systems) in 25 mM Hepes/140 mM NaCl/1 mM CaCl2/5 mM MgCl2/0.2% BSA, pH 7.1. Cells were aspirated onto polyethyleneimine-treated GF/C glass filters (Packard) by using a cell harvester (Packard) and washed twice with 25 mM Hepes/500 mM NaCl/1 mM CaCl2/5 mM MgCl2, pH 7.1. Scintillant was added to dried filters, and cpm was measured on a Packard Topcount Scintillation counter.
Cytoplasmic Calcium Mobilization.
Calcium mobilization was measured by using a Photon Technology International spectrofluorometer (Photon Technology International, Princeton) with excitation at 350 nm and dual-emission recording at 400 and 490 nm. Relative intracellular calcium levels were expressed as the ratio of emission at 400 nm to the emission at 490 nm. Fresh PBN were loaded with 3 μM INDO-1AM (Molecular Probes) for 60 min at 37°C, and then chemokines were added with constant mixing in cuvettes containing 106 cells in 2 ml of Hanks’ balanced salt solution with 25 mM Hepes/1.6 mM CaCl2/5 mM MgCl2. To determine the chemokine concentration giving rise to a half-maximal excitation response (EC50), the maximal amplitudes of the responses (400:490 ratio) were plotted against chemokine concentrations and analyzed by using igor pro software (WaveMetrics, Lake Oswego, OR). As a measure of receptor usage and signaling efficacy, neutrophils were stimulated sequentially with 100 nM chemokines, a concentration typically two orders of magnitude above the EC50 value.
RESULTS
The UL146 Gene Product, vCXC-1, Is Secreted Late During Infection.
Sequence analysis of the nonattenuated CMV strain, Toledo, revealed that UL146 and UL147, which we have designated vCXC-1 and vCXC-2, respectively, had sequence motifs reminiscent of the prototype α chemokine, IL-8. These features include putative signal peptides, cysteine spacing, size, and, in the case of vCXC-1, an ELRCXC motif, known to be important in receptor binding and activation of neutrophils (53) (Fig. 1C). A positional homolog, UL152, is found in the attenuated Towne strain (36) that we now believe is derived from UL146 (Fig. 1C).
We prepared recombinant viruses from strain Toledo cosmids (46) to disrupt expression of gene blocks UL138–UL148 (TolΔ138–148) or UL146–UL148 (TolΔ146–148). We also introduced an in-frame FLAG epitope between amino acid residues 97 and 98 of vCXC-1 (Fig. 1B), creating a recombinant virus (Tol146FLAG) predicted to encode a modified vCXC-1 that would retain native cellular localization patterns. Two independent isolates of Tol146FLAG were generated and behaved similarly in all experiments. All mutants replicated as well as parental Toledo strain in HEL, confirming the disrupted genes were dispensable for replication in vitro. All were subjected to restriction digest pattern and hybridization analyses, suggesting they lacked adventitious mutations (not shown).
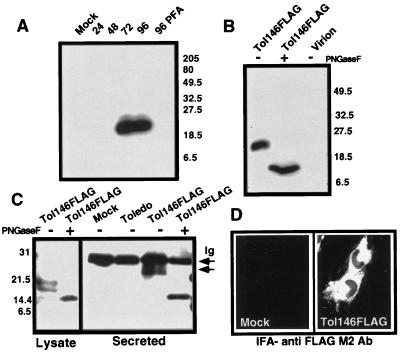
To evaluate localization and kinetics of expression of vCXC-1, Tol146FLAG-infected HEL lysates were prepared at 24, 48, 72, and 96 hr postinfection (hpi), separated by SDS/PAGE, and immunoblotted with a FLAG-specific Ab. A protein with an apparent molecular weight (MW) of 22,000 was detected at 72 and 96 hpi in Tol146FLAG-infected cells, but not in strain Toledo or mock-infected cells. Tagged vCXC-1 is expressed with late (γ2) kinetics, because treatment of the cells with phosphonoformic acid inhibited its expression (Fig. 2A).
Figure 2.
Expression of FLAG-tagged vCXC-1 by Tol146FLAG. (A) Detection of FLAG-tagged vCXC-1 in cell lysates by immunoblot with anti-FLAG M2 Ab. Whole-cell lysates were harvested from uninfected (Mock) or Tol146FLAG-infected cells at 24, 48, 72, and 96 hpi. Cell lysates also were collected at 96 hpi from Tol146 FLAG-infected cells cultured in the presence of 660 μM phosphonoformic acid (96 PFA). (B) Structural analysis of FLAG-tagged vCXC-1. Infected cell lysates and virions, enriched by sucrose gradient, were harvested at 96 hpi and treated with PNGase F as indicated (+). The proteins were immunoblotted with anti-FLAG M2 Ab [Coomassie staining confirmed the composition of the virion preparation: viral (pp65) but not cellular (β-actin) antigens were detected by immunoblotting.] (C) Immunoprecipitation of FLAG-tagged vCXC-1 from medium (Secreted) and cell lysates. Protein was harvested at 96 hpi from mock-infected, Toledo, and Tol146FLAG-infected fibroblasts, treated with PNGase F as indicated (+), and precipitated with anti-FLAG M2 Ab. The arrow labeled “Ig” marks a nonspecific reaction with Ig light chain. The arrow below denotes the position of 25,000- to 27,000-MW secreted form of FLAG-tagged vCXC-1. (D) Mock- and Tol146FLAG-infected fibroblasts were stained with anti-FLAG M2 Ab and goat anti-mouse FITC at 96 hpi. Toledo-infected and antibody controls were negative.
We next evaluated whether vCXC-1 was glycosylated and secreted in a manner similar to chemokines. PNGase F-treated Tol146FLAG-infected cell lysates (96 hpi) contained a species with an apparent MW of 14,000 (Fig. 2B). Two distinct, modified PNGase F-sensitive forms were present in cell lysates (Fig. 2C). Partial PNGase F digestion suggested that at least two of three predicted glycosylation sites were employed during protein maturation (data not shown). The extracellular form of FLAG-tagged vCXC-1 present in medium remained PNGase F-sensitive (Fig. 2C) but migrated with a size (25,000–27,000 MW) that suggested modification with additional carbohydrate residues during secretion (Fig. 2C). Importantly, and consistent with it being a secreted protein, FLAG-tagged vCXC-1 localized to infected cell cytoplasm (Fig. 2D) and was not detected in virion-enriched preparations (Fig. 2B).
vCXC-1 Is a Potent α Chemokine.
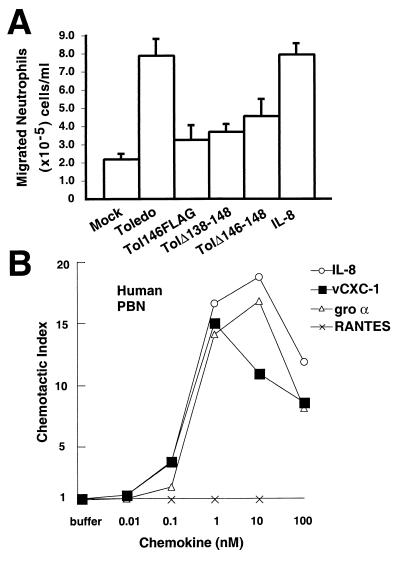
We next examined the ability of virus-encoded vCXC-1 to stimulate chemotaxis. Clarified supernatants from infected cells were separated from freshly prepared PBN by a 3-μm membrane, and their migration was measured (50). Medium collected at 96 hpi from Toledo-infected cells was twice as potent at attracting neutrophils as medium from cells infected with any of the three mutants, TolΔ138–148, TolΔ146–148, and Tol146FLAG (Fig. 3A). This suggests that disruption of vCXC-1, including a 9-aa insertion between residues 97 and 98, was sufficient to disrupt neutrophil attraction, although not to the level exhibited by mock-infected cell medium, suggesting CMV-induced cellular or other CMV-encoded chemokines also contribute to migration (54). Toledo-infected cell medium exhibited potency 4-fold higher than mock-infected cell medium, equivalent to a maximum effective dose of IL-8 added to this medium.
Figure 3.
vCXC-1-induced chemotaxis of human PBN. (A) Chemotaxis of neutrophils toward cell-free supernatants. IL-8 (12 nM) was added to mock-infected cell supernatants as a positive control (IL-8). (Bars = 2 SD.) (B) Induction of migration of PBN by recombinant vCXC-1(T23 form) compared with IL-8 and gro α. RANTES, a CC chemokine, is included as negative control. Chemotactic index represents the fold increase in migration over a basal level in the absence of chemokine.
To determine directly the chemokine activity of vCXC-1, we obtained two forms of recombinant vCXC-1 based on predicted signal peptide cleavage sites, encoding either a 100-aa protein from tyrosine 18 (Y18) or a 95-aa protein from threonine 23 (T23) (Fig. 1C). These were compared with known α chemokines in chemotaxis assays. One of the recombinant forms of vCXC-1, T23, induced potent PBN chemotaxis with activity and dose response virtually indistinguishable from that of IL-8 and gro α (Fig. 3B). Glycosylation, therefore, was not necessary for activity.
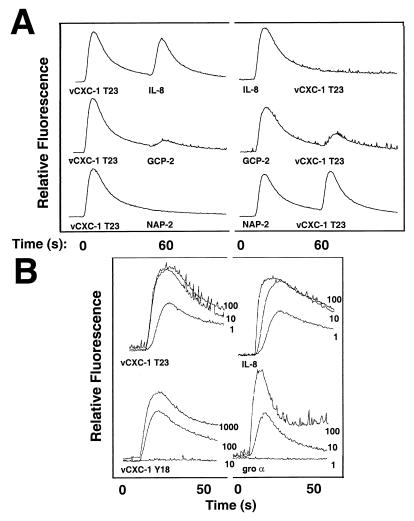
To assess further the functional differences, cytoplasmic calcium-mobilization experiments were carried out on human PBN to measure vCXC-1 Y18- or T23-induced intracellular signaling. The T23 form mobilized cytoplasmic calcium with a dose response similar to that of human IL-8, significantly greater than was achieved with the Y18 form (Fig. 4B). The EC50 differed by approximately 50-fold between the T23 and Y18 forms (not shown). Additional analyses showed vCXC-1 and IL-8 signaling in PBN was coupled to a pertussis toxin-sensitive pathway (not shown), suggesting a common mechanism of signal transduction. T23 (but not Y18) also induced neutrophil degranulation, being nearly equipotent with IL-8 at inducing myeloperoxidase release from cytochalasin B-treated PBN (not shown). Taken together, the signaling, degranulation, and migration data revealed that the T23 form of vCXC-1 exhibited robust chemokine activity, acting as a potent IL-8 mimetic on primary human cells.
Figure 4.
Analysis of active form of recombinant vCXC-1 and its effects on human PBN. (A) Heterologous desensitization profiles of listed chemokines on PBN. (Left) T23 form of vCXC-1 added as initial agonist, followed 60 sec later by other human chemokines, as listed. (Right) Human chemokines added first, followed by the T23 form of vCXC-1. (B) Cytoplasmic calcium induction by Y18 and T23 forms of vCXC-1; comparison with human IL-8 and gro α [chemokine (in nM) is indicated on the right of each data set].
vCXC-1 Desensitizes Signaling to Human α Chemokines.
To assess the relative potency of vCXC-1, a series of heterologous desensitization experiments was performed on PBN. Fig. 4A shows representative assays. vCXC-1 T23 induced robust calcium mobilization and, with two exceptions, left cells desensitized to other human ELRCXC chemokines tested (NAP-2, gro α, gro β, gro γ, and ENA-78; Fig. 4A and data not shown). IL-8 and, to a lesser extent, GCP-2 were able to induce a second calcium mobilization after exposure to vCXC-1 (Fig. 4A Left). When ELRCXC chemokines were added to PBN in advance of vCXC-1, only IL-8 completely blocked the ability of vCXC-1 to induce calcium mobilization. These data show that vCXC-1 is a potent chemokine agonist of neutrophil signaling.
Recombinant vCXC-1 Binds CXCR2.
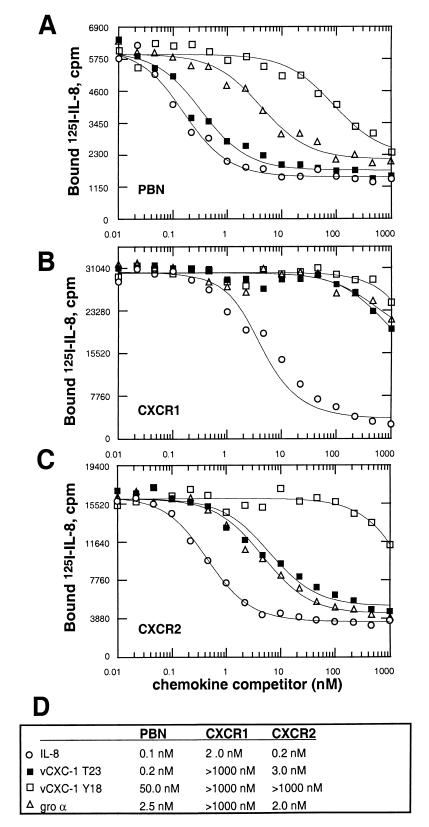
The above data suggest that vCXC-1 binds to endogenous chemokine receptors that normally bind human α chemokines. Therefore, we determined whether vCXC-1 exhibited the ability to bind to CXCR1 (predominantly an IL-8 receptor), CXCR2 (a receptor for all ELRCXC chemokines), or both. A series of equilibrium-binding studies was performed examining the ability of vCXC-1 to compete with (125I) IL-8 by using PBN as well as cells stably expressing transfected CXCR1 or CXCR2. On PBN, the T23 form of vCXC-1, competed for IL-8 binding nearly as well as unlabeled IL-8. The Y18 form was much less able to compete, and the human chemokine gro α exhibited an intermediate profile (Fig. 5A). In transfectant cell lines expressing individual α chemokine receptors, vCXC-1 bound CXCR2 with no indication of binding to CXCR1 (Fig. 5 B and C). Fig. 5D shows competitive binding constants (Ki) for each interaction as derived from Scatchard plot analysis of the data. We do not yet know whether the differences in apparent Ki that we observed in PBN and CXCR2 transfectants (0.2 nM vs. 3.0 nM) represent differences in cellular environment of receptors or suggest that other receptors are involved in vCXC-1 binding. Analysis of binding competition on cell lines expressing single chemokine receptors (CCR1–CCR8, CXCR1–CXCR4, CX3R1, and US28) showed that vCXC-1 bound only CXCR2 (data not shown). The binding data, therefore, showed that vCXC-1 binds with high affinity to PBN, likely via CXCR2, and is able to compete efficiently with human IL-8.
Figure 5.
Equilibrium-binding competition profiles of vCXC-1 on human PBN and individual CXC chemokine receptor-bearing cells. Unlabeled chemokines were incubated along with 0.1 nM 125I-IL-8 by using primary human neutrophils (A) or transfected murine NSO cells stably expressing human chemokine receptors CXCR1 (B) or CXCR2 (C). (D) Competition constants (Ki) for each interaction as derived from Scatchard analysis of the data.
DISCUSSION
Herpesviruses and poxviruses express an array of functions that influence the host immune response, many of which appear to have been transduced from the host genome. The viral β (CC) chemokine homologs (vMIP-1, vMIP-II, and MC148) generally have antagonist properties in contrast to their cellular counterparts (39–42). CMV vCXC-1, the first α (CXC) chemokine to be found in a virus, binds with high affinity to the CXCR2 receptor and, despite its extensive sequence divergence, has retained an agonist profile with potency similar to IL-8. Our studies with FLAG-tagged vCXC-1 suggest that C-terminal conformation may be important for neutrophil attraction. The activity of vCXC-1 documents the importance to this virus of controlling host neutrophil behavior.
Although attraction of host neutrophils seems counterproductive for a virus, vCXC-1 may afford CMV a selective advantage as an IL-8 mimetic. At least two types of benefits can be envisaged. The first would be evasion of the host’s immune response via vCXC-1 disruption of normal cell migration patterns or by preferentially attracting leukocytes that cannot efficiently clear virus. Either of these would allow virus to escape immune clearance and achieve more successful infection. The second would focus on recruitment of leukocyte subsets that selectively migrate in response to vCXC-1 and sustain dissemination during acute infection to ensure access to progenitor leukocyte populations for latency. Leukocytes and their progenitors are prime candidates for dissemination and as sites of latency. Although an understanding of the natural history of CMV infections is far from complete, entry and initial replication often take place at mucosal sites from which virus disseminates systemically to epithelium in the salivary glands and kidney. From these sites, shedding facilitates transmission to new hosts. As the virus gains a foothold, the host mounts an immune response that ultimately controls replication and drives the virus into latency characterized by sporadic reactivation and shedding throughout the lifetime of the host. Viral functions that modulate leukocyte behavior clearly are desirable during all phases of the virus–host interaction. In either of the two scenarios outlined above, expression of vCXC-1 would increase the likelihood of viral dissemination within the host as well as viral transmission between hosts.
Sequence divergence also has been observed within UL146 in a large number of clinical strains (unpublished work), suggesting that variability in this gene may underlie the characteristic pathogenic signature ascribed to different strains in CMV disease (55). It is possible that the absence or divergence of UL146 in AD169 (27) and Towne (36), respectively, contributes to their attenuation.
CMV emerges in immunocompromised individuals when cytotoxic T lymphocyte-mediated immunity fails (11). Under these circumstances, innate or adaptive immunity may contribute to disease progression as the response to viral replication or viral gene products such as vCXC-1 proceeds. In AIDS patients, neutrophil infiltrates have been shown to dominate the immunopathology of CMV-retinitis (21, 22). This immunopathology is not seen in retinal disease of immunocompromised transplant recipients, who lack innate immunity. Thus, progression from infection to disease may depend on vCXC-1 attraction of neutrophils in settings in which innate immunity is intact, such as encephalitis, pneumonitis, carditis, and hepatitis. To the extent that vCXC-1 is a virulence determinant and may control the spread of virus, it should be considered in the design of live-attenuated vaccines and targeted for the development of disease-specific therapeutic drugs.
Active CMV infection in immunocompromised hosts relies on hematologic spread via neutrophils, where a role for vCXC-1 as a potent neutrophil-attracting chemokine seems best supported by clinical and experimental evidence (4, 12, 13). vCXC-1 mimics host IL-8 and affects human neutrophil migration as well as degranulation. The strong neutrophil-stimulatory properties of vCXC-1 would bring neutrophils to sites of viral infection where virus-infected cells would be phagocytosed. vCXC-1 neutrophil-attracting qualities do not fully explain the trafficking of these cells from infected sites back to the bloodstream, although persistent replication and continued expression of late gene products, including vCXC-1, might produce sufficient recruitment that neutrophils spill out of sites of infection. In addition, other viral gene products such as CMV US28, which are made during infection, may influence the migration of infected leukocyte populations away from sites of infection. Recent evidence (56, 57) suggests that this virus-encoded chemokine receptor may influence the migration of infected cells to host β chemokines. The picture that emerges may be one in which the virus uses chemokines and chemokine receptors to dictate migration of host cells before and after they become infected.
Acknowledgments
We thank Monica Tsang, Amy Isaacson, Ryan Olson, Mary Deitz, and John Humphrey of R & D Systems for technical assistance, as well as Richard Spaete and Martin Bryant for their comments on the manuscript. This work was funded partly by Public Health Service Grant AI30363.
ABBREVIATIONS
- CMV
cytomegalovirus
- PBN
peripheral blood neutrophil(s)
- HEL
human embryonic lung fibroblasts
- PNGase F
peptide N-glycosidase F
- hpi
hours postinfection
References
- 1.Taylor-Wiedeman J, Sissons P, Sinclair J. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saltzman R L, Quirk M R, Jordan M C. J Clin Invest. 1988;81:75–81. doi: 10.1172/JCI113313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor-Wiedeman J, Sissons J G, Borysiewicz L K, Sinclair J H. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 4.Dankner W M, McCutchan J A, Richman D D, Hirata K, Spector S A. J Infect Dis. 1990;161:31–36. doi: 10.1093/infdis/161.1.31. [DOI] [PubMed] [Google Scholar]
- 5.Theodossiou C, Temeck B, Vargas H, Yang J, Vargas M, Hahn S, Pass H. Am J Gastroenterol. 1995;90:1174–1176. [PubMed] [Google Scholar]
- 6.Kondo K, Xu J, Mocarski E S. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo K, Kaneshima H, Mocarski E S. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendelson M, Monard S, Sissons P, Sinclair J. J Gen Virol. 1996;77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 9.Soderberg-Naucler C, Fish K N, Nelson J A. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 10.Ho M. Cytomegalovirus: Biology and Infection. 2nd Ed. New York: Plenum; 1991. [Google Scholar]
- 11.Riddell S R, Watanabe K S, Goodrich J M, Li C R, Agha M E, Greenberg P D. Science. 1992;257:238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 12.Revello M G, Percivalle E, Arbustini E, Pardi R, Sozzani S, Gerna G. J Clin Invest. 1998;101:2686–2692. doi: 10.1172/JCI1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundy J E, Lawson K M, MacCormac L P, Fletcher J M, Yong K L. J Infect Dis. 1998;177:1465–1474. doi: 10.1086/515300. [DOI] [PubMed] [Google Scholar]
- 14.Hahn G, Jores R, Mocarski E S. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiala M, Edmondson L, Guze L B. Proc Soc Exp Biol Med. 1973;144:871–875. doi: 10.3181/00379727-144-37701. [DOI] [PubMed] [Google Scholar]
- 16.Martin D C, Katzenstein D A, Yu G S M, Jordan M C. Ann Intern Med. 1984;100:222–225. doi: 10.7326/0003-4819-100-2-222. [DOI] [PubMed] [Google Scholar]
- 17.The T H, van der Ploeg M, van den Berg A P, Vlieger A M, van der Giessen M, van Son W J. Transplantation. 1992;54:193–198. doi: 10.1097/00007890-199208000-00001. [DOI] [PubMed] [Google Scholar]
- 18.van der Bij W, Schirm J, Torensma R, van Son W J, Tegzess A M, The T H. J Clin Microbiol. 1988;26:2531–2535. doi: 10.1128/jcm.26.12.2531-2535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaia J A, Forman S J, Gallagher E, Vanderwal-Urbina E, Blume K G. Transplantation. 1984;37:315–317. doi: 10.1097/00007890-198403000-00021. [DOI] [PubMed] [Google Scholar]
- 20.Turtinen L W, Salzman R, Jordan M C, Haase A T. Microb Pathog. 1987;3:287–297. doi: 10.1016/0882-4010(87)90062-3. [DOI] [PubMed] [Google Scholar]
- 21.Holland G N, Pepose J S, Pettit T H, Gottlieb M S, Yee R D, Foos R Y. Ophthalmology. 1983;90:859–873. doi: 10.1016/s0161-6420(83)80009-8. [DOI] [PubMed] [Google Scholar]
- 22.Pepose J S, Holland G N, Nestor M S, Cochran A J, Foos R Y. Ophthalmology. 1985;92:472–484. doi: 10.1016/s0161-6420(85)34008-3. [DOI] [PubMed] [Google Scholar]
- 23.Duggan M A, Pomponi C, Robboy S J. Diagn Cytopathol. 1986;2:181–186. doi: 10.1002/dc.2840020302. [DOI] [PubMed] [Google Scholar]
- 24.Granter S R, Doolittle M H, Renshaw A A. Am J Clin Pathol. 1996;105:364–366. doi: 10.1093/ajcp/105.3.364. [DOI] [PubMed] [Google Scholar]
- 25.Gerna G, Revello M G, Percivalle E, Zavattoni M, Parea M, Battaglia M. J Clin Microbiol. 1990;28:2681–2688. doi: 10.1128/jcm.28.12.2681-2688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grefte A, Harmsen M C, van der Giessen M, Knollema S, van Son W J, The T H. J Gen Virol. 1994;75:1989–1998. doi: 10.1099/0022-1317-75-8-1989. [DOI] [PubMed] [Google Scholar]
- 27.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A I, Kouzarides T, Martignetti J A, et al. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 28.Neote K, DiGregorio D, Mak J Y, Horuk R, Schall T J. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 29.Browne H, Smith G, Beck S, Minson T. Nature (London) 1990;347:770–772. doi: 10.1038/347770a0. [DOI] [PubMed] [Google Scholar]
- 30.Leong C C, Chapman T L, Bjorkman P J, Formankova D, Mocarski E S, Phillips J H, Lanier L L. J Exp Med. 1998;187:1681–1687. doi: 10.1084/jem.187.10.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert M J, Riddell S R, Plachter B, Greenberg P D. Nature (London) 1996;383:720–722. doi: 10.1038/383720a0. [DOI] [PubMed] [Google Scholar]
- 32.Jones T R, Hanson L K, Sun L, Slater J S, Stenberg R M, Campbell A E. J Virol. 1995;69:4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinnan G, Jr, Delery M, Rook A H, Frederick W R, Epstein J S, Manischewitz J F, Jackson L, Ramsey K M, Mittal K, Plotkin S A, et al. Ann Intern Med. 1984;101:478–483. doi: 10.7326/0003-4819-101-4-478. [DOI] [PubMed] [Google Scholar]
- 34.Plotkin S A, Starr S E, Friedman H M, Gonczol E, Weibel R E. J Infect Dis. 1989;159:860–865. doi: 10.1093/infdis/159.5.860. [DOI] [PubMed] [Google Scholar]
- 35.Brown J M, Kaneshima H, Mocarski E S. J Infect Dis. 1995;171:1599–1603. doi: 10.1093/infdis/171.6.1599. [DOI] [PubMed] [Google Scholar]
- 36.Cha T A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baggiolini M, Dewald B, Moser B. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 38.Premack B A, Schall T J. Nat Med. 1996;2:1174–1178. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 39.Kledal T N, Rosenkilde M M, Coulin F, Simmons G, Johnsen A H, Alouani S, Power C A, Luttichau H R, Gerstoft J, Clapham P R, et al. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 40.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, et al. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 41.Krathwohl M D, Hromas R, Brown D R, Broxmeyer H E, Fife K H. Proc Natl Acad Sci USA. 1997;94:9875–9880. doi: 10.1073/pnas.94.18.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Damon I, Murphy P M, Moss B. Proc Natl Acad Sci USA. 1998;95:6403–6407. doi: 10.1073/pnas.95.11.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacDonald M R, Li X Y, Virgin H T., III J Virol. 1997;71:1671–1678. doi: 10.1128/jvi.71.2.1671-1678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dairaghi D, Greaves D R, Schall T J. Semin Virol. 1998;8:377–385. [Google Scholar]
- 45.Spaete R R, Mocarski E S. J Virol. 1985;56:135–143. doi: 10.1128/jvi.56.1.135-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kemble G, Duke G, Winter R, Spaete R. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hopp T P, Prickett K S, Price V, Libby R T, Marhc C J, Cerretti P, Urdal D L, Conlon P J. Biotechnology. 1988;6:1205–1210. [Google Scholar]
- 48.Britt W J. Virology. 1984;135:369–378. doi: 10.1016/0042-6822(84)90193-4. [DOI] [PubMed] [Google Scholar]
- 49.Penfold M E, Armati P, Cunningham A L. Proc Natl Acad Sci USA. 1994;91:6529–6533. doi: 10.1073/pnas.91.14.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroder J M, Mrowietz U, Morita E, Christophers E. J Immunol. 1987;139:3474–3483. [PubMed] [Google Scholar]
- 51.Bazan J F, Bacon K B, Hardiman G, Wang W, Soo K, Rossi D, Greaves D R, Zlotnik A, Schall T J. Nature (London) 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 52.Markert M, Andrews P C, Babior B M. Methods Enzymol. 1984;105:358–365. doi: 10.1016/s0076-6879(84)05048-5. [DOI] [PubMed] [Google Scholar]
- 53.Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. J Biol Chem. 1991;266:23128–23134. [PubMed] [Google Scholar]
- 54.Craigen J L, Yong K L, Jordan N J, MacCormac L P, Westwick J, Akbar A N, Grundy J E. Immunology. 1997;92:138–145. doi: 10.1046/j.1365-2567.1997.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shepp D H, Match M E, Ashraf A B, Lipson S M, Millan C, Pergolizzi R. J Infect Dis. 1996;174:184–187. doi: 10.1093/infdis/174.1.184. [DOI] [PubMed] [Google Scholar]
- 56.Michelson S, Dal Monte P, Zipeto D, Bodaghi B, Laurent L, Oberlin E, Arenzana-Seisdedos F, Virelizier J L, Landini M P. J Virol. 1997;71:6495–6500. doi: 10.1128/jvi.71.9.6495-6500.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vieira J, Schall T J, Corey L, Geballe A P. J Virol. 1998;72:8158–8165. doi: 10.1128/jvi.72.10.8158-8165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]