Abstract
hrp genes are reportedly required for pathogenicity in Pseudomonas syringae pv. syringae (Pss) and other phytopathogenic bacterial species. A subset of these genes encodes a type III secretion system through which virulence factors are thought to be delivered to plant cells. In this study, we sought to better understand the role that hrp genes play in interactions of Pss with its host as they occur naturally under field conditions. Population sizes of hrp mutants with defects in genes that encode components of the Hrp secretion system (ΔhrcC∷nptII and hrpJ∷ ΩSpc) and a protein secreted via the system (ΔhrpZ∷nptII) were similar to B728a on germinating seeds. However, phyllosphere (i.e., leaf) population sizes of the hrcC and hrpJ secretion mutants, but not the hrpZ mutant, were significantly reduced relative to B728a. Thus, the Hrp type III secretion system, but not HrpZ, plays an important role in enabling Pss to flourish in the phyllosphere, but not the spermosphere. The hrcC and hrpJ mutants caused brown spot lesions on primary leaves at a low frequency when they were inoculated onto seeds at the time of planting. Pathogenic reactions also were found when the hrp secretion mutants were co-infiltrated into bean leaves with a non-lesion-forming gacS mutant of B728a. In both cases, the occurrence of disease was associated with elevated population sizes of the hrp secretion mutants. The role of the Hrp type III secretion system in pathogenicity appears to be largely mediated by its requirement for growth of Pss in the phyllosphere. Without growth, disease does not occur.
Pseudomonas syringae pv. syringae (Pss) is a common inhabitant of leaves and other aerial parts of a diversity of plant species (1). On snap bean plants (Phaseolus vulgaris L.), the bacterium causes bacterial brown spot disease (2). Foliar symptoms are necrotic spots frequently surrounded by a narrow chlorotic margin. Lesions on pods lower the quality of the product and cause economic loss. Although disease causation is of central importance from the perspective of humans, it is only one of several interactions that occur between Pss and its host. In the field, the bacterium is able to multiply and survive for many generations on leaf surfaces of host and nonhost plants, similar to the numerous nonpathogenic bacteria that exist as epiphytes in the phyllosphere (1, 3, 4).
Epiphytic populations of Pss provide inoculum for outbreaks of disease. Two separate studies, using very different approaches, demonstrated that population sizes of Pss on bean leaves are predictive of amounts of subsequent disease (5, 6). Establishment of the large pathogen population sizes that frequently lead to disease is associated with the occurrence of intense rains (7, 8). Rains with sufficient momentum appear to trigger the onset of rapid multiplication of the bacterium (8). In the absence of intense rains, population sizes of Pss tend to decline, or at least do not increase significantly, and little to no brown spot disease develops.
Substantial progress has been made in the identification of genes in Pss that are reportedly required for pathogenicity (cf. 9–11). Many of the genes participate in one of two major regulons—the gacS (global activator sensor kinase; formerly lemA) (9) and hrp (hypersensitive reaction and pathogenicity) (10, 11) regulons. A phenotype shared in common by mutants bearing defects in hrp genes and in gacS, gacA, and salA [key components of the gacS regulon (12–15)] is the inability to cause disease in susceptible hosts in laboratory leaf infiltration assays. hrp, but not gacS, gacA, or salA mutants, are additionally impaired in their abilities to cause the plant defense-associated hypersensitive response in nonhost plants and to grow when infiltrated into leaves of susceptible plants (9, 12, 16).
hrp genes in Pss and in other major genera of Gram-negative plant pathogenic bacteria are found as clusters of genes spanning ≈25 kilobases (kb) (cf. ref. 11). Putative products of hrp genes include components of a type III secretion system similar to that found in animal pathogens such as Yersinia, Shigella, and Salmonella spp., proteins that are secreted via the pathway, and regulators that control transcription of hrp and avr genes (reviewed in ref. 11). In animal systems, virulence effector molecules are exported via the type III secretory pathway and may pass directly into the cytoplasm of the target host cells after pathogen contact (cf. refs. 17 and 18). Similarly, effector molecules required for elicitation of the hypersensitive response and virulence apparently traverse the Hrp type III secretion pathway in the plant systems (19–22).
Most of the research on hrp genes has been from the perspective of their function in elicitation of the hypersensitive response, an incompatible reaction that is believed to be an important component of disease resistance. Little is known about the ways in which the Hrp secretion system and its associated effector molecules contribute to pathogenicity. The possibilities that the requirement of the hrp cluster for pathogenicity may be attributable to either the inability of hrp mutants to grow in host plants or their inability to produce factors necessary for symptom development first were suggested by Lindgren et al. in 1986 (16). Subsequent findings confirmed that hrp mutants do not grow well in leaves of susceptible hosts (16, 23). However, the possibility that the effect on growth may explain the requirement of hrp genes for pathogenicity has been largely overlooked. Additionally, what is known about the plant-associated phenotypes of hrp mutants is based on laboratory assays in which the bacteria are infiltrated into leaves. Such assays circumvent a significant portion of the interactions that occur between bacteria and plants in the field. Thus, the overall objective of this study was to determine the role(s) that hrp genes play in interactions of Pss with its host plant in the field. Our data indicate that hrp mutants of Pss strain B728a with defects in the Hrp type III secretion system (ΔhrcC∷nptII and hrpJ∷ΩSpc) are significantly affected in their ability to grow in association with leaf habitats but, interestingly, not in the spermosphere. Our data also demonstrate that, under certain conditions, hrp secretion mutants are able to cause brown spot lesions.
MATERIALS AND METHODS
Bacterial Strains.
Pss strain B728a (wild type), isolated from a snap bean leaflet in Wisconsin, is a highly virulent and field-competent strain. B728a carries a spontaneous mutation to resistance to rifampicin. B728aCm is a derivative of the wild-type strain in which an Ω-interposon conferring resistance to chloramphenicol (Cm) (24) was inserted in a null site in the intergenic region between the gacS and cysM genes (13, 25). Fitness of B728a and B728aCm was indistinguishable in the field (25).
The B728a hrp mutants examined included a hrpZ, hrcC, and hrpJ operon mutant. hrpZ encodes a protein that is secreted via the Hrp type III secretion system (26). On the basis of sequence similarity to the yscC gene in Yersinia and biochemical studies, hrcC could code for a protein that functions as an outer membrane component of the Hrp secretion system (27, 28). One or more of the genes in the hrpJ operon appears to function as inner membrane component(s) of the secretion system (28).
The B728a hrpZ mutant (ΔhrpZ∷nptII) carries a nonpolar mutation in which the entire hrpZ ORF was deleted by oligonucleotide-directed PCR amplification of flanking sequences in a subclone from pHIR11 as described (29). A 1.5-kb nptII [neomycin phosphotransferase II conferring resistance to kanamycin (Kan)] cartridge was inserted into the site of deletion such that the downstream genes are driven by the nptII promoter. A polar mutation in hrpJ (hrpJ∷ΩSpc) was constructed by marker exchange of pCPP2319 (28) into the chromosome of B728a. pCPP2319 contains a BamHI-SacI 8.6-kb fragment from pHIR11 with an ΩSpcr interposon [spectinomycin (Spc) resistance] inserted into an EcoRI site in hrpJ, the first of five genes in the operon. The hrcC mutant (ΔhrcC∷nptII) carries a nonpolar, ≈0.5-kb deletion mutation in hrcC, the third of five genes in the operon. Two subclones from pHIR11 (i.e., a 3.5-kb KpnI-EcoRI fragment containing most of the hrcC gene and a 1.3-kb KpnI-Eco0109I fragment containing the downstream region and 3′ portion of hrcC) were used to construct the deletion. The nptII cartridge was inserted into the deleted site. All mutant constructs were confirmed by Southern blot analysis.
The strains were cultured on King’s Medium B (30) supplemented with rifampicin (50 μg/ml) (KBR) and the antibiotic specific to the strain. The antibiotics were Kan (30 μg/ml) for the hrcC and hrpZ mutants, Spc (50 μg/ml) for the hrpJ mutant, Cm (30 μg/ml) for B728aCm, and nalidixic acid (Nal, 50 μg/ml) for B728aNal (Nalr spontaneous B728a mutant).
Field Plot Design.
Field experiments were conducted in 1995 and 1997 at the University of Wisconsin Arlington Experiment Station using snap bean cultivar Eagle (Asgrow Seed, Kalamazoo, MI). Treatments were arranged in a randomized complete block design with either three (1995) or four (1997) blocks. In 1995, the treatments were single inoculations of B728a, the hrpZ mutant, and the hrpJ mutant, and co-inoculations of ≈1:1 mixtures of B728aNal with either the hrpZ or hrpJ mutant. In 1997, the treatments were single inoculations with B728aCm, the hrpZ, hrpJ, and hrcC mutants. Each plot was 8 m (12 rows) × 8 m in 1995 and 5.4 m (8 rows) × 8 m in 1997. Rows were spaced 76 cm apart with ≈10 plants/m within a row. The plots were separated from each other by 5 m of bare ground. Seeds were inoculated with the bacterial strains by immersion of the seeds for 1 minute in a bacterial suspension [≈107 colony-forming units (cfu)/ml in water] immediately before planting (31).
Sampling, Enumeration, and Data Analysis of Bacterial Population Sizes.
Eight to ten samples were collected at random from each plot at each sampling time, six to eight of which were processed by dilution plating for enumeration of bacterial population sizes. Samples collected before plant emergence were dug from the ground with sterile plastic spoons. The first set of samples after seedling emergence consisted of all above-ground parts (i.e., cotyledons and emerging primary leaves). Subsequent samples were individual primary leaves or leaflets of trifoliolate leaves. Each sample was placed in a test tube containing 9 ml of sterile potassium phosphate buffer (10 mM, pH 7.0) and was stored at −20°C for subsequent processing as described (7).
Population sizes of the bacterial strains were determined by dilution plating of leaflet or seed homogenates as described (7, 31). Each sample and appropriate dilutions were spiral plated (Spiral Biotech, Bethesda, MD) onto four to five different media to enumerate the specific strain (or combination of strains in 1995) inoculated onto the seeds and to detect possible interplot spread of the strains. The samples were additionally plated onto P medium, a semiselective medium for Pss (32), to determine total P. syringae population sizes.
Bacterial counts were log10-transformed and expressed as log cfu per sample before calculation of population statistics (33). Censored observations were assigned the limit of sensitivity of the plating assay (1.95 log cfu/sample for seeds collected immediately after planting; 2.57 log cfu/sample for all other samples). Statistical analyses, including tests for lognormality of bacterial population sizes, analysis of variance, and repeated measures ANOVA (34), were performed with minitab (Minitab, State College, PA) and sas (SAS Institute, Cary, N.C.).
Brown Spot Disease Assessment and Determination of Lesion Occupancy.
Rough estimates of disease incidence were made by noting whether brown spot lesions were present on leaves collected for enumeration of bacterial population sizes. In 1997, all plants were additionally scrutinized for the presence of lesions at 14 days after planting (DAP). To determine lesion occupancy, additional leaves were collected, and individual lesions were removed with a 5-mm cork borer. The diseased tissues were individually surface-sterilized with 70% ethanol, were rinsed with sterile water, were macerated in 1 ml of potassium phosphate buffer (10 mM, pH 7.0), and were dilution-plated onto P, KBR, KBRCm, KBRKan, and KBRSpc media. For each sample, two well separated colonies were taken from plates containing KB plus antibiotics. Additionally, mass streakings were made from the P plates seeded with the undiluted homogenate to detect the presence of naturally occurring conspecifics that may have been present at smaller numbers than the mutants. The isolates were tested for pathogenicity by using a leaf infiltration assay.
Growth Chamber Assays.
Leaf infiltration pathogenicity and in planta growth assays were conducted with cultivar Bush Blue Lake 274 (Rogers Seed, Boise, ID). For most experiments, maximum and minimum temperatures were set at 24°C (14-hr light period) and 20°C (10-hr dark period), respectively. Cells from 2-day-old cultures (grown on KBR plus antibiotic) were suspended in sterile water. For routine pathogenicity assays, suspensions of 106 and 105 cfu/ml were infiltrated into primary leaves of 10- to 11-day-old plants by using a needleless syringe. The resulting doses were ≈2.5–3.5 × 103 and 2.5–3.5 × 102 cfu per infiltrated site of ≈30–40 mm2. Strains (e.g., wild-type B728a) that caused necrosis over the entire infiltrated area in 3–4 days at both doses were considered pathogenic.
For assessing in planta growth, bacterial suspensions diluted to 106 cfu/ml were infiltrated as described above. A single site was infiltrated on as many leaves as necessary to allow the collection of five samples per sampling time. A cork borer was used to remove a 6-mm (diameter) disk from the infiltrated site. Immediately after inoculation, each disk contained ≈3 × 103 cfu. The leaf disks were macerated and dilution-plated as described above.
RESULTS
Laboratory Characterization of the hrp Mutants.
The hrcC and hrpJ mutants did not elicit the hypersensitive response on tobacco nor cause disease on bean when infiltrated into leaves in growth chamber assays, expected phenotypes of hrp mutants (data not shown). In contrast, the hrpZ mutant was indistinguishable from B728a in these assays. Immunoblot analyses of culture supernatants and cellular fractions using anti-HrpZ antibodies (26, 35) confirmed that the hrcC and hrpJ mutants produced but did not secrete the HrpZ protein whereas the hrpZ mutant did not produce the HrpZ protein (data not shown). The phenotypes of the hrcC mutant were restored by pNCHU421, a plasmid that contains the hrcC gene from Pss strain 61 (28). The entire hrpJ operon carried on cosmid clones was required to restore the phenotypes of the hrpJ mutant.
Population Sizes of the Mutants on Preemergent Seedlings.
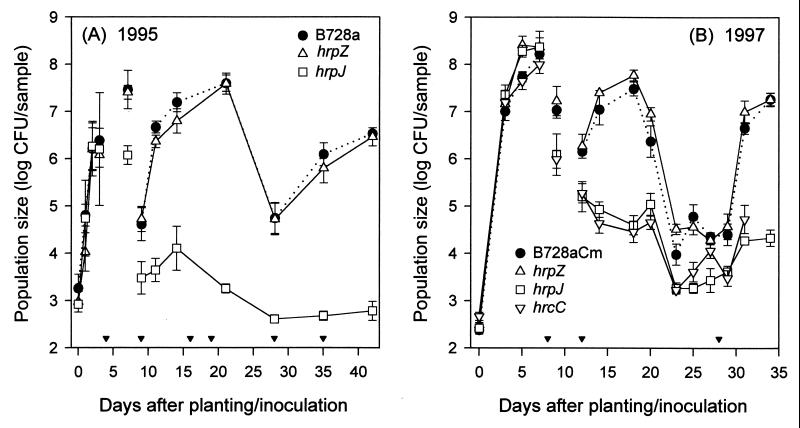
In each of two field experiments, population sizes of the hrpJ, hrcC, and hrpZ mutants on germinating seedlings increased rapidly and were comparable to those of B728a or B728aCm (Fig. 1 A and B). In the first experiment, the ≈1,000-fold increases by 3 DAP (Fig. 1A) were typical of increases in bacterial numbers measured in previous experiments (31). In the second experiment, plant emergence was delayed by ≈2 days, presumably because of below-normal temperatures, and bacterial population sizes attained on below-ground samples were unusually large (Fig. 1B). By 3 DAP, bacterial population sizes had increased nearly five orders of magnitude. At 7 DAP, the seedlings were still below ground, and mean population sizes of the hrp mutants and B728aCm were ≈108 cfu/sample. There were no significant differences among population sizes of B728a and the mutants on below-ground bean samples collected in 1995 (P = 0.59) or in numbers of B728aCm and the mutants in the 1997 experiment (P = 0.19). The indistinguishable behavior of the mutants relative to the wild type suggests that the products encoded by hrpZ, hrcC, and the hrpJ operon did not affect growth of Pss B728a on preemergent seedlings.
Figure 1.
Population sizes of Pss B728a hrp mutants (ΔhrpZ∷nptII, hrpJ∷ΩSpcr, ΔhrcC∷nptII) in association with field-grown bean plants. The bacterial strains were inoculated onto seeds immediately before planting. Samples were seeds or germinating seedlings collected between 0 and 3 DAP (A) and 0 and 7 DAP (B); cotyledons and primary leaves were collected at 7 (A) and 9 DAP (B); primary leaves were collected between 9 and 14 (A) and 12 and 20 DAP (B); and single leaflet from trifoliolate leaves were collected at all other times. Each datum point represents the mean log cfu/sample and SE based on three (A) or four (B) replicate plots with six or eight individual samples per plot. Inverted solid triangles indicate rain events with sustained rates >1 mm/min.
Population Sizes of the Mutants in Association with Leaves.
Leaf population sizes of the hrpZ mutant were not significantly different from B728a (P = 0.56, Fig. 1A) or B728aCm (P = 0.13, Fig. 1B) throughout the two experiments. The mutant and wild type both responded to the environment as expected for Pss (7, 8). However, leaf population sizes of the hrpJ (Fig. 1 A and B) and hrcC (Fig. 1B) mutants were significantly lower than those of B728a (P < 0.0002). The absence of substantial population increases during rainy periods favorable for growth suggests that hrcC and genes in the hrpJ operon play critical roles in enabling Pss to grow in the phyllosphere.
Detection of Brown Spot Lesions in Field Plots.
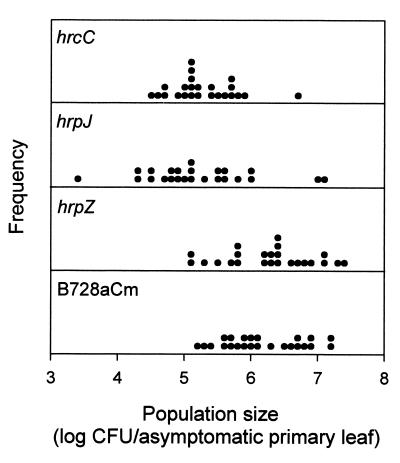
As expected, disease was detected in both field experiments in which large numbers of B728a and the hrpZ mutant were present. The hrpZ mutant was isolated from lesions, as were B728a and B728aCm. In 1997, relatively large population sizes of the hrcC and hrpJ mutants were detected on individual primary leaves at 12 DAP (Fig. 2). Indeed, some of the leaves harbored population sizes that were sufficiently large (e.g., >106 cfu/leaf) to be predictive of disease for strains capable of causing brown spot lesions (6). By 14 DAP, ≈4% (n = 64) of the leaves that were collected from the hrcC and hrpJ mutant plots for enumeration of bacterial population sizes had brown spot lesions (Fig. 3). At this time, disease incidences were 63 and 75% for B728aCm and the hrpZ mutant, respectively.
Figure 2.
Distribution of population sizes of B728aCm and the hrp mutants in association with individual asymptomatic primary leaves collected at 12 DAP in 1997. Mean bacterial population sizes for the sets of individual leaves are shown in Fig. 1B.
Figure 3.
Brown spot lesions caused by the B728a ΔhrcC∷nptII mutant under field conditions. Lesions were detected 14 DAP in 1997.
Additional leaves with brown spot lesions were collected from the hrcC and hrpJ plots for determination of lesion occupancy. Bacterial population sizes estimated from counts on P medium used to monitor populations of total Pss were similar to those on KBRKan and KBRSpc for lesions collected from the hrcC and hrpJ inoculated plots, respectively. The results indicated that the mutants were the dominant components within the lesions. None of the bacterial isolates recovered either as single colony isolates from the KBRKan and KBRSpc plates or as mass isolates from the P plates (to detect naturally occurring Pss) caused a pathogenic reaction when subsequently tested in the growth chamber leaf infiltration assay at routine doses of ≈3 × 103 and ≈3 × 102 cfu/infiltrated site. When genomic digests from 10 of these isolates were probed with either the nptII or ΩSpc fragment, the patterns obtained were indistinguishable from those of the hrcC or hrpJ mutant (data not shown). The results strongly suggest that the lesions detected in the plots inoculated with the hrcC and hrpJ mutants were caused by these mutants and not by other pathogenic Pss. Further, the mutants had not reverted during passage on plants in the field.
Growth Chamber Assays to Reevaluate the Pathogenic Potential of the hrp Secretion Mutants.
To determine whether lesion causation by the hrcC and hrpJ mutants in the field could be reproduced under controlled conditions, the mutants and B728a were inoculated onto bean seeds as was done in the field. Inoculated seeds were germinated under conditions of saturated relative humidity and temperatures mimicking those of the 1997 field experiment (13°C and 22°C during the night and day hours, respectively). Under these conditions, brown spot lesions, similar to those observed in the field, developed on primary leaves. Disease incidences were 2.4% (n = 163 leaves) for the hrpJ mutant, 4.4% (n = 159 leaves) for the hrcC mutant, and 30% (n = 149 leaves) for B728a. The experiment was repeated with similar results.
Additional experiments were conducted to address the apparent paradox that the mutants were able to cause lesions on primary leaves when inoculated onto seeds but not when infiltrated into leaves. To test the effect of temperature, routine doses of ≈3 × 103 and ≈3 × 102 cfu/infiltrated site were inoculated into plants grown and maintained under the conditions used for the seed-inoculation assay (i.e., lower temperatures than normally used for the leaf infiltration assay). The hrcC and hrpJ mutants failed to cause a pathogenic reaction under these conditions. To test the effect of dose, the mutants were infiltrated at doses 10- to 100-fold greater (i.e., ≈3 × 104 to ≈3 × 105 cfu/infiltrated site) than routinely used. The hrcC and hrpJ mutants caused some necrosis when infiltrated with the larger doses (data not shown). The results indicate that the secretion mutants are able to cause a pathogenic reaction in the leaf infiltration assay, albeit reduced relative to wild type, when sufficient doses are used to overcome their inability to grow to numbers necessary to cause disease.
Behavior of the hrpJ Mutant in Co-Inoculation Treatments.
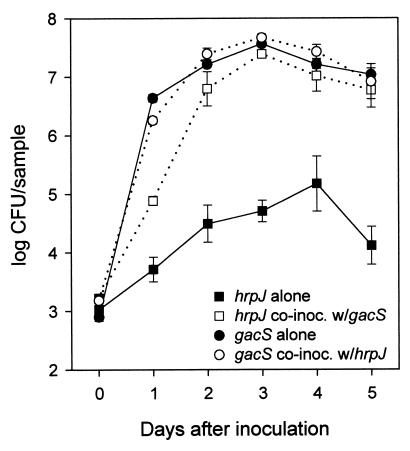
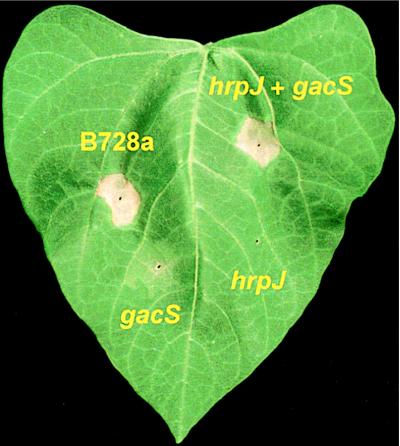
When the hrpJ mutant was co-inoculated with B728aNal in field experiments and with B728aCm in growth chamber assays, population sizes of the mutant were substantially larger than when inoculated alone (data not shown). Any possible contribution of the hrp mutant to disease development could not be ascertained because the B728a derivatives were present in the inoculum and are able to cause disease. We next tested whether growth of the hrpJ mutant could be complemented by the non-lesion-forming B728a gacS mutant NPS3136 (gacS1:Tn5) (12, 13). GacS is similar to the transmembrane histidine kinase protein of a family of bacterial two-component regulatory systems (13). When co-infiltrated with the gacS mutant, population sizes of the hrpJ mutant were significantly larger than when inoculated alone (Fig. 4). Of interest, a pathogenic reaction developed when the mutants were co-infiltrated into leaves but not when each was inoculated alone (Fig. 5). Co-inoculation of a gacA [cognate response regulator of gacS (14)] mutant with the hrpJ mutant also produced a pathogenic reaction (data not shown). Similar results were obtained when the hrcC mutant was co-inoculated with the gacS and gacA mutants. When the hrpJ and hrcC mutants were co-inoculated into leaves, growth was comparable to that measured when they were each inoculated alone.
Figure 4.
Rescued growth of the B728a hrpJ∷ΩSpc mutant by a non-lesion-forming B728a gacS1∷Tn5 mutant. The mutants were infiltrated into primary leaves of growth chamber-grown bean plants either alone or in a 1:1 mixture. Approximately 3 × 103 cfu were deposited initially in each of the 6-mm-diameter disks removed for sampling. Samples inoculated with the mixture were dilution-plated onto both KBRSpc and KBRKan to enumerate population sizes of the hrpJ and gacS mutants, respectively. Each datum point represents the mean log cfu/sample and SE based on dilution plating of five individual samples.
Figure 5.
Leaf infiltration assay for pathogenicity. Necrosis indicative of brown spot disease developed at sites infiltrated with either B728a or a 1:1 mixture of the hrpJ and gacS mutants. Neither mutant caused the pathogenic reaction when inoculated alone. The dose was ≈103 cfu for B728a and the mutants when inoculated alone. In the co-inoculation treatment, the dose for each mutant was ≈5 × 102 cfu.
DISCUSSION
Pss may be transmitted from one generation of its host to the next via seeds. Hence, inoculation of bean seeds with relatively small numbers of bacteria at the time of planting was selected as a way to naturally initiate the plant–bacterial interaction in the field. The apparently normal growth of the type III secretion mutants on preemergent bean plants was unexpected given the in planta growth defects of hrp mutants observed in laboratory experiments (16, 23). After plant emergence, however, leaf population sizes of the hrcC and hrpJ mutants were significantly lower than B728a. Numbers of the mutants tended to remain constant or decline, even under conditions of intense rains, when population sizes of B728a and the hrpZ mutant increased significantly. The hrcC and hrpJ mutants behaved similarly, although the specific genes mutated and the nature of the mutations differed in the two constructs. Thus, mutations in hrp genes that affected the Hrp secretion system substantially reduced growth and, possibly, survival of Pss B728a in the phyllosphere. If the behavior of the secretion mutants is attributable primarily to defects in the Hrp type III secretion apparatus, then effector molecules traversing the pathway may be necessary for growth in association with leaves but, interestingly, not with preemergent seedlings.
Our method to remove bacteria from leaves for enumeration precluded a direct assessment of the effect of hrp genes on epiphytic versus intercellular growth of Pss. However, several lines of evidence suggest that the diminished population sizes of the hrcC and hrpJ mutants resulted from their inabilities to grow on the surfaces as well as intercellular spaces of leaves. Roughly 80–99% of naturally occurring populations of Pss on asymptomatic leaves of field-grown plants are sensitive to a leaf surface sterilization treatment (36), suggesting that a large proportion of the populations is present in unprotected sites on leaves (i.e., epiphytic populations). Hence, if the hrcC and hrpJ mutants were able to grow epiphytically, we would have expected larger populations than those obtained. Additional evidence for the role of the type III secretion system in epiphytic growth has been obtained by Romantschuk et al. (37). After spray inoculation onto tomato plants in the field, epiphytic populations (based on dilution plating of leaf sonicates) of hrp mutants of P. syringae pv. tomato DC3000 were significantly reduced relative to wild type. Models describing the functions of hrp genes in interactions of plant pathogenic bacteria with susceptible hosts should be consistent with the scenario that hrp genes play a fundamental growth-enabling role that is likely not limited to the intercellular spaces of leaves.
The behaviors of the hrpZ mutant and B728a were similar in the spermosphere and the phyllosphere and with regard to lesion formation. Thus, although HrpZ is secreted via the Hrp type III secretory system, and hrpZ is located within an operon of the hrp cluster, mutation of hrpZ had no measurable effect on interactions of B728a with bean plants in the field. The role of HrpZ in interactions of B728a with its susceptible host remains unknown.
In 1997, conditions were highly conducive for growth of the strains before plant emergence. As a result, population sizes of the hrcC and hrpJ mutants on some newly emerged primary leaves were large enough to be predictive of disease. These relatively large population sizes may have resulted from growth of the mutants while the developing leaves were still enclosed between the cotyledons on the pre- and very early postemergent seedlings. Lesion occupants from the field were recovered and identified as the hrp mutants that had been applied to the seed on the basis of their failure to cause a pathogenic reaction in a standard laboratory leaf infiltration assay, their antibiotic resistance marker profiles, and by Southern hybridization analysis. The possibility that naturally occurring pathogenic Pss caused the lesions was eliminated by our inability to isolate bacteria from the lesions that caused a pathogenic reaction when infiltrated into leaves of growth-chamber grown plants. The finding that the hrcC and hrpJ mutants are capable of causing brown spot lesions on primary leaves after seed inoculation was confirmed in growth chamber experiments.
Necrosis developed when relatively large doses of the hrcC and hrpJ mutants were infiltrated into leaves of growth chamber-grown plants. Hence, when the requirement for growth was partially alleviated, the secretion mutants were able to produce a pathogenic reaction in the leaf infiltration assay, although reduced relative to B728a. The observation is consistent with the explanation that the apparent requirement for the type III secretion system for pathogenicity in Pss is largely mediated through its role in enabling growth of the pathogen.
Population sizes of the hrp secretion mutants were larger when co-inoculated with B728a or the non-lesion-forming gacS mutant than when inoculated alone. Because B728a and the gacS mutant have functional Hrp systems, it may be that factor(s) secreted by these strains complemented the deficiency in the hrcC and hrpJ mutants. A similar finding was reported by Kamoun and Kado (38). In their study, growth of a hrpXc mutant of the black rot bacterial pathogen, Xanthomonas campestris pv. campestris, was rescued when the mutant was co-inoculated with its pathogenic parent in cauliflower and radish leaves. An unexpected finding in our experiments was the development of a pathogenic reaction when leaves were co-inoculated with the gacS (or gacA) and the hrpJ or hrcC mutants. An explanation for the reaction is that, when co-inoculated, the gacS and hrp mutants contributed to the pathogenic reaction by providing functions necessary for lesion formation that each alone lacked. Another is that rescued growth of the hrp mutant by the gacS mutant (either through a direct effect on the mutant or by some plant-mediated mechanism) led to the development of population sizes of the hrp mutant that were sufficient to cause disease. Although neither explanation has been ruled out, the second hypothesis is consistent with the findings from field and laboratory experiments that, when growth of the hrp mutants in association with plants (preemergent seedlings) leads to establishment of sufficient numbers of the hrp secretion mutants in/on primary leaves, brown spot lesions developed.
Although current models for the function of hrp genes suggest that pathogenicity or virulence factors may traverse the Hrp type III pathway, none have yet been identified for which effects on disease development have been demonstrated to be distinct from effects on growth in or on leaves. Thus, we cannot rule out the possibility that the role of the type III secretion system in pathogenicity is completely mediated by its requirement for growth of Pss in the phyllosphere. Genes that play a more direct role in symptom development in the Pss-brown spot-snap bean system may reside in the gacS/gacA regulon identified by Willis and colleagues (12–15, 39). The role of the Hrp type III secretion system in growth but not symptom development per se in Pss is consistent with the recent finding of Shea et al. (40) on the role of the type III secretion system within Salmonella typhimurium pathogenicity island 2 (SPI2). A mutation in ssaJ, a gene encoding a component of the SPI2 type III secretion system, affected growth but not survival of the pathogen in a mouse model system (40).
Acknowledgments
We thank the personnel at the University of Wisconsin-Madison Arlington Experiment Station for assistance in field plot preparation and Asgrow and Rogers Seed companies for providing the bean seeds used in this research. This research was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (Grant 9601097) and the Agricultural Research Service, U.S. Department of Agriculture.
ABBREVIATIONS
- Pss
Pseudomonas syringae pv. syringae
- DAP
days after planting
- cfu
colony-forming unit
- kb
kilobases
- Cm
chloramphenicol
- Kan
kanamycin
- Spc
spectinomycin
References
- 1.Hirano S S, Upper C D. Annu Rev Phytopathol. 1990;28:155–177. [Google Scholar]
- 2.Hirano S S, Rouse D I, Clayton M K, Upper C D. Plant Dis. 1995;79:1085–1093. [Google Scholar]
- 3.Ercolani G L, Hagedorn D J, Kelman A, Rand R E. Phytopathology. 1974;64:1330–1339. [Google Scholar]
- 4.Lindemann J, Arny D C, Upper C D. Phytopathology. 1984;74:1329–1333. [Google Scholar]
- 5.Lindemann J, Arny D C, Upper C D. Phytopathology. 1984;74:1334–1339. [Google Scholar]
- 6.Rouse D I, Nordheim E V, Hirano S S, Upper C D. Phytopathology. 1985;75:505–509. [Google Scholar]
- 7.Hirano S S, Clayton M K, Upper C D. Phytopathology. 1994;84:934–940. [Google Scholar]
- 8.Hirano S S, Baker L S, Upper C D. Appl Environ Microbiol. 1996;62:2560–2566. doi: 10.1128/aem.62.7.2560-2566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willis D K, Rich J J, Kinscherf T G, Kitten T. In: Genetic Engineering: Principles and Methods. Setlow J K, editor. Vol. 16. New York: Plenum; 1994. pp. 167–193. [PubMed] [Google Scholar]
- 10.Alfano J R, Collmer A. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindgren P B. Annu Rev Phytopathol. 1997;35:129–152. doi: 10.1146/annurev.phyto.35.1.129. [DOI] [PubMed] [Google Scholar]
- 12.Willis D K, Hrabak E M, Rich J J, Barta T M, Lindow S E, Panopoulos N J. Mol Plant–Microbe Interact. 1990;3:149–156. [Google Scholar]
- 13.Hrabak E M, Willis D K. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rich J J, Kinscherf T G, Kitten T, Willis D K. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitten T, Kinscherf T G, McEvoy J L, Willis D K. Mol Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 16.Lindgren P B, Peet R C, Panopoulos N J. J Bacteriol. 1986;168:512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M-P, Stainier I. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alfano J R, Collmer A. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He S Y. Annu Rev Phytopathol. 1998;36:363–392. doi: 10.1146/annurev.phyto.36.1.363. [DOI] [PubMed] [Google Scholar]
- 21.Collmer A. Curr Opin Plant Biol. 1998;1:329–335. doi: 10.1016/1369-5266(88)80055-4. [DOI] [PubMed] [Google Scholar]
- 22.Mudgett M B, Staskawicz B J. Curr Opin Microbiol. 1998;1:109–115. doi: 10.1016/s1369-5274(98)80150-1. [DOI] [PubMed] [Google Scholar]
- 23.Huang H-C, Hutcheson S W, Collmer A. Mol Plant–Microbe Interact. 1991;4:469–476. doi: 10.1094/mpmi-6-515. [DOI] [PubMed] [Google Scholar]
- 24.Fellay R, Frey J, Krisch H. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 25.Hirano S S, Willis D K, Upper C D. Phytopathology. 1997;87:S42. [Google Scholar]
- 26.He S Y, Huang H-C, Collmer A. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 27.Huang H-C, He S Y, Bauer D W, Collmer A. J Bacteriol. 1992;174:6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charkowski A O, Huang H-C, Collmer A. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alfano J R, Bauer D W, Milos T M, Collmer A. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- 30.King E O, Ward M K, Raney D E. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 31.Hirano S S, Ostertag E M, Savage S A, Baker L S, Willis D K, Upper C D. Appl Environ Microbiol. 1997;63:4304–4312. doi: 10.1128/aem.63.11.4304-4312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohan S K, Schaad N W. Phytopathology. 1987;77:1390–1395. [Google Scholar]
- 33.Hirano S S, Nordheim E V, Arny D C, Upper C D. Appl Environ Microbiol. 1982;44:695–700. doi: 10.1128/aem.44.3.695-700.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Littell R C, Milliken G A, Stroup W W, Wolfinger R D. SAS System for Mixed Models. Cary, NC: SAS Institute; 1996. [Google Scholar]
- 35.Yuan J, He S Y. J Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock J D, Hirano S S. Phytopathology. 1991;81:1222. [Google Scholar]
- 37.Romantschuk M, Roine E, He S-Y, Wilson M. Phytopathology. 1997;87:S82–S83. [Google Scholar]
- 38.Kamoun S, Kado C I. J Bacteriol. 1990;172:5165–5172. doi: 10.1128/jb.172.9.5165-5172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rich J J, Willis D K. J Bacteriol. 1997;179:2247–2258. doi: 10.1128/jb.179.7.2247-2258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shea J E, Beuzon C R, Gleeson C, Mundy R, Holden D W. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]