Abstract
Bacteriophage χ is known to infect motile strains of enteric bacteria by adsorbing randomly along the length of a flagellar filament and then injecting its DNA into the bacterial cell at the filament base. Here, we provide evidence for a “nut and bolt” model for translocation of phage along the filament: the tail fiber of χ fits the grooves formed by helical rows of flagellin monomers, and active flagellar rotation forces the phage to follow the grooves as a nut follows the threads of a bolt.
Keywords: Escherichia coli, Salmonella typhimurium, phage infection, flagellar rotation
Bacteriophage χ infects motile strains of the genera Escherichia, Salmonella, and Serratia (1–3). Schade et al. (4) demonstrated translocation of phage along flagellar filaments by electron microscopy: phage heads tended to be full when attached to a filament but empty when attached to its base. When it was realized that flagellar filaments rotate, the suggestion was made that a phage moves along the filament “like a nut on a bolt” (5). This observation was consistent with the finding that mutants with straight filaments, while nonmotile, remain fully sensitive to χ (6). Ravid and Eisenbach (7) found that the number of phage particles adsorbed by cells, i.e., removed from the supernatant fraction, correlated only with the fraction of the population of cells whose flagella rotated incessantly; it did not correlate with the direction of rotation. This finding was not consistent with a nut and bolt model. Yamaguchi et al. (8) found conditions in which unflagellated bacteria are sensitive to χ-phage: drops of concentrated phage cleared spots on the surfaces of hard agar plates. However, this sensitivity might have resulted simply from the large multiplicity of phage. Attempts to visualize phage translocation by dark-field or differential-interference-contrast microscopy have not been successful: one can see χ-phage attached to filaments but not their travel in either direction (R.M. Macnab, personal communication; ref. 9). However, these experiments required low flagellar rotation rates. The prevailing view seems to be that phage particles adsorb and desorb, stepping along the filament in a one-dimensional random walk.
If flagellar rotation drives χ translocation in the manner of a nut on a bolt, then a bacterial strain’s sensitivity to χ-phage has three mechanical requirements: flagellar rotation, the correct direction of flagellar rotation, and the correct pattern of grooves on the surface of the flagellar filament. By measuring infectivity directly, we asked whether these mechanical constraints are determinants of sensitivity to χ-phage.
MATERIALS AND METHODS
Bacteria, Phage, Media.
Strains are listed in Table 1. E. coli strain HCB8 is a derivative of AW405 made by P1 transduction of the fliK allele from strain YK4161 of the 4101 series of ref. 18. HCB626 is a derivative of RP437 that contains a deletion of cheY and harbors a lambda-lysogen that expresses cheY under control of the lac promoter.
Table 1.
Bacterial strains and plasmid used in this study
| Strain | Relevant genotype | Ref. or source |
|---|---|---|
| E. coli | ||
| AW405 | Wild type | 10 |
| RP437 | Wild type | 11 |
| HCB5 | AW405, fliC | 12 |
| HCB8 | AW405, fliK | This study |
| HCB626 | RP437, ΔcheY, λcI857(ts)[plac-cheY] | D. F. Blair |
| MS5037 | AW405, motA | 13 |
| S. typhimurium | ||
| SJW1103 | Wild type (f2) | 14 |
| SJW1655 | SJW1103, fliCA449V (f11) | 15 |
| SJW1660 | SJW1103, fliCG426A (f0) | 15 |
| SJW1664 | SJW1103, fliCG426A, A449T (f1) | 15 |
| SJW2868 | SJW1103, fliCA427V (f5/6*) | 16 |
| HCB1280 | SJW1660, ΔcheY | This study |
| HCB1281 | SJW1660, fliGΔ169-171 (ΔPAA) | This study |
| Plasmid | ||
| pDFB36 | plac-motA | 17 |
This allele is curly, but whether it is f5 or f6 has not been determined.
Salmonella strains SJW1655, 1660, 1664, and 2868 each contain one or two point mutations in fliC that alter the polymorphism of the flagellar filament. The amino acid change conferred by each mutation, noted in Table 1, was determined by ref. 16. The f-numbers refer to the number of short protofilaments in the flagellum (19). These flagellar variants derive from the same parental strain (SJW1103); therefore, they have the same serotype (i).
HCB1280 contains an in-frame deletion of the cheY locus. The deletion was created by PCR amplification of two fragments containing portions of the cheY ORF. The first fragment was amplified by using the following primers: cheR, 5′-CGGTGTAT GAGCTCAGT AAGGATAAG-3′ and cheYup, 5′-CCTCTTCC ACATTGTTG GATCCAAGC TCTTTTAAGAG-3′. The second fragment was amplified by using cheYdown, 5′-GATGGTCACG GCGGGATCCA AAAAAGAGAA TATTATCG-3′ and cheYdnpvu, 5′-CTTGACAGCT GGTTGCATCA TCATCGC-3′. Fragment 1 was subcloned into the SacI and BamHI sites of pMAK705 (20), creating pMAKcheY1. Fragment 2 was subcloned into the BamHI and PvuI sites of pMAKcheY1, creating pMAKΔcheY. When situated in this manner, fragments 1 and 2 generate an in-frame deletion within the cheY ORF. This deletion was recombined onto the chromosome of SJW1660 by the method of ref. 20. The presence of the deletion was confirmed by sizing a PCR product.
HCB1281 contains a three-codon deletion (codons 169–171) within fliG that confers a super-tumbly phenotype on Salmonella. This fliG allele was PCR-amplified from SJW2811 (21) by using the following primers: fliG1, 5′-GTGGAAAGCCGGATCCAACGCCGTATC-3′ and fliG2, 5′-GCGCGTCAAGCTTGTTCTGAAACTCGC-3′. The amplified product was subcloned into the BamHI and HindIII sites of pMAK705. The deletion then was introduced into the chromosome of SJW1660 by the method of ref. 20. The presence of the deletion was confirmed by sequence analysis.
Fructose minimal medium was 0.2% (wt/vol) fructose in M63 salts (22) supplemented with 100 μg/ml thiamine, 1 mM MgSO4, and 1 mM each of the l-amino acids threonine, leucine, histidine, and methionine.
Assay for Infectivity.
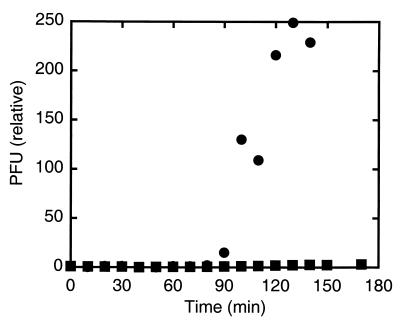
We monitored the number of plaque-forming units (PFUs) in a cell suspension as a function of time. The bacterial cultures were grown on fructose minimal medium (a medium that does not catabolite-repress flagellar synthesis) in a shaking incubator at 33°C. We inoculated a suspension of bacterial cells at early exponential phase (OD650≈0.2) with χ-phage (ATCC 9841-B1) at a multiplicity of infection (the ratio of phage to cells) of ≈0.01. At intervals of 15–20 min after inoculation and for up to 180 min after inoculation, a sample (100 μl) was taken and diluted by fixed amounts between 104- and 106-fold. The diluted samples were combined with bacteria known to be lysed by the phage [100 μl of wild-type strain AW405, grown to saturation on tryptone broth (TB), diluted 1:100, and then grown for another 5 h]. The resulting mixtures were combined with top agar (TB, 0.5% agar) and plated on the surfaces of hard agar plates (TB, 1% agar). These plates were incubated overnight at 33°C. The next day, the numbers of plaques on the plates were counted. The data were normalized by dividing by the PFUs observed about 1 min after inoculation. If the cells to which χ-phage were added are sensitive to the phage, the phage replicate, and the number of PFUs increases by a large factor. Under our conditions, this increase began about 90 min after addition of phage and was effectively all or none (Fig. 1).
Figure 1.
Quantitative plaque assays of suspensions of HCB626 with flagellar filaments rotating CCW (●) or CW (■). Suspensions were inoculated with phage at t = 0. PFUs were normalized by dividing the PFU at all times by the PFU at t = 1.
RESULTS
Requirement for Flagellar Rotation.
Schade and Adler (23) found two kinds of resistant derivatives of a strain that is sensitive to χ, mutants without flagella and mutants with paralyzed flagella. They concluded that only motile strains of E. coli are sensitive to χ. We started this investigation by confirming these results. First, we compared strains of E. coli that are isogenic except for a single mutation in fliC, the gene encoding flagellin. The wild-type strain AW405 has normal flagellar filaments and motors (10); HCB5 has normal flagellar motors but lacks flagellar filaments, although it does have the short hook structure that ordinarily couples the filament to the basal body. We found that AW405 is sensitive to infection whereas HCB5 is resistant. Next we asked whether flagellar rotation, and not the mere presence of flagella, promotes infection. We used a strain of E. coli (MS5037) that has paralyzed flagella owing to a defect in the gene motA. MS5037’s flagellar filaments and basal bodies are normal; however, its motors cannot generate torque. Flagellar rotation can be restored in MS5037 by inducing transcription of wild-type motA fused to the lac promoter carried on a plasmid, pDFB36 (Table 1). We found that when MS5037 carrying the plasmid is grown in the presence of the inducer isopropyl β-d-thiogalactopyranoside, the flagella rotate, and the strain is sensitive to χ-phage. On the other hand MS5037 with paralyzed flagella is resistant to χ-phage. Thus active flagellar rotation is required for infection by χ-phage.
Dependence on Direction of Rotation.
The restored motors of MS5037 turn alternately clockwise (CW) and counterclockwise (CCW). But directionality is critical to the proposed mechanism. So we studied strain HCB626 that has normal flagellar motors and filaments but in which the gene cheY is controlled by the lac promoter. CheY is a chemotaxis signaling protein that interacts with the flagellar motor, decreasing the CCW bias. When HCB626 is grown without isopropyl β-d-thiogalactopyranoside (IPTG), cells swim smoothly because of a large CCW bias; in this case, we found that the cells are sensitive to χ-phage, as shown in Fig. 1. When HCB626 is grown with 2.5 mM IPTG, cells tumble incessantly because of a small CCW bias; in this case, the cells are resistant to χ-phage (Fig. 1). The rotational biases in the two cases were confirmed by observing tethered cells. Thus CCW rotation is a constraint on sensitivity to χ-phage. Because IPTG has opposite effects in our experiments, i.e., confers sensitivity with MS5037 and confers resistance with HCB626, we also can rule out any artifact caused by its presence. An alternative interpretation is that swimming motility promotes infection and that CCW rotation is only its indirect cause. This interpretation can be ruled out because a Salmonella mutant with straight filaments that rotate alternatively CCW and CW is sensitive to χ-phage (6).
Dependence on Hook Length.
The composition of the “bolt” is intrinsic to the proposed mechanism. The hook is also essential, because after travelling the length of the filament, the phage must traverse the hook. Yamaguchi et al. (8) found that when χ-phage was spotted at high concentrations on bacterial lawns it could infect both mutants lacking filaments and mutants with polyhooks (elongated hooks owing to a defect in FliK, a factor that regulates hook length). Using the same spot assay, Kagawa et al. (24) found one resistant polyhook strain that had paralyzed motors. The proposed model readily explains this result; sensitivity to χ-phage requires active motor rotation. However, because strains without flagellar filaments are resistant to χ-phage (2, 4), the sensitivity of the hook and polyhook mutants observed in this work probably reflects the use of the spot assay.
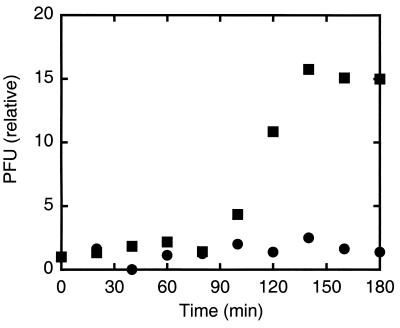
Hooks are so short (≈50 nm long) that they cannot capture diffusing phage particles from the surrounding fluid with an efficiency comparable to that of flagellar filaments (several μm long). Therefore, hook elongation should promote sensitivity to χ-phage in suspension. The only difference between E. coli strain HCB5 and the polyhook strain HCB8 is the hook’s elongation to ≈1 μm. We found that HCB8 is sensitive to χ-phage, whereas HCB5 is not (Fig. 2). We note a significant difference in relative increase in PFUs between HCB8 (≈20) and HCB626 (Fig. 1, ≈200). Phage burst sizes can vary strongly with genetic background (25). The observed difference is partly the result of this effect, because the parent strain of HCB8 is AW405, which has a comparable burst size (≈40). HCB8’s polyhooks are shorter than filaments, so its smaller burst size also might reflect a lower efficiency of adsorption. HCB8 does not swim: we reiterate the fact that swimming does not directly promote infection.
Figure 2.
Quantitative plaque assays of suspensions of HCB5 that has hooks (●) and HCB8 that has polyhooks (■).
Dependence on Filament Structure.
That χ-phage attacks the flagella of different species, as well as both polyhooks and flagellar filaments, implies a lack of binding specificity. The nut-and-bolt hypothesis implies that the helical grooves between the flagellin or hook monomers serve as threads, but CCW rotation will only pull the phage toward the cell body if the phage slides along a right-handed groove. Electron micrographs reveal two kinds of right-handed helical grooves, a right-handed six-start helix and a right-handed one-start helix (26). The flagellar hook surface shares these helical grooves (27). However, the flagellar filament is polymorphic and the pitch of these grooves depends on the polymorphic form. Therefore, sensitivity to χ likewise could depend on polymorphic form.
The flagellar filament is a cylinder with a wall composed of 11 helical protofilaments. Each protofilament can have one of two conformational states, called L and R. If all protofilaments are either L or R, then the flagellar filament is straight. Alternatively, if some protofilaments are L and others R then the flagellar filament contorts into a superhelix. Assuming that the protofilaments are grouped together, there can be 12 distinct polymorphic forms called f0 through f11 depending on the number of R-protofilaments present (19). Both the overall shape and the surface groove patterns of the flagellar filaments vary continuously progressing from f0 to f11 (28, 29). Polymorphism bears on the proposed model in two ways. Because the phage interacts with the surface of the filament, the model predicts that susceptibility to phage depends on filament surface structure, not overall flagellar shape. Also, because the normal flagellar filament is f2, the model requires that if other polymorphic forms support phage translocation, they will be those with surface structure nearest to that of f2.
A number of flagellar mutants with filaments locked into different polymorphic shapes are available in S. typhimurium. Because the binding affinity of χ-phage might vary with serotype, we studied flagellar variants derived from one parent strain (Table 1). We found that normal motors driving f0, f1, or f2 flagella promote infection by χ-phage whereas strains with f5/6 or f11 flagella resist infection. The model is consistent with this partitioning: the surface structures of f0 and f1 are closer to that of f2 than to either f5/6 or f11. Both the six- and one-start helical grooves of the f11 flagellar filament have smaller pitch than the corresponding helical grooves of the f0 filament; in other words, increasing the fraction of R- protofilaments tightens the threads of the six- and one-start helical grooves. We suggest that the tail fiber of χ-phage only fits in the looser grooves.
Dependence on Direction of Rotation (Revisited).
Because f0 and f11 are both straight rods but only f0 promotes infection, only the structure of the helical grooves matters, not overall flagellar shape. Moreover, unlike normal filaments, straight filaments do not undergo torsion-induced flagellar polymorphism. Thus the f0 filament offers a more rigorous test of the constraints of directionality. We found that strain HCB1280 with f0 flagellar filaments with a large CCW bias is sensitive to χ-phage infection whereas strain HCB1281 with f0 flagellar filaments with a small CCW bias is resistant. Therefore, CCW rotation of the flagellar filament is required for sensitivity to χ-phage. The fact that CW rotation confers resistance is not an artifact of directionality-dependent polymorphism. Both HCB1280 and HCB1281 were derived from SJW1660 by the introduction of single mutant alleles, so there is no artifact caused by strain background.
DISCUSSION
Both CCW flagellar rotation and the correct structure of the helical grooves on the flagellar surface are essential to infection by χ-phage. We suspect that the reason that wild-type strains with normal motors that turn alternately CCW and CW are sensitive to χ-phage is because the time required for translocation of the phage is less than the time of a CCW interval, which has a mean length of about 1 sec. Because the groove of the six-start helix has a pitch of ≈50 nm and normal flagellar rotation is ≈100 Hz, χ could travel a few microns in 1 sec, provided that the friction of the tail within the groove is small. The screw motion should be more efficient when other filaments block the phage head, which occurs when normal f2 flagella spin CCW and form a bundle. However, this blockage is not necessary, because neither straight filaments nor polyhooks form stable bundles.
We have not determined whether the phage travels along the six- or the one-start helix, although the six-start helix runs more deeply into the surface of the filament and thus might be more likely. Direct observation of phage translocation (along rapidly rotating filaments) or structural studies of the fiber-groove interaction might clarify this issue.
The accumulated evidence supports a model in which active flagellar rotation drives translocation of the bacteriophage along the flagellar filament in the manner of a nut on a bolt.
Acknowledgments
We thank L. Turner, A. Kay, and D. F. Fung for helpful discussions, and S.-I. Aizawa, S. Yamaguchi, M. I. Simon, and R. M. Macnab for gifts of strains. P.N.D. gratefully acknowledges fellowship support from the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation. This work was supported by National Institutes of Health Grant AI16478 and the Rowland Institute for Science.
ABBREVIATIONS
- CW
clockwise
- CCW
counterclockwise
- PFU
plaque-forming unit
References
- 1.Sertic V, Boulgakov N A. C R Soc Biol. 1936;123:887–888. [Google Scholar]
- 2.Meynell E W. J Gen Microbiol. 1961;25:253–290. doi: 10.1099/00221287-25-2-253. [DOI] [PubMed] [Google Scholar]
- 3.Iino T, Mitani M. J Virol. 1967;1:445–447. doi: 10.1128/jvi.1.2.445-447.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schade S Z, Adler J, Ris H. J Virol. 1967;1:599–609. doi: 10.1128/jvi.1.3.599-609.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg H C, Anderson R A. Nature (London) 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- 6.Iino T, Mitani M. J Gen Microbiol. 1967;49:81–88. doi: 10.1099/00221287-49-1-81. [DOI] [PubMed] [Google Scholar]
- 7.Ravid S, Eisenbach M. J Bacteriol. 1983;154:604–611. doi: 10.1128/jb.154.2.604-611.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi S, Fujita H, Kuroiwa T, Iino T. J Gen Microbiol. 1977;99:209–214. [Google Scholar]
- 9.Fahrner K A. Ph.D. thesis. Cambridge, MA: Harvard University; 1995. [Google Scholar]
- 10.Armstrong J B, Adler J, Dahl M M. J Bacteriol. 1967;93:390–398. doi: 10.1128/jb.93.1.390-398.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkinson J S. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scharf B E, Fahrner K A, Turner L, Berg H C. Proc Natl Acad Sci USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverman M, Simon M. Nature (London) 1976;264:577–580. doi: 10.1038/264577a0. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. J Gen Microbiol. 1984;130:255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]
- 15.Kamiya R, Hotani H, Asakura S. Symp Soc Exp Biol. 1982;35:53–76. [PubMed] [Google Scholar]
- 16.Kanto S, Okino H, Aizawa S-I, Yamaguchi S. J Mol Biol. 1991;219:471–480. doi: 10.1016/0022-2836(91)90187-b. [DOI] [PubMed] [Google Scholar]
- 17.Blair D F, Berg H C. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 18.Komeda Y, Kutsukake K, Iino T. Genetics. 1980;94:277–290. doi: 10.1093/genetics/94.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calladine C R. J Mol Biol. 1978;118:457–479. [Google Scholar]
- 20.Hamilton C M, Marti A, Washburn B K, Babitzke P, Kushner S R. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Togashi F, Yamaguchi S, Kihara M, Aizawa S-I, Macnab R M. J Bacteriol. 1997;179:2994–3003. doi: 10.1128/jb.179.9.2994-3003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silhavy T J, Berman M L, Enquist L W. Experiments with Gene Fusions. Plainview, NY: Cold Spring Harbor Lab. Press; 1984. p. 219. [Google Scholar]
- 23.Schade S Z, Adler J. J Virol. 1967;1:591–598. doi: 10.1128/jvi.1.3.591-598.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kagawa H, Ono N, Enomoto M, Komeda Y. J Bacteriol. 1984;157:649–654. doi: 10.1128/jb.157.2.649-654.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delbruck M. J Bacteriol. 1945;50:131–135. doi: 10.1128/JB.50.2.131-135.1945. [DOI] [PubMed] [Google Scholar]
- 26.O’Brien E J, Bennett P M. J Mol Biol. 1972;70:133–152. doi: 10.1016/0022-2836(72)90168-4. [DOI] [PubMed] [Google Scholar]
- 27.Wagenknecht T, DeRosier D J, Aizawa S-I, Macnab R M. J Mol Biol. 1982;162:69–87. doi: 10.1016/0022-2836(82)90162-0. [DOI] [PubMed] [Google Scholar]
- 28.Namba K, Vonderviszt F. Q Rev Biophys. 1997;30:1–65. doi: 10.1017/s0033583596003319. [DOI] [PubMed] [Google Scholar]
- 29.Hasegawa K I, Yamashita I, Namba K. Biophys J. 1998;74:569–575. doi: 10.1016/S0006-3495(98)77815-4. [DOI] [PMC free article] [PubMed] [Google Scholar]