Abstract
Nerve damage is the hallmark of Mycobacterium leprae infection, which results from M. leprae invasion of the Schwann cell of the peripheral nervous system. We have recently shown that the laminin-2 isoform, specially the G domain of laminin α2 chain, on the Schwann cell–axon unit serves as an initial neural target for M. leprae. However, M. leprae surface molecules that mediate bacterial invasion of peripheral nerves are entirely unknown. By using human α2 laminins as a probe, a major 28-kDa protein in the M. leprae cell wall fraction that binds α2 laminins was identified. After N-terminal amino acid sequence analysis, PCR-based strategy was used to clone the gene that encodes this protein. Deduced amino acid sequence of this M. leprae laminin-binding protein predicts a 21-kDa molecule (ML-LBP21), which is smaller than the observed molecular size in SDS/PAGE. Immunofluorescence and immunoelectron microscopy on intact M. leprae with mAbs against recombinant (r) ML-LBP21 revealed that the protein is surface exposed. rML-LBP21 avidly bound to α2 laminins, the rG domain of the laminin-α2 chain, and the native peripheral nerve laminin-2. The role of ML-LBP21 in Schwann cell adhesion and invasion was investigated by using fluorescent polystyrene beads coated with rML-LBP21. Although beads coated with rML-LBP21 alone specifically adhered to and were ingested by primary Schwann cells, these functions were significantly enhanced when beads were preincubated with exogenous α2 laminins. Taken together, the present data suggest that ML-LBP21 may function as a critical surface adhesin that facilitates the entry of M. leprae into Schwann cells.
Nerve damage in leprosy results from Mycobacterium leprae invasion of Schwann cell of the peripheral nerves and is responsible for most of the deformity and disability of this disease (1, 2). Although patients can be cured of infection by multidrug therapy, the immunopathological sequelae responsible for the characteristic deformities of leprosy can continue during and even after antimicrobial therapy (3). Such therapeutic intervention has so far prevented only one third of infected individuals from suffering new disabilities (3). Therefore, interventions targeted directly to block the early interaction of M. leprae with peripheral nerve is important for the prevention of nerve infection and subsequent peripheral neuropathy.
M. leprae invasion of Schwann cells of the peripheral nervous system represents an early crucial step leading to the nerve damage (1, 2). However, the surface molecules of M. leprae that mediate bacterial binding to and invasion of peripheral nerve are entirely unknown. As an important step toward identifying this invasion process, we have recently established that the laminin-2 isoform, specifically the G domain of laminin α2 chain, on the Schwann cell–axon unit serves as an initial neural target of M. leprae (4). Laminins, which consist of α, β, and γ chains, are major matrix proteins of basal laminae and play a crucial role in variety of cellular functions, including normal nerve functions (5–10). Laminins exist in at least 11 different isoforms, each with restricted tissue distribution (5). In Schwann cell basal lamina, the tissue-restricted laminin variant is laminin-2, which comprises α2, β1, and γ1 chains (9). Because laminin-2 is expressed on the outermost surface of and surrounds completely the Schwann cell–axon unit (4, 11), laminin-2 serve as an initial target molecule that M. leprae might encounter before Schwann cell invasion. Therefore, surface proteins of M. leprae cell wall that bind laminin-2 are crucial for bacterial targeting to the peripheral nerves.
In the present study, we describe the identification and molecular characterization of a surface protein of M. leprae, ML-LBP21, that binds peripheral nerve laminin-2. We provide evidence that ML-LBP21 is a major surface protein of M. leprae and is involved in Schwann cell invasion via laminin-2-dependent pathway.
MATERIALS AND METHODS
Bacteria.
In vivo-grown M. leprae was purified from armadillo liver and spleen as described (12). M. leprae were tested for their acid-fast property and M. leprae-specific phenolic glycolipid-1 before the experiments. Preparation of the cell-wall fraction of M. leprae has been described (13). M. leprae and cell wall fraction were provided by P. J. Brennan (Colorado State University, Fort Collins, CO).
Identification of α2 Laminin-Binding Protein (LBP).
M. leprae cell wall fraction was separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted to poly(vinylidene difluoride) (PVDF) membranes as described (4, 17). The membrane was first blocked with 1% acidified BSA, 0.05% gelatin, and 0.5% Tween 20 in 0.5 M Tris⋅HCl and then probed with 125I-labeled human α2 laminins (merosin) for 4 hr. α2-Laminins purified from human placenta (GIBCO/BRL) consist of a mixture of laminin-2 (α2, β1, and γ1 chains) and laminin-4 (α2, β2, and γ1 chains) with tissue-restricted α2 chain common to both isoforms (Fig. 1B). After washing steps, blots were autoradiographed by exposure to Kodak X-Omat AR film. In parallel, a part of M. leprae cell wall blots was subjected to amido black staining, and the protein band corresponding to the LBP was excised from the PVDF membrane and the N-terminal amino acid sequence was determined at The Rockefeller University Protein/Biotechnology facility.
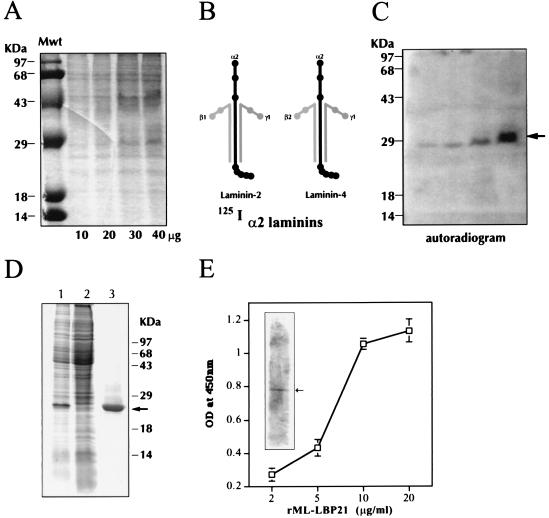
Figure 1.
Identification, expression, and characterization of ML-LBP21. (A–C) Identification of α2-laminin-binding protein from M. leprae cell wall fraction. (A) Increasing concentration of M. leprae cell wall fraction (10–40 μg/ml per lane) was separated by SDS/10% PAGE and stained with Coommassie brilliant blue. Molecular mass markers (Mwt) are indicated on the extreme left lane. (B) Diagram of human α2-laminins (a mixure of laminin-2 and laminin-4) and their chain specificity are shown. (C) Identical gels containing M. leprae cell wall fraction (10–40 μg/ml) was transferred to PVDF membrane and probed with 125I-labeled α2-laminins, and bound laminin was detected by autoradiography. The arrow indicates the binding of α2-laminins to a 28-kDa band in the cell wall fraction in a concentration-dependent manner. (D) Expression and purification of recombinant ML-LBP21. ML-LBP21 was expressed as His-tagged fusion protein, purified on a Ni-nitrilotriacetic acid resin, and separated by SDS/PAGE. The gels were stained with Coomassie brilliant blue. Lane 1, lysates of isopropyl β-d-thiogalactoside (IPTG)-induced culture; lane 2, uninduced culture; lane 3, affinity-purified recombinant protein. Molecular mass markers (kDa) are shown on the right, and the arrow indicates the purified rML-LBP21. (E) Characterization of rML-LBP21 binding to α2-laminins. The binding of biotinylated α2-laminins to rML-LBP21 as detected by ELISA and immunoblotting (Inset) by using streptavidin method. The arrow in the inset indicates the rML-LBP21-bound α2-laminins.
Cloning and Sequencing of LBP Gene of M. leprae.
DNA manipulation was carried out by using standard procedures (14). Based on the N-terminal amino acid sequence and homology search of M. leprae genome sequence project (EMBL accession no. Z99263), PCR primers AWG1 (5′-GTTTACCGTGTCCATTAGTC-3′) and AWG2 (5′-TAACACG CAGATACCCTCCG-3′) were designed, and the DNA region encoding putative LBP and its flanking region were amplified from M. leprae chromosomal DNA (kindly provided by P. J. Brennan) by PCR. The PCR product was purified and both strands of DNA were directly sequenced with dye-termination method (15). The sequences were analyzed with the genetyx-mac program, version 7.3 and sequence similarity searches were performed with GenBank sequences by using blast network service.
Construction and Purification of Recombinant Fusion Protein.
Construction and purification of histidine (His)-tagged fusion protein was performed by using the QiaExpress Kit (Qiagen, Chasworth, CA). PCR was performed by using primers A1 (5′-CTCAGGATCCACAGAAATT-GGGCTCGGACC-3′) and A5 (5′-CCCCAGATCAATTTTTCTTTACTTGC-GACC-3′), which constitute an in-frame BamHI and BglII restriction sites at the 5′ end and 3′ end, respectively. The PCR consisted of denaturation at 94°C for 1 min, annealing at 50°C for 30 sec, and extension at 72°C for 1 min performed for 30 cycles. The amplified gene product was digested with BamHI and BglII, and then ligated with the expression vectors pQE-51 and pQE-16 (Qiagen) to generate pYS6.
Preparation of mAbs.
BALB/c mice were immunized subcutaneously with approximately 100 μg of purified recombinant (r) ML-LBP21 in incomplete Freund’s adjuvant followed by a booster immunization. Spleen cells from immunized mice were fused with SP20 BALB/c myeloma cell line by using standard procedure. Antibody-producing hybridomas were screened by ELISA and immunoblotting using rML-LBP21 and M. leprae cell-wall fraction. Selected hybridomas were cloned by limiting dilution to establish stable hybridomas, which were then grown in bulk cultures. mAb-containing culture supernatants were partially purified by using 40% ammonium sulfate precipitation, and the characteristics of these mAbs were determined by using immunoblotting, immunofluorescence, and immunogold labeling as described (16).
Immunoblotting.
Various antigen preparations were electrophoresed in 10% and 3–12% gradient SDS/PAGE (4, 17) and transferred to nitrocellulose or PVDF membranes and processed for immunoblotting as described (17) by using mAbs raised against rML-LBP21 and laminin α2 and β/γ chains. Bound antibodies were detected by using either enhanced chemiluminescence (ECL; Amersham Pharmacia) or alkaline phosphatase methods.
Preparation of the rLNα2G Domain and rML-LBP21 Binding Assays.
Preparation of rLNα2G with a baculovirus expression system has been described (4). rML-LBP21 binding to the rLNα2G domain was performed by ELISA using the alkaline phosphatase method. Briefly, a preevaluated concentration of rML-LBP21 (25 μg/ml; 1.25 μg per well) was coated on ELISA plates (Nunc) by using bicarbonate buffer (pH 9.6). BSA was used as a negative control. After washing, increasing concentrations of rLNα2G (0.5–5.0 μg/ml; 0.025–0.25 μg per well) was added to the wells and incubated for 1hr at 37°C. rML-LBP21-bound rLNα2G was detected by affinity-purified rabbit antibody against rLNα2G (4) followed by monoclonal anti-rabbit immunoglobulins conjugated to alkaline phosphatase (Sigma). Alternatively, we also tested the binding of rML-LBP21 to rLNα2G-coated plates by using mAbs against rML-LBP21.
Biotinylated α2-laminins were used to detect the laminin binding to rML-LBP21 in ELISA and immunoblotting by using the streptavidin method.
Preparation of the Bovine Peripheral Nerve Membrane Fraction (PNMF).
Bovine PNMF, which contains laminin-2 and other matrix proteins in addition to the membrane proteins (17, 18), was prepared as described (17). PNMF was suspended at a protein concentration of 5 mg/ml in 50 mM Tris⋅HCl (pH 7.4) containing a mixture of protease inhibitors: benzamidine (0.75 mM), phenylmethylsulfonyl fluoride (0.1 mM), pepstatin A (0.7 mM), aprotinin (76.8 nM), and leupeptin (1.1 mM). The suspension was titrated to pH 12.0, extracted by stirring vigorously for 1 hr at room temperature, and centrifuged at 140,000 × g for 30 min at 25°C. The supernatants were collected by decanting, cooled to 4°C, and titrated to pH 7.4.
Overlay Assay for rML-LBP21 Binding to Endogenous Bovine Peripheral Nerve Laminin-2.
The nitrocellulose transfers of rML-LBP21 or M. leprae cell wall were blocked with 10 mM triethanolamine (pH 7.6), 140 mM NaCl, 1 mM CaCl2, and 1 mM MgCl2 (LBB) containing 5% nonfat dry milk (MLBB) and then incubated with 10 ml bovine PNMF (5 mg/ml) in the presence of 1 mM CaCl2 and 1 mM MgCl2 for 48 hr at 4°C. After washing three times for 10 min at room temperature in MLBB, the endogenous peripheral nerve laminin that bound to rML-LBP21 or the native LBP21 in the M. leprae cell wall was detected with either mAbs 2D9 against the G domain of human laminin α2 chain or affinity-purified rabbit antibody against mouse EHS (Engelbreth-Holm-Swarm) tumor laminin (Sigma) as described (17, 18).
Primary Schwann Cell Cultures.
Schwann cells were isolated from neonatal rat sciatic nerve and purified as described (19, 20). The purity of these primary Schwann cells was determined by anti-S100 antibody counterstained with Hoechst nuclear labeling and showed that all nucleated cells are positive for S-100 antigen.
Assays for rML-LBP21-Coated Beads Adherence to and Invasion of Schwann Cells.
To test the role of ML-LBP21 in Schwann cell adhesion and invasion, fluorescent polystyrene beads (Polysciences) were coated with rML-LBP21 and examined for adhesion to and invasion of primary rat Schwann cells in the presence and the absence of exogenous α2-laminins. Beads (1-μm diameter) were coated with increasing concentrations (5–100 μg/ml) of rML-LBP21 in bicarbonate buffer (pH 9.6) for 2 hr at 27°C. Control beads were incubated with bicarbonate buffer alone. All beads were washed twice in PBS and blocked with 2% BSA in PBS at 27°C for 2–3 hr. After washing, beads were used either alone or preincubated with various concentrations of α2-laminins. Trypsinized Schwann cells were washed and suspended in DMEM without serum or growth factors at a concentration of 6 × 104 cells per ml. Coated beads were incubated with Schwann cells in suspension (300:1 ratio) under constant rotation at 37°C for 30 min. Cell suspensions were centrifuged by using a single pulse of 500 × g to obtain the pellet containing cell-bound beads. Under these conditions, beads that were not bound to cells remained in the supernatant. After removing the supernatant, the cell pellet was washed three times and suspended in fresh Schwann cell media. For the adhesion assay, we used flow cytometric analysis on FACScan (Beckton Dickinson) by using the built-in property of the fluorescence of the cell-bound beads. The median fluorescence intensity values were chosen to quantify the number of Schwann cells bearing rML-LBP21-coated beads.
For the invasion assay, instead of processing Schwann cell-bound beads for FACScan, the cell suspension was seeded onto polylysine coated eight-well LabTek chamber slides (Nunc) and incubated for 12 hr at 37°C in a standard humidified CO2 incubator. Under these conditions, only the cell-bound beads were allowed to invade. Cells were fixed with 2.5% glutaraldehyde and mounted with Citifluor mounting media containing Hoechst/bisbenzimide (Sigma). Schwann cell-invaded beads were evaluated and quantified by using fluorescence microscopy. Bead invasion of Schwann cells was further confirmed by electron microscopy using standard procedures.
RESULTS
Identification LBP21 from M. leprae Cell Wall.
Because M. leprae bind to the α2 chain of laminin-2 isoform on Schwann cell basal lamina (4), we have used human α2-laminins (merosin), which consist of the mixture of laminin-2 (α2, β1, and γ1 chains) and laminin-4 (α2, β2, and γ1 chains) with common α2 chain (Fig. 1B) as a probe to identify the LBP from M. leprae cell wall. 125I-labeled human α2 laminins strongly bound to a single protein band with a 28-kDa molecular mass in the cell wall fraction in a concentration-dependent manner (Fig. 1 A–C). N-terminal sequence analysis of the α2-laminin-reactive band in the cell wall blot revealed a single protein with a sequence of KAELIDVLTQKLG. This sequence matched with 100% identity a protein encoded by a gene (MLCB637.34) with unknown function in the M. leprae genome database.
Characterization of the Putative LBP21 Gene of M. leprae.
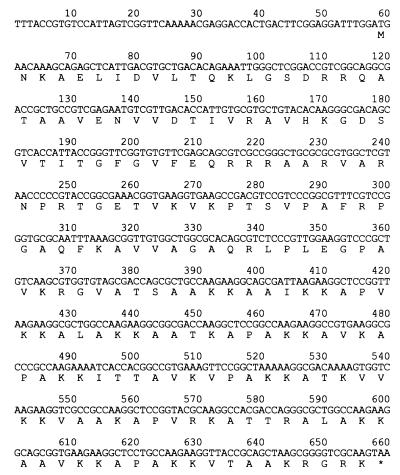
To sequence the putative LBP gene, PCR primers were designed on the basis of the sequence in the genome database. By using M. leprae DNA as a template, LBP gene and its flanking regions were amplified by PCR, and both strands of the DNA were sequenced. The nucleotide sequence was 100% identical to a gene region (nucleotides 38,586–39,317) of M. leprae cosmid MLCB637 in the GenBank database (data not shown). Sequence analysis of the putative LBP revealed that it encodes a protein of 200 aa with a deduced molecular mass of 20,960 Da. On the basis of its laminin-2-binding function and the calculated molecular mass of the deduced amino acid sequence, we designated this protein the 21-kDa laminin-2-binding protein of M. leprae (ML-LBP21). ML-LBP21 was found to lack a potential signal-peptidase cleaving site, predicted by the method of von Heijne (21). The interesting features of ML-LBP21 are (i) an unusually high contents of Ala (22%) and Lys (18.5%) residues distributed mainly in the C-terminal region of the protein, with many repeats of the sequence XKKX in which X is usually Ala (Fig. 2) and (ii) a high content of positively charged amino acids (a total of 53 positively charged residues). However, it is unlikely that the ML-LBP21-binding to laminin-2 depending on charge, because laminin-2 is also a highly positively charged molecule. Comparison of the deduced amino acid sequence of ML-LBP21 with known protein sequences deposited in databases revealed that it shows ≈78% identity over 214 aa residues with a probable histone-like DNA-binding protein (hupB gene product) of Mycobacterium tuberculosis (EMBL accession no. Z83018). Moreover, sequence homology was also found with many histone H1 proteins from microorganisms, plants, and animals (data not shown). This similarity is due to the high content of AKKA repeats and positively charged residues that are shared by both ML-LBP21 and histone proteins. However, our results show no evidence of DNA-binding activity of ML-LBP21 as demonstrated by Southwestern assay (data not shown).
Figure 2.
Nucleotide sequence of the ML-LBP21 gene and upstream region from M. leprae. The deduced amino acid sequence is indicated in single-letter code under the nucleotide sequence. ∗ denotes the stop codon.
Construction and Purification of Recombinant ML-LBP21.
To produce the rLBP21 protein, the DNA fragment encoding the mature ML-LBP21 was amplified from M. leprae chromosome, and the resultant PCR fragment was ligated with pQE expression vectors. The recombinant plasmid was transformed into Escherichia coli M15, and His-tagged fusion protein was purified by chromatography on Ni-nitrilotriacetic acid resins. Purified protein was analyzed by SDS/PAGE and Western immunoblot analyses. The molecular mass of the rML-LBP21 was estimated to be 27 kDa, which is larger than the calculated molecular mass of the protein as deduced from the nucleotide sequence (Fig. 1D). As shown in Fig. 1E, purified rML-LBP21 bound to α2-laminins in both ELISA and immunoblotting.
Characterization of ML-LBP21.
The surface proteins of M. leprae are likely to be crucial for the initial interaction with peripheral nerve. Therefore, we determined whether ML-LBP21 is localized on the surface of intact M. leprae. To test this, we have raised a panel of stable hybridomas against purified rML-LBP21. Hybridomas producing mAbs 1C3, 3H2, 3E11, 5F11, and 5F5 (all IgG class) have been characterized by using ELISA, immunoblotting, and immunofluorescence. All mAbs reacted with both recombinant and native cell wall ML-LBP21. A representative example is shown in Fig. 3B. Because ML-LBP21 contains highly positively charged residues, it is possible that this protein has a slower electrophoretic mobility on SDS/PAGE, resulting in the apparent molecular mass of recombinant protein (27 kDa) slightly above the calculated 21-kDa molecular mass. Whereas mAbs detected a single 28-kDa protein in the M. leprae cell wall fraction, all mAbs reacted much strongly with rML-LBP21 that appeared slightly lower to 28-kDa band in SDS/PAGE (Fig. 3B). This difference may be attributed to posttranslational modification of this protein, because several mycobacterial proteins have been identified as glycoproteins (22–24). Taken together, we concluded that the native 28-kDa M. leprae cell wall protein recognized by α2-laminins and mAbs is the same as rML-LBP21. In addition, ML-LBP21 shows no sequence homology to previously reported 28- to 30-kDa region mycobacterial proteins (23, 24), indicating that ML-LBP21 is a previously unidentified protein in M. leprae.
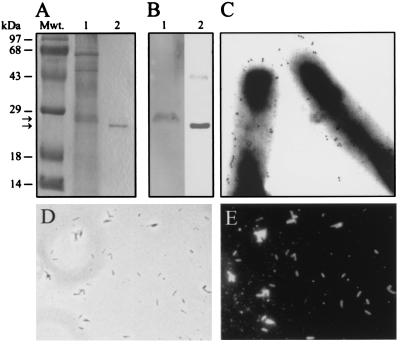
Figure 3.
ML-LBP21 is a major cell wall-associated and surface protein of M. leprae. (A) Coommassie blue-stained SDS/polyacrylamide gel of M. leprae cell wall fraction (lane 1) and rML-LBP21 (lane 2). Molecular mass markers (Mwt) in kDa are indicated above the left lane. (B) Corresponding immunoblot of M. leprae cell wall fraction (lane 1) and rML-LBP21 (lane 2) was labeled with mAb raised against rML-LBP21. mAb-reactive single 28-kDa protein band (B, lane 1) is a major protein in the cell-wall fraction (A, lane 1) that corresponds to rML-LBP21 (B, lane 2). (C) Immunoelectron microscopy on intact M. leprae with mAb against rML-LBP21. Colloidal gold particles represent the localization of ML-LBP21 on the surface of M. leprae. (D and E) Indirect immunofluorescence staining of intact M. leprae with mAb to rML-LBP21 demonstrating the intense labeling in all bacilli in the field (E). Corresponding phase contrast image is shown in D.
Immunofluoresence labeling of whole intact M. leprae by mAbs specific for ML-LBP21 suggests the presence of this protein epitopes on bacterial surface in high density. Immunoelectron microscopy further revealed the gold labeling, which is represented by the numerous black dots of 1-nm gold particle, on the surface of M. leprae (Fig. 3C). Control experiments with irrelevant mAbs show no bacterial labeling in either immunofluorescence or immunogold labeling (data not shown). In addition, ML-LBP21 could be detected as a strong band in the M. leprae cell wall fraction (Fig. 3A). These data suggest that ML-LBP21 is a major surface-exposed antigen of M. leprae and are thus likely to serve as an adhesin for the interaction with peripheral nerve.
Binding of ML-LBP21 to Endogenous Peripheral Nerve Laminins and rLNα2G Domain.
To determine the binding of ML-LBP21 to native peripheral nerve laminin-2, we have isolated the bovine PNMF, which consists of Schwann cell basal lamina. Because laminin subunits in humans and other mammalian species showed >90% amino acid sequence similarities (7, 10, 25), bovine peripheral nerve laminins corresponds closely to human laminins. As we have reported previously (17), immunoblot analysis with mAbs specific for different laminin chains revealed that laminin subunits, including laminin α2, β1, and γ1 chains, which forms the laminin-2 isoform, were extracted from the PNMF at pH 12.0 (Fig. 4 A and B). To determine whether the endogenous bovine peripheral nerve laminin-2 binds to ML-LBP21, we overlaid the PVDF transfers of rML-LBP21 and the M. leprae cell wall fraction with the pH 12.0 extracts of the native PNMF and detected the bound endogenous laminin with antibodies directed to laminin α2 and β1/γ1 chains. The endogenous bovine peripheral nerve laminin bound to rML-LBP21 (Fig. 4D, lane 2) and also bound to a broad diffuse band at ≈28 kDa in the M. leprae cell wall fraction (Fig. 4D, lane 1) that appears to correspond to the native ML-LBP21 in the cell wall fraction (Fig. 4C; Fig. 3 A and B). By using mAbs against different laminin chains, the endogenous bovine peripheral nerve laminin that bound to rML-LBP21 was identified to contain the laminin α2, β1, and γ1 chains (Fig. 4B).
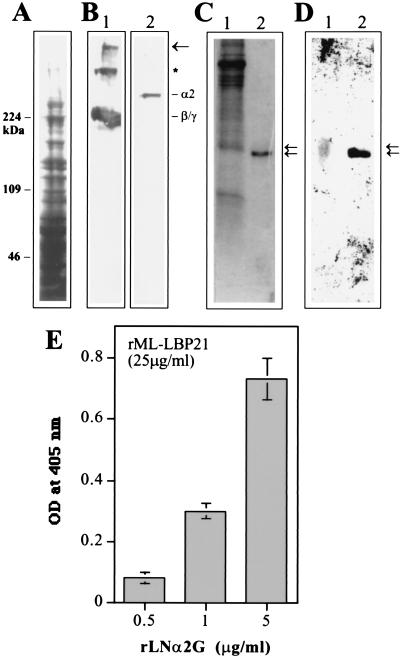
Figure 4.
rML-LBP21 binds to the endogenous peripheral nerve laminin. (A) Coommassie blue-stained SDS/polyacrylamide gel of the pH 12.0 extracts of the bovine PNMF. Molecular mass markers (Mwt) in kDa are indicated on the left. (B) Corresponding immunoblot reacted with polyclonal anti-laminin antibody (lane 1) and monoclonal anti-laminin α2 chain antibody (lane 2). The high-molecular-mass band indicated by the arrow corresponds to the laminin heterotrimer that was not dissociated by SDS/PAGE. The band indicated by ∗ appears to correspond to the heterodimer composed of β1 and γ1 chain that were not dissociated by SDS. Also shown are the laminin β1, γ1 (lane 1), and α2 chains (lane 2). (C and D) The nitrocellulose transfers of rML-LBP21 and the M. leprae cell wall fraction were incubated with the pH 12.0 extracts of the peripheral nerve membrane fraction. (D) Bound peripheral nerve laminin was detected by polyclonal anti-laminin antibody. Arrows indicate the peripheral nerve laminin bound to native cell wall ML-LBP21 (lane 1) and r ML-LBP21 (lane 2) with slight difference in molecular masses. (C) Corresponding Coommassie blue-stained SDS/polyacrylamide gel containing M. leprae cell wall fraction and rML-LBP21. Arrows indicate to show the corresponding laminin-reactive bands. (E) Binding of rML-LBP21 to the rG domain of laminin α2 chain in ELISA. Plates coated with 25 μg/ml of rML-LBP21 were incubated with increasing concentration of rLNα2G domain and detected by affinity-purified antibody against rLNα2G.
The binding of rML-LBP21 to laminin α2 chain was confirmed by the strong reactivity of rML-LBP21 to the rLNα2G domain in ELISA (Fig. 4E). Taken together, these data suggest the involvement of the α2 chain in the binding of rML-LBP21 to endogenous peripheral nerve laminin-2.
Role of ML-LBP21 in Schwann Cell Adhesion and Invasion.
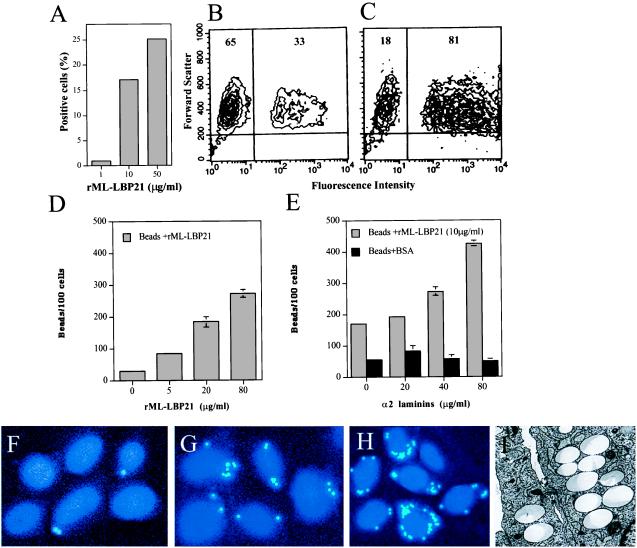
Because ML-LBP21 appears to be a major protein on the surface of M. leprae, we studied its role in Schwann cell adhesion and invasion. To test this, a method was established that uses the built-in property of fluorescent beads coupled to rML-LBP21 in flow cytometry. The rML-LBP21-coated beads were analyzed for their capacity to attach to primary Schwann cell in suspension. Fluorescence intensity was considered a measure of cell-bound beads. Beads coated with rML-LBP21 alone adhered to Schwann cells significantly after 30 min of incubation as compared with BSA-coated beads in a concentration dependent manner (Fig. 5A). The scattergram in Fig. 5B shows a population of Schwann cells carrying rML-LBP21-coated beads, as indicated by a significant shift of fluorescence intensity. Cells with strong fluorescence intensity would indicate a high number of beads per cell. The level of binding of rML-LBP21-coated beads to Schwann cells was significantly increased when beads were preincubated with exogenous α2-laminins (Fig. 5C), suggesting that the augmented attachment is due to the exogenous α2-laminins. Moreover, individual cells with high fluorescence intensity suggest that α2-laminins also increased the number of beads per cells.
Figure 5.
Role of rML-LBP21 in Schwann cell adherence and invasion. (A) Adherence of rML-LBP21-coated fluoresceinated beads to primary Schwann cells in suspension in the absence of α2 laminins as analyzed by flow cytometry. A representative example showing the percentage of positive cells carrying fluoresceinated beads coated with increasing concentration of rML-LBP21. (B and C) Representative FACS plots of Schwann cell-bound rML-LBP21-coated beads in the absence (B) and the presence of α2 laminins (C). Numbers indicate the percentage of total Schwann cells within the indicated gate. (D–I) Schwann cell invasion of rML-LBP21-coated beads in the absence and the presence of exogenous α2 laminins as determined by fluorescence and electron microscopy. (D) Quantification of Schwann cell invasion of beads coated with increasing concentration of rML-LBP21 alone as determined by fluorescence microscopy. (E) Schwann cell invasion of rML-LBP21- and BSA-coated beads in the presence of increasing concentration of exogenous α2 laminins. Fluorescence microscopic images of Schwann cell-invaded beads coated with BSA (F) rML-LBP21 (G) and rML-LBP21 + α2 laminins (H). Intracellular beads and Schwann cell nuclei are shown in light blue (dots) and blue, respectively. (I) Electron micrograph of Schwann cells showing the intracellular location of beads coated with rML-LBP21 + α2 laminins.
To assess the role of ML-LBP21 in Schwann cell invasion, cell-bound beads were allowed to invade as described in Materials and Methods. rML-LBP21-coated beads entered Schwann cells in a concentration-dependent manner after 12 hr as analyzed by using fluorescence microscopy (Fig. 5 D and G). This time point was selected for the invasion assay because all adhesin-bound beads were detected intracellularly by electron microscopy. Most of these beads were found within vacuoles in the cytoplasm. However, preincubation of rML-LBP21-coated beads, but not BSA-coated beads, with α2-laminins significantly augmented the Schwann cell invasion (Fig. 5 E and H). The invasive capacity of rML-LBP21-coated beads (10 μg/ml) was increased by more than 2-fold when beads were preincubated with 80 μg/ml α2-laminins. Electron microscopic studies further confirmed the invasion of rML-LBP21-coated beads in the presence of α2-laminins (Fig. 5I), suggesting the involvement of a laminin-dependent pathway for the Schwann cell invasion of the rML-LBP21-coated beads.
DISCUSSION
The binding of M. leprae to endoneural laminin-2 isoform on the Schwann cell basal lamina appears to be crucial in targeting the bacteria to the peripheral nerve (4). M. leprae surface proteins that bind laminin-2 play an important role in mediating this interaction. In this study, we described the identification and molecular characterization of a surface protein of M. leprae, ML-LBP21, that binds peripheral nerve laminin. We have also shown that ML-LBP21, when coated on beads, has the capacity to induce Schwann cells to ingest the beads and that exogenous α2-laminins enhance this ingestion.
By using human α2-laminins as a probe, we have identified a major 28-kDa laminin-binding protein in the cell wall of M. leprae. After N-terminal sequence analysis followed by cloning and expression in E. coli, the purified recombinant protein, rML-LBP21, also bound strongly to α2-laminins. Whereas the deduced amino acid sequence predicts a 21-kDa protein, it migrates in SDS/PAGE as a ≈6-kDa-larger molecule than the calculated molecular mass. This difference seems to be because of the high content of Lys and Arg residues, which make ML-LBP21 a highly positively charged molecule, thus allowing it to migrate slowly in SDS/PAGE. Several lines of evidence indicate that rML-LBP21 and the native 28-kDa protein in the M. leprae cell wall are the same protein: (i) the N-terminal sequence of native cell wall 28-kDa protein matched 100% with rML-LBP21; (ii) mAbs raised against rML-LBP21 specifically reacted with a single 28-kDa band in the cell wall fraction, exactly corresponding to the laminin-reactive 28-kDa protein; and (iii) endogenous peripheral nerve laminin in overlay assays reacted with both rML-LBP21 and a broad band at 28 kDa in the M. leprae cell wall fraction. On the other hand, the difference in migration between recombinant and native ML-LBP21 in SDS/PAGE is probably because of the glycosylation of the native cell wall protein during posttranslational modification. Although glycosylation is not common in bacteria, there is mounting evidence for the existence of glycosylation in many bacteria, including mycobacteria (22, 24).
Such posttranslational modifications of cell wall ML-LBP21 are not required for binding to laminins because purified rML-LBP21 strongly bound to α2-laminins.
M. leprae surface proteins are likely to play a crucial role in the initial bacterial interaction with peripheral nerves. We have demonstrated that ML-LBP21 is a major surface protein of leprosy bacilli. This was supported by demonstrating the abundance of ML-LBP21 in the cell wall fraction by immunoblotting and on the surface of intact M. leprae by immunofluorescence and immunogold labeling by using mAbs raised against rML-LBP21. Despite the surface location of this protein, amino acid sequence analysis revealed the lack of a signal peptide. It is known that some exported proteins of mycobacteria do not possess signal sequence, although the exact mechanisms of exporting such proteins are unknown (23, 24).
In the peripheral nerves, the laminin-2 isoform deposits on the Schwann cell–axon units. Laminin-2 is composed of the α2 heavy chain together with the β1 and γ1 light chains. Whereas the α2 chain has a tissue-restricted distribution, predominantly express in the basal lamina of Schwann cells and muscles, the β1 and γ1 chains have a wide range of tissue distribution (9, 10). These laminin chains are highly conserved among mammalian species, and thus bovine laminin does not differ much from human or rodent laminins (4, 10, 25). We have shown that endogenous bovine peripheral nerve laminins, which contain α2, β1, and γ1 chains bound to both the recombinant and native cell wall ML-LBP21. The binding of ML-LBP21 to the M. leprae-targeted laminin α2 chain was confirmed by the reactivity of rML-LBP21 with rG domain of human laminin α2 chain. Because M. leprae uses laminin-2 for the interaction with bovine peripheral nerve (4), ML-LBP21-binding to endogenous peripheral nerve laminin strengthens the hypothesis that ML-LBP21 may be involved in M. leprae interaction with peripheral nerves. Additionally, the binding of the endogenous peripheral nerve laminin to rML-LBP21 was not affected by EDTA or heparin (data not shown). On the other hand, EDTA and heparin affected the binding activity of laminins to their eukaryotic cell receptors such as dystroglycan to varying degrees (20, 26). This suggests that the nature of laminin-2 binding to ML-LBP21 is different from that of mammalian laminin receptors.
The inability to culture M. leprae in vitro and the highly complex nature of the M. leprae cell wall has made the analysis of the pathogenic role of individual surface proteins of M. leprae extremely difficult. Moreover, we also believe that the presence of multiple laminin binding antigens on the surface of M. leprae are likely to make such analysis complicated. The property of fluorescent beads coupled to purified rML-LBP21 and their use in flow cytometry and fluorescence microscopy has enabled us to identify the role played by the ML-LBP21 in Schwann cell adhesion and invasion. Our data showed that beads coated with rML-LBP21 adhered to and entered primary Schwann cells. Because Schwann cells secrete laminin-2 constitutively, it is possible that endogenous laminin-2 directly mediates the binding of rML-LBP21-coated beads to Schwann cells. On the other hand, preincubation of rML-LBP21-coated beads with exogenous α2-laminins significantly increased the bead’s adhesion and invasion. This result suggests that the exogenous α2-laminins bridge rML-LBP21 to unoccupied laminin receptors on Schwann cells and subsequently mediates the ingestion. Taken together, our data suggest that ML-LBP21 may function as a critical surface adhesin that facilitates the entry of M. leprae into Schwann cells, and this invasion involves a laminin-dependent pathway, possibly through laminin receptors on Schwann cells. More recently, we have shown the involvement of α-dystroglycan, a non-integrin laminin receptor, as a Schwann cell receptor for M. leprae (20). Whether α-dystroglycan plays a role in ML-LBP21-mediated Schwann cell invasion remains to be investigated.
Acknowledgments
We gratefully acknowledge Patrick J. Brennan, James L. Salzer, George Zanazzi, Steve Terlow, Fumiaki Saito, and Peter D. Yurchenco for their valuable contribution, material supply, and expertise. A.R. thanks Patricia Ryan for the initial 125I labeling experiments and for the encouragement of this study. We also thank Clara Eastby for the assistance with preparing mAbs, Helen Shio for electron microscopy, and Ines Chen for Southwestern assay. Supported by grants from the UNDP/World Bank/World Health Organization Special Program for Research in Tropical Diseases (TDR) and the National Institutes of Health (ROI Grant AI45816) to A.R.
ABBREVIATIONS
- ML-LPB21
laminin-2-binding protein of M. leprae
- PNMF
peripheral nerve membrane fraction
- LNα2G
G domain of the laminin-α2 chain
- LBP
laminin-binding protein
- r
recombinant
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB022517).
References
- 1.Job C K. Int J Lepr. 1989;57:532–539. [PubMed] [Google Scholar]
- 2.Stoner G L. Lancet. 1979;10:994–996. doi: 10.1016/s0140-6736(79)92564-9. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. WHO Weekly Epidemiological Record, 20 September. 1995. p. 269. [Google Scholar]
- 4.Rambukkana A, Salzer J L, Yurchenco P D, Tuomanen E I. Cell. 1997;88:811–821. doi: 10.1016/s0092-8674(00)81927-3. [DOI] [PubMed] [Google Scholar]
- 5.Burgeson R E, Chiquet M, Deutzmann R, Ekblom P, Engel J, Kleinman H, Martin G R, Meneguzzi G, Paulsson M, Sanes J, et al. Matrix Biol. 1994;14:209–211. doi: 10.1016/0945-053x(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 6.Yurchenco P D, O’ Rear J J. Curr Opin Cell Biol. 1994;6:674–681. doi: 10.1016/0955-0674(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 7.Engvall E, Wewer U M. J Cell Biochem. 1996;61:493–501. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C493::AID-JCB2%3E3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 8.Anton E S, Sandrock A W, Matthew W D. Dev Biol. 1994;164:133–146. doi: 10.1006/dbio.1994.1186. [DOI] [PubMed] [Google Scholar]
- 9.Leivo I, Engvall E. Proc Natl Acad Sci USA. 1988;85:1544–1548. doi: 10.1073/pnas.85.5.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanes J R, Engvall E, Butkowsky R, Hunter D D. J Cell Biol. 1990;111:1685–1699. doi: 10.1083/jcb.111.4.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornbrooks C J, Carey D J, McDonald J A, Timpl R, Bunge R P. Proc Nat Acad Sci USA. 1983;80:3850–3854. doi: 10.1073/pnas.80.12.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. Purification of M. leprae. Vol. 5. WHO document TDR/SWG/IMMLEP; 1980. p. 80. [Google Scholar]
- 13.Hunter S W, Rivoire B, Mehra V, Bloom B R, Brennan P J. J Biol Chem. 1990;265:14065–14068. [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rambukkana A, Das P K, Burggraaf J D, Yong S, Faber W R, Thole J E, Harboe M. Infect Immun. 1992;60:5172–5181. doi: 10.1128/iai.60.12.5172-5181.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito F, Yamada H, Sunada Y, Hori H, Shimizu T, Matsumura K. J Biol Chem. 1997;272:26708–26713. doi: 10.1074/jbc.272.42.26708. [DOI] [PubMed] [Google Scholar]
- 18.Yamada H, Chiba A, Endo T, Kobata A, Anderson L V, Hori H, Fukuta-Ohi H, Kanazawa I, Campbell K P, Shimizu T. J Neurochem. 1996;66:1518–1524. doi: 10.1046/j.1471-4159.1996.66041518.x. [DOI] [PubMed] [Google Scholar]
- 19.Einheber S, Milner T A, Giancotti F, Salzer J L. J Cell Biol. 1993;123:1223–1236. doi: 10.1083/jcb.123.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rambukkana A, Yamada H, Zanazzi G, Mathus T, Salzer J L, Yurchenco P D, Campbell K P, Fischetti V A. Science. 1998;282:2076–2079. doi: 10.1126/science.282.5396.2076. [DOI] [PubMed] [Google Scholar]
- 21.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garbe T, Harris D, Vordermeier M, Lathigra R, Ivanyi J, Young D. Infect Immun. 1993;61:260–267. doi: 10.1128/iai.61.1.260-267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thole J E, Wieles B, Clark-Curtiss J E, Ottenhoff T H, de Wit T F. Mol Microbiol. 1995;18:791–800. doi: 10.1111/j.1365-2958.1995.18050791.x. [DOI] [PubMed] [Google Scholar]
- 24.Menozzi F D, Bischoff R, Fort E, Brennan M J, Locht C. Proc Natl Acad Sci USA. 1998;95:12625–12630. doi: 10.1073/pnas.95.21.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vuolteenaho R, Nissinen M, Sainio K, Byers M, Eddy R, Hirvonen H, Shows T B, Sariola H, Engvall E, Tryggvason K. J Cell Biol. 1994;124:381–394. doi: 10.1083/jcb.124.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada H, Denzer A J, Hori H, Tanaka T, Anderson L V B, Fujita S, Fukutaohi H, Shimizu T, Ruegg M A, Matsumura K. J Biol Chem. 1996;271:23418. doi: 10.1074/jbc.271.38.23418. [DOI] [PubMed] [Google Scholar]