Abstract
ClC-type anion-selective channels are widespread throughout eukaryotic organisms. BLAST homology searches reveal that many microbial genomes also contain members of the ClC family. An Escherichia coli–derived ClC Cl− channel homologue, “EriC,” the product of the yadQ gene, was overexpressed in E. coli and purified in milligram quantities in a single-step procedure. Reconstitution of purified EriC into liposomes confers on these membranes permeability to anions with selectivity similar to that observed electrophysiologically in mammalian ClC channels. Cross-linking studies argue that EriC is a homodimer in both detergent micelles and reconstituted liposomes, a conclusion corroborated by gel filtration and analytical sedimentation experiments.
Keywords: yadQ, liposome, chromatography, glutaraldehyde, ultracentrifugation
INTRODUCTION
The phylogenetically ubiquitous Cl− channel proteins of the ClC family are responsible for a multitude of physiological functions in organisms as varied as mammals, elasmobranchs, yeast, and green plants (Jentsch et al. 1995, Jentsch et al. 1999). In humans, these channels are intimately involved in electrical excitability of skeletal muscle and neurons, in epithelial electrolyte homeostasis, and in cellular volume regulation. The ClC channels make up the only known molecular family of voltage-gated Cl− channels, but very little is understood about their molecular architecture or about the roles their conserved sequences play in gating or permeation. Our ignorance of ClC mechanisms stems from the molecular asymmetry of the Cl− permeation pathway, a necessary consequence of the unusual construction of these channels, in which each subunit of a homodimeric complex forms an independent pore (Middleton et al. 1994a, Middleton et al. 1996; Ludewig et al. 1996). This architecture demands that the pore is lined by protein residues scattered throughout the primary sequence rather than in localized “hot spots” (Middleton et al. 1996). In this sense, ClC channels differ fundamentally from many well-studied ion channels, such as voltage-gated Na+, K+, and Ca2+ channels, cyclic nucleotide–gated channels, inward rectifiers, neurotransmitter-activated channels, and connexins, which are all built on a “barrel-stave” plan with the ion conduction pore formed by repetition of a transmembrane subunit around an axis of four-, five-, or sixfold symmetry (Hille 1992). Just as the symmetry of these familiar channels has contributed greatly to their tractability in sequence-function studies, its absence seriously hinders efforts to discern molecular features of ClC channels by the classical, but indirect, means of mutagenesis and functional analysis.
Recently emergent genome sequences have revealed in prokaryotes many homologues of ion channels hitherto considered strictly eukaryotic (MacKinnon and Doyle 1997; Jentsch et al. 1999). The biological functions of these bacterial genes are entirely unknown. Nevertheless, membrane proteins from prokaryotes have been uniquely successful in heterologous overexpression, and so we were inspired to search for prokaryotic members of the ClC family. This study examines sequences of nine putative ClC channel genes from bacteria and archaea and pursues one of these from Escherichia coli, which can be overexpressed and functionally reconstituted as a Cl− channel. This prokaryotic ClC channel, which we name “EriC,” forms a functional homodimer, a result implying that dimeric quaternary structure is general to the ClC family.
MATERIALS AND METHODS
Materials
All chemicals were reagent grade. 36Cl was purchased from DuPont NEN as a 0.5–1 M HCl solution and was neutralized with NaOH. Working stock solutions were 50–60 mM NaCl, 26–31 μCi/ml. 3H-Glutamic acid was also obtained from DuPont NEN. Dowex 1×4–100, from Sigma Chemical Co. was converted to the glutamate form and stored in water. E. coli lipids (polar lipid extract) were obtained from Avanti Polar Lipids. Glutaraldehyde was Grade I from Sigma Chemical Co.
ClC Searches
Searches for ClC homologues were performed using BLAST 2.0 (Altschul et al. 1997) or WU-BLAST2 at the following sites on the indicated protein database: National Center for Biotechnology Information (NCBI; nonredundant data base), Swiss Institute of Bioinformatics (SIB; completed bacterial genomes and EMBL prokaryotes), The Institute for Genomic Research (TIGR; microbial genomes). Initial searches using ClC-0, ClC-3, or ClC-7 as query retrieved eight prokaryotic sequences with high statistical scores for similarity to ClCs; further searches using prokaryotic sequences as query retrieved one additional sequence (Methanococcus).
Recombinant DNA and General Biochemical Methods
Oligonucleotides corresponding to flanking regions of the E. coli ClC gene yadQ (Blattner et al. 1997) were used to PCR-amplify the gene from E. coli genomic DNA. The PCR product was blunt-end ligated into the ZeroBlunt vector (Invitrogen Corp.) and subcloned into pASK90 (Skerra 1994), downstream from the tetracycline promoter. The sequence of the open reading frames of the products of two independent PCR reactions matched the sequence in the E. coli TIGR database. A hexahistidine tag was added immediately after the initiating methionine codon using PCR mutagenesis, and the resulting construct, pEriC, was used in all experiments.
All SDS-PAGE experiments were run under reducing conditions (2% β-mercaptoethanol) using standard Laemmli solutions (Laemmli 1970). Proteins were visualized using Coomassie Brilliant Blue R-250.
Expression and Purification
E. coli JM83 cells were transformed with pEriC, grown overnight at 37°C, and diluted 50-fold into room-temperature Terrific Broth (Sambrook et al. 1989), containing 100 μg/ml ampicillin. Cells were grown at room temperature (23–25°C) to A550 of 1.0, and then induced overnight by addition of 0.2 mg/liter anhydrotetracycline (added from a 0.2 mg/ml stock in dimethylformamide). Cells were harvested and kept at 4°C during all subsequent steps. After washing with Buffer A (95 mM NaCl, 5 mM KCl, 50 mM MOPS-NaOH, pH 7.0) and resuspension at 50 ml per liter culture, the cells were disrupted by sonication in the presence of leupeptin (1 μg/ml), pepstatin (1.4 μg/ml), PMSF (0.17 mg/ml), and β-mercaptoethanol (6 mM). Debris was discarded after centrifugation (8,000 g, 25 min), and membranes were collected from the resulting supernatant (110,000 g, 45 min). The membranes were resuspended in Buffer B (95 mM Na-Pi, 5 mM KCl, pH 7.0), supplemented with 250 mM sucrose and then stored at −80°C.
Membranes were extracted for 2 h (at a concentration equivalent to A280 = 20 in 0.5% SDS) in Buffer BK (95 mM K-Pi, 5 mM KCl, pH 7.0) containing 15 mM dodecylmaltoside (DDM)1. Insoluble material was removed by centrifugation (110,000 g, 45 min). For binding, 1/10 volume of Buffer E (400 mM imidazole-HCl in Buffer BK, 1 mM DDM) was added and the pH was adjusted to 7.8. This extract was incubated overnight with Ni-NTA beads (0.25 ml beads/liter culture; QIAGEN Inc.). The beads were transferred into a column and washed with ∼40 vol wash buffer (40 mM imidazole-HCl, 95 mM K-Pi, 5 mM KCl, 1 mM DDM, pH 7.8) until A280 < 0.02. EriC was then eluted with Buffer E, pH 7.0. The 40-mM imidazole present during binding and wash steps suppresses the binding of contaminating E. coli proteins and thereby increases the purity (while decreasing the final yield) of the EriC preparations.
Protein concentration was measured from the absorbance at 280 nm, using a mass extinction coefficient (∈ = 0.85 cm2/mg) calculated from the sequence (Creighton 1984), or with the Coomassie Plus protein microassay (Pierce), calibrated using purified EriC dialyzed free of imidazole. Some experiments used the bacterial K+ channel protein KcsA, for which expression and purification were carried out as described (Heginbotham et al. 1997).
Protein Reconstitution
Reconstituted vesicles were formed by combining EriC with lipid-detergent–mixed micelles, and then removing detergent by gel filtration. Except where noted, all reconstitution procedures and flux assays were performed at room temperature. E. coli lipid was dried under N2 in a glass tube, resuspended in pentane, and redried. The lipid was suspended at 20 mg/ml by sonication in Buffer R [450 mM KCl, 20 mM morpholino ethanesulfonic acid (MES)-NaOH, pH 6.2], 34 mM 3-[(3-cholamido-propyl)dimethylammonio]-1-propanesulfonate (CHAPS) was added, and the suspension was sonicated again. After a 2-h incubation, EriC was added to the desired concentration (0.09–5 μg/mg lipid), along with enough Buffer R to bring the lipid concentration to 10 mg/ml. In control samples, equivalent volumes of Buffer E were added in place of EriC. After 20 min, detergent was removed and vesicles were retrieved using Sephadex G-50 spin columns as follows. Columns (1.5-ml bed volume) equilibrated with Buffer R were prespun in a clinical centrifuge at 1,000 g, 15 s to remove excess solution. A 95-μl sample of the reconstitution mix was applied to each column, and vesicles were collected by centrifuging 700 g, 1 min. Recovery from the spin varied in the range 100–150 μl. The samples were frozen in a dry ice/acetone bath and stored overnight in a frost-free freezer at −20°C. Before use, samples were removed from the freezer, cooled in a dry ice-acetone bath, thawed at room temperature, and then sonicated 5–10 s in a cylindrical bath sonicator.
Concentrative 36Cl− Uptake Assay
Influx of 36Cl− against a concentration gradient was assayed by a three-step procedure, essentially as described (Goldberg and Miller 1991; Middleton et al. 1994a,Middleton et al. 1994b). First, extravesicular Cl− was removed by spinning reconstituted vesicles (100 μl) through Sephadex G-50 columns equilibrated in Buffer F (400 mM sorbitol, 20 mM MES-NaOH, pH 6.2). Second, 36Cl− uptake was initiated by adding 5 μl of the working 36Cl− stock to 100 μl of each vesicle sample (final extravesicular concentrations: 2.4–2.8 mM NaCl, 1.2–1.5 μCi/ml 36Cl−). Finally, after the desired time of uptake, 36Cl− trapped inside the liposomes was measured after applying the sample to a Dowex-glutamate column (1.5-ml bed volume) and eluting into a scintillation vial with of 1.5 ml 400 mM sorbitol. Just before the assay, Dowex columns were prewashed with 400 mM sorbitol–5 mg/ml bovine serum albumin (2 ml) followed by 400 mM sorbitol (2 ml). To test for conductive release of 36Cl−, valinomycin (1 μg/ml) was added after uptake had reached steady state. In some experiments, the internal anion was varied by replacing KCl with the potassium salt of the desired anion in Buffer R. In other experiments, external test anions were added from stock solutions in Buffer F immediately before addition of 36Cl−.
3H-Glutamate Trapped-Volume Assay
EriC-reconstituted liposomes were prepared as described above with the following variations. Proteoliposomes were formed in Buffer L (20 mM KCl, 20 mM K-glutamate, 20 mM MES-KOH, pH 6.0), and stock solutions of 36Cl− and 3H-glutamate were added to the vesicles (final concentrations of 2 and 7 μCi/ml, respectively). Intra- and extra-vesicular solutions were equalized by freezing and thawing the samples twice, and then sonicating the suspension 5–10 s. Passive equilibrium-exchange efflux was initiated by diluting the loaded vesicles into Buffer L lacking radioactive tracers. After 40–60 min, intravesicular content was determined by spinning through Buffer L–equilibrated G-50 columns as above. Separate experiments under these conditions demonstrated: (a) that insignificant efflux of Cl− occurs in protein-free liposomes, (b) that full equilibration of Cl− is achieved with EriC-containing liposomes, and (c) that negligible efflux of glutamate occurs for liposomes with or without EriC. The crucial measurement in this assay is the fraction of the intraliposomal volume that is inaccessible to external Cl−. This parameter is determined from the fractional trapped Cl− space, f, measured by double-label counting of internal and external tracer concentrations (dpm per gram lipid and per cubic-centimeter solution, respectively, shown in ):
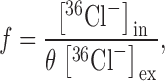 |
1a |
where θ, the total intravesicular volume (cm3/g lipid), is measured from 3H-glutamate ():
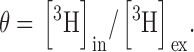 |
1b |
For vesicles that have been diluted infinitely, the Cl−-inaccessible fraction, f 0 is ():
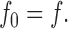 |
2a |
For practical purposes, the samples were diluted not infinitely, but 10-fold, and f 0 was correspondingly normalized, by :
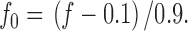 |
2b |
This assay can be used to estimate the fraction, s, of EriC protein that is functionally active as a Cl− channel. The analysis assumes that the protein distributes randomly according to a Poisson distribution into spherical liposomes of uniform size, and that Cl− is retained only in those liposomes that contain no EriC molecules. As the mass, m E, of EriC reconstituted in a fixed mass, m L, of liposomes increases, this Cl−-inaccessible fraction will decrease according to Goldberg and Miller 1991:
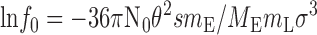 |
3 |
where No is Avogadro's number, θ is the intravesicular volume (cm3/g lipid), M E is the molecular mass of the EriC channel (51,000 g/mol per subunit), and σ is the surface area of lipid in a bilayer (cm2/g).
Chemical Cross-linking
Glutaraldehyde-mediated cross-linking of EriC was performed at room temperature. EriC (6 μg) was diluted to 27.5 μl in Buffer X (150 mM NaCl, 50 mM Na-Pi, 1 mM DDM, pH 7.0), and glutaraldehyde (0.5 μl, 18 mM final concentration) was added. The reaction was quenched with 19 mM Tris for 10 min. Sodium dodecylsulfate (SDS)–loading buffer was added, and the sample was analyzed by SDS-PAGE. In some experiments, we varied concentrations of protein (5–500 μg/ml), DDM (1–5 mM), or glutaraldehyde (14–160 mM).
Gel Filtration
Samples were chromatographed at 1 ml/min on a Superdex 200 column (10 × 300 mm, 24 ml bed volume; Pharmacia LKB Biotechnology Inc.) equilibrated in buffer G (100 mM KCl, 50 mM Na-Pi, 1 mM DDM, pH 7.0). Elution profiles were monitored at 280 nm.
Analytical Ultracentrifugation
EriC was dialyzed overnight against Buffer U (155 mM KCl, 95 mM K-Pi, 1 mM DDM, pH 7.0) containing 5 mM dithiothreitol (DTT). For control experiments, KcsA protein was dialyzed into Buffer U lacking DTT. To ensure proper blank subtraction, for each dialyzed sample, the solution outside the dialysis cell at equilibrium was collected as the blank. Samples and buffer blanks were loaded into charcoal-Epon two-sector cells. Sedimentation velocity was analyzed using an Optima XL-A analytical ultracentrifuge (Beckman Instruments, Inc.) at 44,000 rpm, 20°C; scans at 280 nm were taken every 5 min. Data were fit to the modified Fujita-Macosham function (Philo 1997) using Svedberg 5.01 software. Protein partial-specific volumes, calculated from amino acid composition (Cohn and Edsall 1943; McMeekin and Marshall 1952), were 0.743 and 0.724 cm3/g for EriC and KcsA, respectively.
RESULTS
For the past few years, it has been appreciated that ClC channels are represented in microbial genomes (Jentsch et al. 1999). By sequence identity, the prokaryotic ClCs lie in the twilight zone for homology modeling (∼15–25%), but the statistical scores obtained from BLAST alignments indicate a high probability that the prokaryotic proteins are true ClC homologues (Abagyan and Batalov 1997; Brenner et al. 1998). Manual scrutiny of the prokaryotic sequences supports this conclusion. Several stretches of ClC sequence are invariant among the known eukaryotic family members, and these appear throughout the entire membrane-spanning region, ∼400 residues, and only in this region. In these regions, the prokaryotic proteins display high sequence identity to their eukaryotic counterparts (Fig. 1). The roles that these conserved sequences play in channel function have not been experimentally established; however, certain of these, when mutated, lead to changes in both gating and permeation (Fahlke et al. 1997b,Fahlke et al. 1997d; Ludewig et al. 1997), as though they either directly mediate these functions or are necessary for proper folding and assembly of ClC channels.
Figure 1.
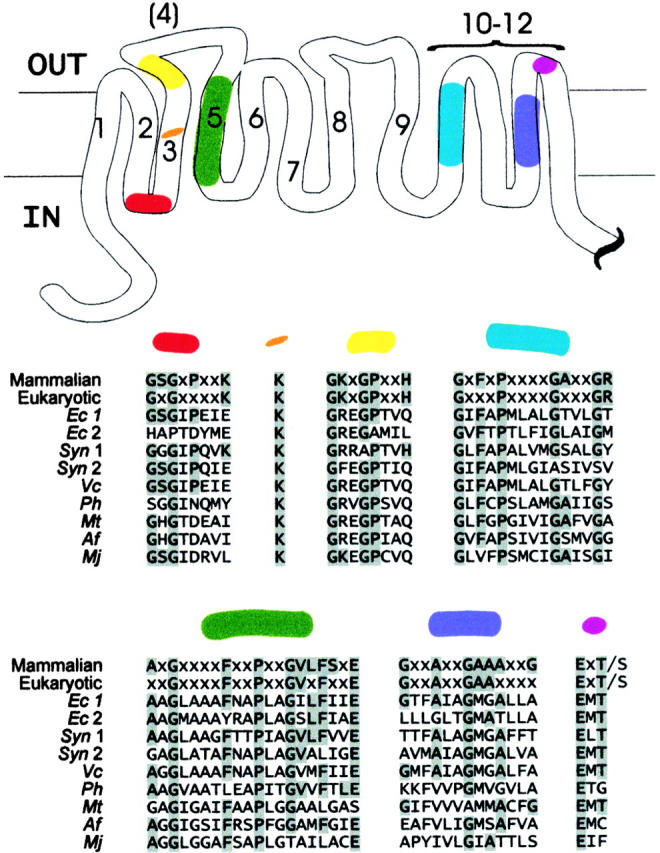
Conservation of prokaryotic ClC channels. Regions of protein that contain residues invariant among mammalian or all eukaryotic ClC channels are color-coded and mapped onto the ClC transmembrane topology model of Schmidt-Rose and Jentsch 1997. The topology of transmembrane stretches 5–7 is currently unresolved (Fahlke et al. 1997c; Jentsch et al. 1999). The COOH-terminal cytoplasmic domain, which contains no invariant regions, ranges from ∼170 to 420 residues in the mammalian ClCs and 6 to 475 residues in the prokaryotes (35 residues in EriC). Corresponding sequences of the prokaryotic ClC genes identified by BLAST searches are shown below the eukaryotic and mammalian consensus sequences (x, variant residue). Prokaryotic species and sequence references are as follows. Ec, Escherichia coli (Blattner et al. 1997); Syn, Synechocystis sp. strain PCC 6803 (Kaneko et al. 1996); Vc, Vibrio cholerae (TIGR, preliminary sequence data); Ph, Pyrococcus horikoshii (Kawarabayasi et al. 1998); Mt, Mycobacterium tuberculosis (Cole et al. 1998); Af, Archaeoglobus fulgidus (Klenk et al. 1997); Mj, Methanococcus jannaschii (Bult et al. 1996). EriC corresponds to Ec 1.
Overexpression, Purification, and Reconstitution of EriC
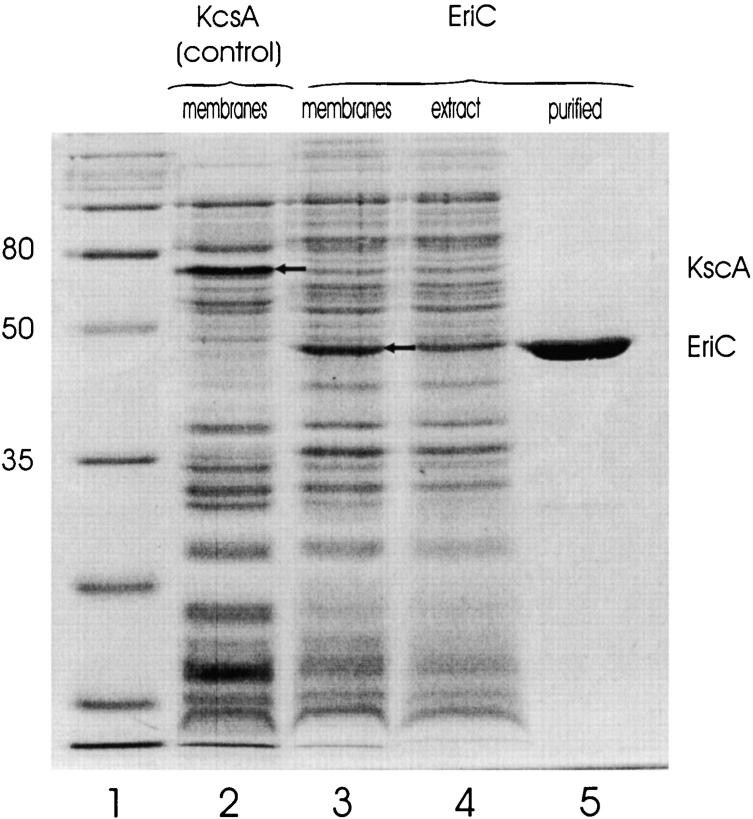
We cloned one of the above ClC genes into an E. coli expression vector and added an NH2-terminal hexahistidine sequence for large-scale production and straightforward purification of EriC, a putative ClC-type channel product of the yadQ gene. After an overnight room-temperature induction of E. coli bearing the pEriC plasmid, a protein band of ∼38 kD, absent in uninduced controls, was observed on SDS-PAGE of bacterial membrane fractions (Fig. 2). This protein was detergent-extracted from the bacterial membrane fraction and purified on a Ni-chelate column. The apparent molecular mass of ∼38 kD is smaller than that calculated from the sequence (51 kD), but the integrity of the full-length protein is indicated by two observations. First, robust interaction with the metal-affinity column indicates that the NH2-terminal hexahistidine tag is present; second, an EriC construct carrying an eight-residue, COOH-terminal 1D4 epitope (Molday and MacKenzie 1983), showed a similar apparent molecular mass, detected on immunoblots (data not shown). Typical yields of purified EriC are 0.5–1 mg/liter culture.
Figure 2.
Overexpression and purification of EriC. Protein samples were run on a 13% polyacrylamide gel. (1) Molecular mass markers (broad range; Bio-Rad Laboratories), (2) control membranes (600 μg protein, estimated as 0.6 A280-ml) from E. coli induced to express KcsA, (3) membranes from E. coli expressing EriC (600 μg protein), (4) clarified membrane extract (equivalent to 600-μg membranes), (5) 9 μg purified EriC.
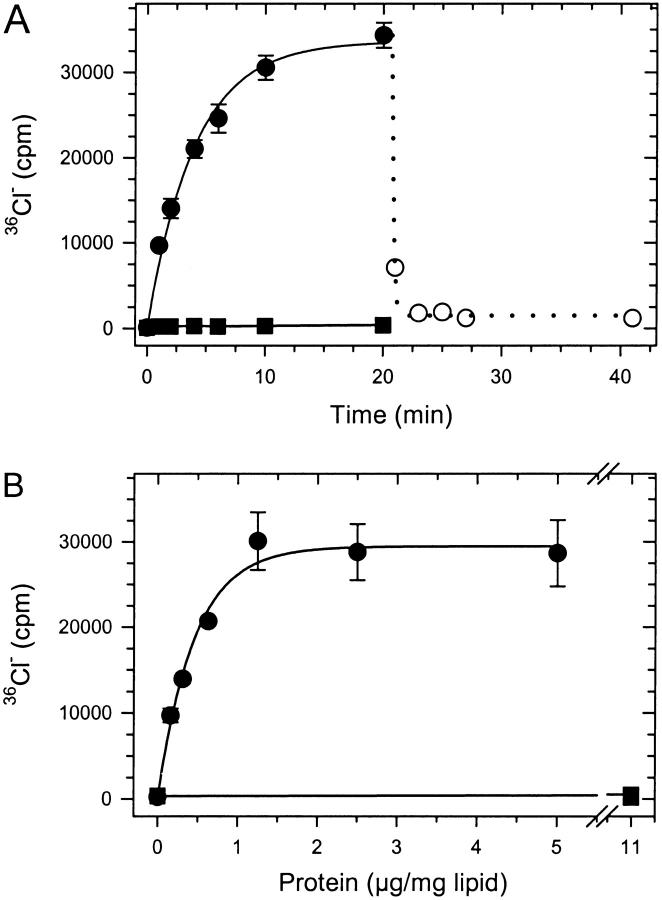
EriC is unambiguously a ClC-family protein at the level of primary sequence, but does it function as an ion channel? To assess this protein's ability to catalyze passive transmembrane flux of 36Cl−, we reconstituted EriC into liposomes and performed a concentrative influx assay (Goldberg and Miller 1991). Vesicles were loaded with a high concentration (450 mM) of Cl−, and 36Cl− influx was measured at low (2.5 mM) external Cl−. The resulting Cl− gradient will polarize any Cl−-selective liposome membrane (positive potential inside), and 36Cl− will accordingly accumulate inside the liposome. Liposomes that have failed to incorporate Cl− channels or have sprung nonselective leaks will not accumulate 36Cl− in this assay. The results of Fig. 3 demonstrate that EriC is a Cl−-selective channel; tracer Cl− accumulates into EriC-containing vesicles with a time constant of ∼4 min. Permeabilization of the liposomes to K+ with valinomycin causes rapid loss of Cl−, a result demonstrating that the movement of 36Cl− is conductive, not electroneutral. The final level of tracer rises in a saturating fashion with the amount of EriC reconstituted, as the population of channel-containing liposomes increases. Control protein-free vesicles or vesicles reconstituted with KcsA, a bacterial K+ channel, fail to transport 36Cl−, although the latter vesicles accumulate 86Rb+ under similar conditions (Heginbotham et al. 1998).
Figure 3.
36Cl− flux through reconstituted EriC. Concentrative uptake of 36Cl− was followed as in materials and methods. (A) Time course of accumulation of 36Cl− in vesicles reconstituted with 4.5 μg EriC/mg lipid (•, mean ± SEM, n = 4) or without protein (▪, n = 1). 36Cl− release was measured after addition of valinomycin at 21 min (○, n = 1). (B) Protein concentration-dependent accumulation of 36Cl− into vesicles reconstituted with EriC (○, •) or KcsA (▪). Uptake was measured at 20 min (mean ± SEM, n = 3).
Absolute Estimation of Functional Activity
It is desirable to know if the observed flux activity is due to a major, minor, or minuscule fraction of the purified EriC protein. However, the observed influx kinetics are inadequate to quantify activity in our preparations because the absolute rate of Cl− uptake catalyzed by a single EriC channel is unknown. Instead, we employ an assay that does not require knowledge of absolute flux rates, but merely relies on the assumption that any liposome permeable to Cl− will equalize internal and external concentrations of this ion at thermodynamic equilibrium. Vesicles are loaded to equilibrium with two tracers: 36Cl−, which is permeant exclusively to EriC-containing liposomes, and 3H-glutamate, a trapped volume marker that is impermeant to all liposomes on the experimental time scale. The loaded vesicles are then diluted into tracer-free solution of identical ionic composition, and 36Cl− is allowed to flow out (in exchange for unlabeled Cl−) until equilibrium is achieved. Any vesicle containing one or more functionally active EriC molecules releases 36Cl− and retains impermeant glutamate; vesicles devoid of EriC channels will trap both 36Cl− and 3H-glutamate. Glutamate is thus used as an internal standard to measure the total intravesicular volume to which the Cl−-accessible volume can be compared.
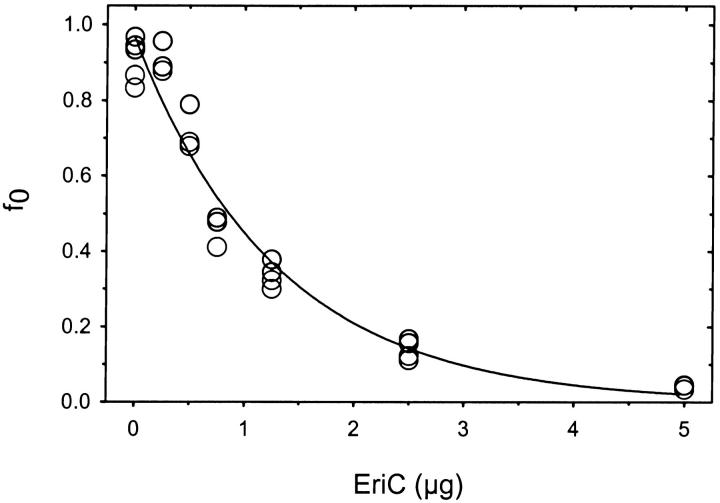
As EriC is reconstituted at increasing protein/lipid mass ratio, the fraction of Cl−-impermeable volume, f 0, falls exponentially from a protein-free value of unity (Fig. 4), as demanded by a Poisson distribution governing the random incorporation of NE EriC channels into NL liposomes (Goldberg and Miller 1991). The absolute value of intravesicular volume, θ, does not change systematically over this range of protein concentration (data not shown), which roughly spans the range of 0.2–4 channels per liposome. The protein concentration dependence of f 0 quantitatively obeys the expectations of , where the crucial fit quantity, the fraction of active protein, is s = 1.9. Since we have based our analysis on a utopian model in which protein molecules distribute perfectly into uniform spherical vesicles, it is perhaps not surprising that we reach a physically impossible conclusion of 190% activity. As discussed below, several assumptions in this analysis are expected to lead to uncertainty in the estimation of EriC activity, so we can easily rationalize this absurd value. The important point of this calculation is that a major fraction of the purified EriC protein—perhaps 100%—is responsible for the functional activity detected in the assay.
Figure 4.
Quantification of EriC activity. The fraction of vesicles containing no active Cl− channels, f 0, was determined for vesicles (3.8 mg lipid) reconstituted with the indicated amount of EriC. The data were fit to as described in the text. Parameters of the fit were: ME = 102,000 g/mol; θ = 5.2 cm3/g (±0.2, n = 26); σ = 2.3 × 106 cm2/g. This latter parameter was calculated from molecular surface area, 61 Å2 (Rand and Parsegian 1989), an estimated molecular weight of the phospholipid (750 g/mol), and a factor of 0.93 to account for liposome curvature.
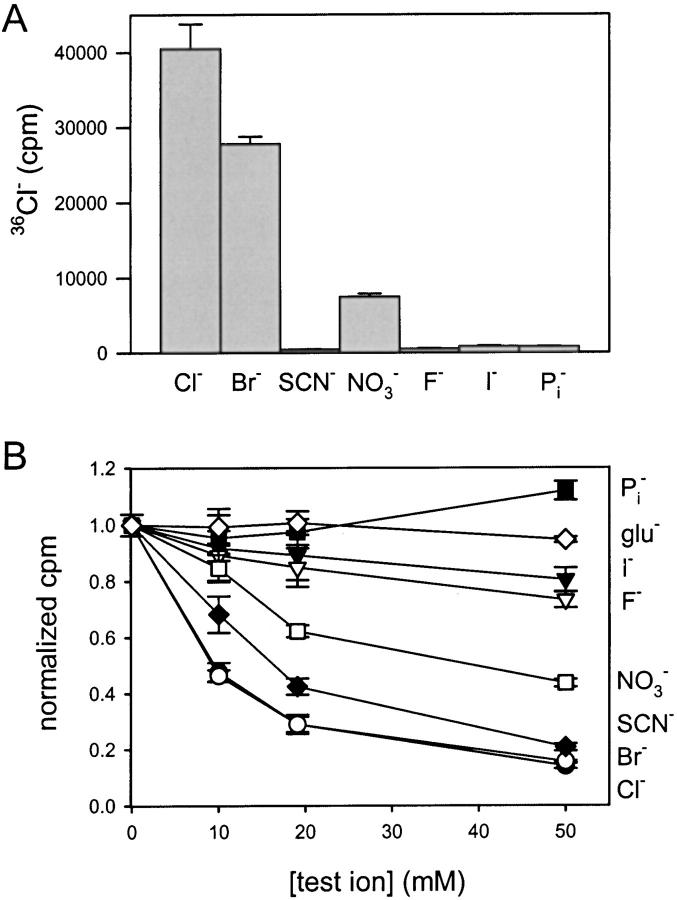
Ion Selectivity of EriC
The concentrative uptake assay was used in two ways to gauge the ionic selectivity of EriC. In the first set of experiments, internal Cl− was replaced with various test anions. Permeant anions support concentrative uptake, while impermeant anions do not. Of the anions tested (Fig. 5 A), only Cl−, Br−, and NO3 − are permeant by this criterion. In the second set of experiments, we added test ions to the external solution to see which of these would collapse the liposome membrane potential and thereby impede influx. In this assay, SCN−, and to a lesser extent I− and F− score as permeant in addition to Cl−, Br−, and NO3 −. The discrepancy observed with SCN− is not disconcerting; SCN− both blocks and permeates some eukaryotic ClCs (White and Miller 1981; Fahlke et al. 1997b; Rychov et al. 1998) and, if acting in such a fashion here, would also inhibit influx. In other words, the flux assay in which the test anion is applied externally does not distinguish a permeant ion from a strong blocker. In any case, the two assays taken together demonstrate (a) that EriC-mediated fluxes are highly selective for anions over cations, and (b) that among anions the selectivity sequence is roughly similar to that found for ClC-0 and ClC-1 (Rychov et al. 1998; Jentsch et al. 1999): Cl−, Br− > NO3 − > I−, F− >> H2PO4 −, glutamate−. It is worth noting, however, that these selectivity assays are performed under ionic conditions and with methods very different than in electrophysiological measurements, and that no single interanionic selectivity sequence applies to all eukaryotic ClC channels.
Figure 5.
Ion selectivity of EriC. (A) Effect of internal ion on uptake of 36Cl−. Vesicles were reconstituted with 1.9 μg EriC/mg lipid in 450 mM KX, 20 mM Na-MES, pH 6.2. Uptake was measured at 13 min; data points represent mean ± SEM of three or range of two measurements. (B) Effect of external ions on 36Cl− uptake. Vesicles were reconstituted with 3.7 (10 mM test ion) or 3.0 (19 and 50 mM test ion) μg EriC/mg lipid. Uptake was measured at 13 min. Data (mean ± SEM, n = 3), are normalized to uptake with no test ion added. glu−, glutamate.
EriC Is a Homodimer
The most extensively studied eukaryotic ClC channels, ClC-0 and ClC-1, are both homodimers, a quaternary structure underlying their double-barreled behavior (Middleton et al. 1994b, Middleton et al. 1996; Ludewig et al. 1996; but see Fahlke et al. 1997a). But is this dimeric characteristic merely an idiosyncrasy of these two ClCs, which populate the same muscle-type subfamily, or is it a general property of the entire ClC family? Since EriC is evolutionarily distant from the eukaryotic ClC subfamilies, its quaternary structure would provide valuable insight into this question. We used three different techniques to assess the oligomeric state of EriC in detergent micelles: cross-linking, gel filtration, and velocity sedimentation.
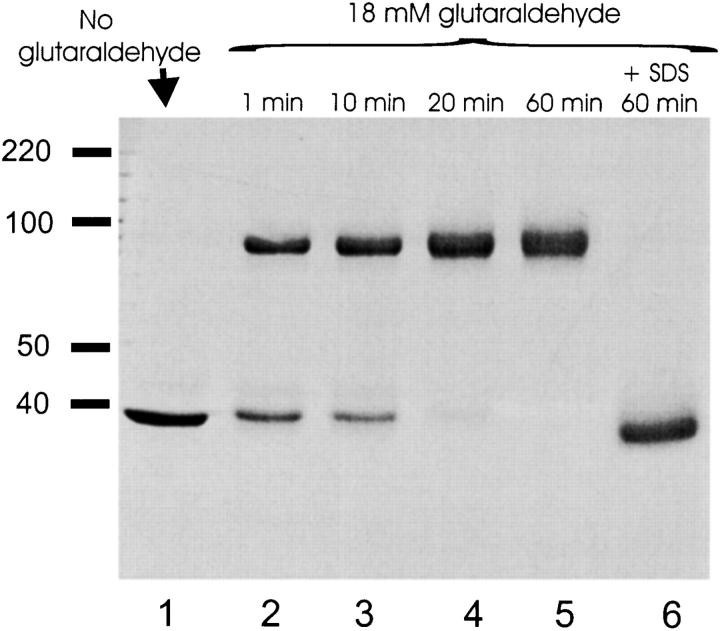
Glutaraldehyde, a nonspecific cross-linking agent, has been used convincingly to report the oligomeric state of several membrane proteins (Canals 1992; Craig 1982a,Craig 1982b; Rabon et al. 1990). Reaction of glutaraldehyde with EriC gives a clean result (Fig. 6). Within 10 min of reaction, the 38-kD band is quantitatively converted into a band migrating at ∼90 kD; no conversion to higher mass species occurs over the next 60 min. The possibility of intermolecular cross-linking is largely eliminated by the control experiment showing no cross-linking of EriC dispersed under denaturing conditions in SDS. In addition, variation of the protein-detergent ratio over two orders of magnitude failed to alter the cross-linking, a result that rules out artifactual dimerization resulting from multiple protein molecules sharing the same micelle (Fleming et al. 1997). Moreover, similar results were obtained with EriC reconstituted at very low density (0.09 μg/mg lipid), where most liposomes have either one EriC channel or none. Cross-linking was robust to varying glutaraldehyde or protein concentrations; while the rates of cross-linking increased with these variables, the overall pattern, particularly the absence of high aggregates, was unchanged.
Figure 6.
Glutaraldehyde cross-linking of EriC. EriC was treated with glutaraldehyde as in materials and methods, and the samples were run on a 10% polyacrylamide gel.(1) 6 μg EriC, no glutaraldehyde, (2–5) 6 μg EriC cross-linked for 1, 10, 20, or 60 min, respectively, (6) 6 μg EriC incubated with 1% SDS before reaction with glutaraldehyde for 60 min.
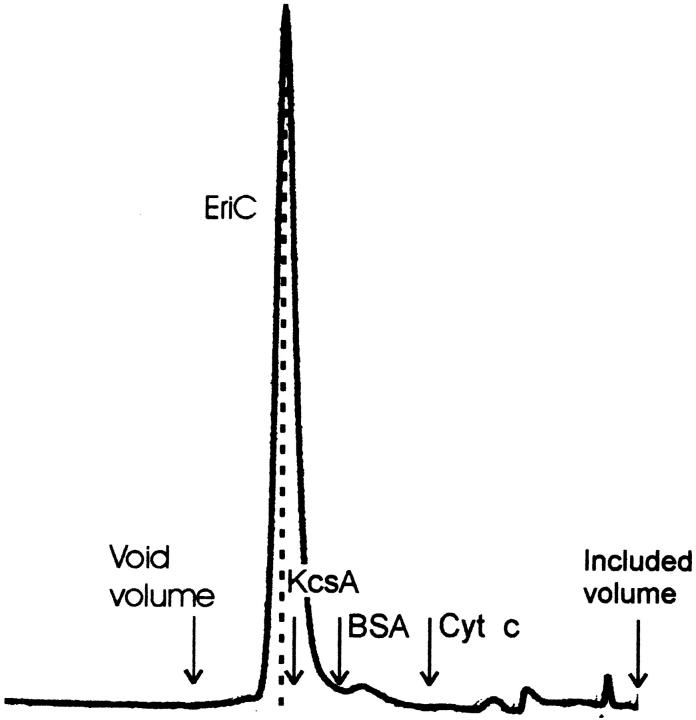
To complement the cross-linking experiments, we analyzed EriC by gel filtration chromatography and compared its migration to a reference membrane protein of known size, the K+ channel KcsA, a 74-kD homotetramer (Heginbotham et al. 1997). EriC runs slightly ahead of KcsA on a Superdex gel filtration column (Fig. 7). This result suggests that the EriC channel is substantially larger than its monomer molecular mass of 51 kD.
Figure 7.
Superdex gel filtration. Purified EriC (230 μg) was applied to a Superdex FPLC column, and elution was monitored by 280-nm absorbance. A dashed line is drawn under the EriC peak; arrows mark the peak positions of a void volume marker (blue dextran), KcsA (74 kD), serum albumin (67 kD), cytochrome c (13 kD), and an included volume marker (imidazole).
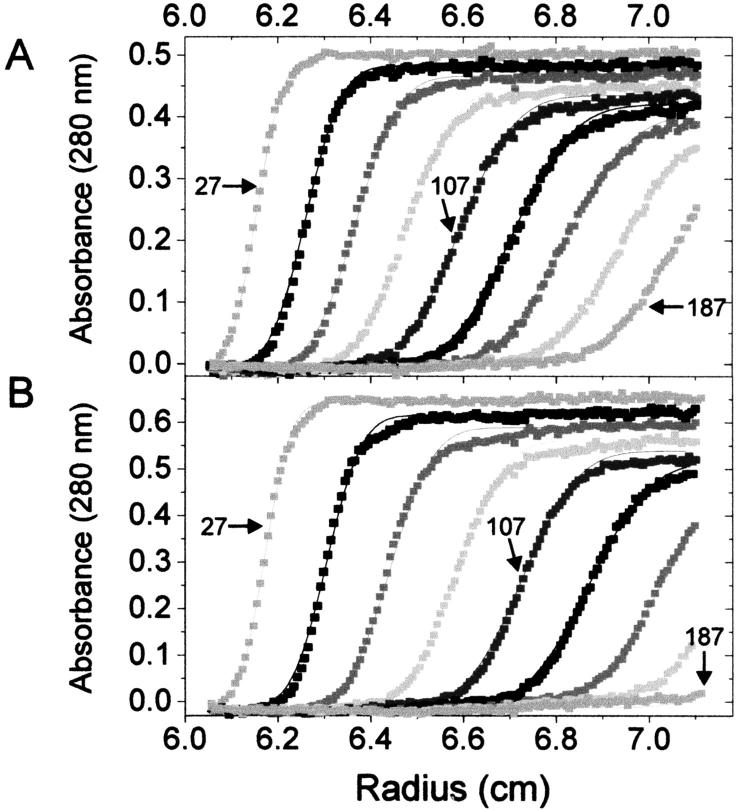
Since these gel filtration results cannot distinguish dimers from higher-order oligomers, we also analyzed velocity sedimentation profiles of EriC and again used KcsA as a membrane protein size standard. Qualitatively, EriC sediments more rapidly than KcsA (Fig. 8). A fit of the sedimentation data to a Fujita function (Williams 1972), which determines both the sedimentation and diffusion coefficients and hence molecular mass, yields 92 kD for KcsA and 126 kD for EriC. These estimates are both ∼25% higher than the formula size of tetrameric KcsA and dimeric EriC, a result easily rationalized by bound detergent. A value of 126 kD for EriC would be difficult to reconcile with a trimer or higher oligomer.
Figure 8.
Velocity sedimentation of Eric. A280 in each sample cell is plotted as a function of radius from the axis of rotation. Each scan was made at a different time during the centrifugation run; the scans made at 27, 107, and 187 min are specifically labeled. Fits to the Fujita-MacCosham function yields values for the sedimentation coefficient, s, and diffusion coefficient, D, which in turn yield estimates of molar mass, Mw. (A) KcsA: s = 6.73 S, D = 6.55 × 10−7 cm2/s, Mw = 92 kD. (B) EriC: s = 8.43 S, D = 6.47 × 10−7 cm2/s, Mw = 126 kD.
We attempted to carry out equilibrium sedimentation experiments in neutral-density detergents, to eliminate rigorously the contribution of bound detergent to the measured mass of the protein (Reynolds and Tanford 1976; Reynolds and McCaslin 1985; Fleming et al. 1997). These experiments failed, however, since EriC was unstable in all neutral-density detergents tested, as indicated by rapid aggregation accompanied by high-order cross-linking (data not shown). Nevertheless, the three independent lines of evidence presented here argue powerfully that the functionally active form of EriC used for reconstitution is a homodimer.
DISCUSSION
At the current early stage of understanding the chemistry of integral membrane proteins, prokaryotes have served as the singular bearers of high-resolution structural information about ion channels. Porins, K+ channels, and mechanosensitive channels from bacteria have been expressed at high levels in E. coli as a prelude to crystallization and structure determination (Weiss et al. 1991; Doyle et al. 1998; Chang et al. 1998). No eukaryotic ion channel has been successfully expressed in any heterologous systems at levels high enough even to contemplate crystallization. It is therefore encouraging that the ClC family of Cl− channels is represented widely in prokaryotes.
We have overexpressed a prokaryotic ClC channel, EriC, the product of one of the two ClC genes in E. coli. The biological role of this channel is unknown, but the present results make it clear that it is in fact a ClC-type Cl− channel. The purified protein promotes the passive, conductive flux of Cl− and other anions across liposome membranes, and the ionic selectivity of this effect is reminiscent of vertebrate ClC channel electrophysiology. We attempted but failed to observe single-channel current fluctuations induced by EriC in planar lipid bilayers; this negative result is disappointing but not surprising in light of the extremely low conductances of many eukaryotic ClC channels (Jentsch et al. 1999). In the liposome flux assay, the time scale (seconds) of EriC-catalyzed 36Cl− release is substantially lower than expected (milliseconds) for a channel of conventional unitary current (1 pA) open all the time. In the absence of direct electrophysiological information, we have no way of knowing whether the low fluxes reflect low channel current, low open probability, or both.
From a biochemical standpoint, perhaps the most important property to quantify for any new protein preparation, even more important than purity or yield, is functional competence. This measurement is difficult for uncharacterized ion channels reconstituted into liposomes, and our value implying 190% activity (Fig. 4) is certainly disconcerting. We have used a limiting Poisson method of “counting” the fraction of liposomes that carry no channels in a sample containing NL liposomes and NE EriC channels, a fraction s of which are functionally active. If the insertion of channels into a uniform population of liposomes is random, then this fraction, f 0, must obey a Poisson distribution ():
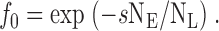 |
4 |
We estimate f 0 directly from the fraction of Cl−-impermeable liposomes, and NE is known. The experimental samples consist of a known mass of lipid corresponding to a fixed liposome surface area, and the total intravesicular volume is directly measured. If the liposomes were uniform spheres of known lipid surface density, this information would be sufficient to determine NL and hence to derive a functional activity value s.
However, estimation of the number of liposomes is subject to several sources of error. First, the estimated value of activity varies with the cube of the bilayer surface area per mass of lipid, σ, in . The reconstituted liposomes used here are formed from a phosphatidylethanolamine-rich, undefined, complex mix of lipids. Molecular surface areas of phosphatidylethanolamines vary substantially (55–72 Å2) depending on hydrocarbon chain, lipid composition in mixed bilayers, and other factors (Rand and Parsegian 1989); if used in these would lead to an approximately twofold range of estimated activity values (average 190%, range 140–320%). Second, large unilamellar liposomes such as these are not expected to be spherical in general; ignoring this kind of nonideality always overestimates activity in the trapped-Cl− assay since any departure from a spherical shape increases the surface-to-volume ratio. For example, if the liposomes were on average approximated by a squat cylinder (height = radius), this effect alone would lower the activity estimated above into the range 80–180%. Unfortunately, we have no experimental data on liposome morphology, so we cannot rigorously quantify the expected errors in our estimate of s. Nevertheless, this uncertainty provides a plausible rationale for an activity value that is “impossibly” high. Another source of ambiguity arises from the assumption that the liposomes are uniform in size, which is certainly false (Mimms et al. 1981); however, calculations indicate that this effect is relatively minor, producing errors of <20% in either direction, depending on the size distribution of the liposome population. Together, the magnitude of these uncertainties dictates that we cannot estimate the value of s to better than two- to threefold accuracy. Nonetheless, the key inference of the experiments—that the flux activity we observe is mediated by a nontrivial fraction of the purified EriC—is preserved. The hydrodynamic properties of EriC further argue that a major, not a minor, fraction of the protein is active; in both gel filtration and analytical centrifugation experiments, purified EriC behaves as a properly folded, monodisperse macromolecule.
EriC behaves, as do the muscle-type channels ClC-0 and ClC-1 (Middleton et al. 1994a; Fahlke et al. 1997a), as a homodimer in both micellar and bilayer environments. None of the three methods used to assess quaternary structure is in itself rigorous. Nevertheless, a dimeric structure for EriC is strongly supported by the consistency of these three independent, complementary techniques. Gel filtration and velocity sedimentation behavior argue that Eric is larger than a monomer; velocity sedimentation additionally indicates that it is smaller than a trimer; chemical cross-linking marks it as a dimer. These results underscore the structural homology between EriC and the eukaryotic ClCs and thereby identify homodimeric architecture as a general hallmark of the ClC family rather than a peculiarity of the muscle-type subfamily.
ClC channels carry out many crucial biological functions, but conventional mutagenesis studies have provided only limited glimpses of ClC molecular architecture. Since EriC is functionally active as a Cl− channel and may be obtained in milligram quantities, this protein is an excellent candidate for future structural studies.
Acknowledgments
We thank Lise Heginbotham for E. coli genomic DNA, the KcsA gene in pASK90, and the waiver of consulting fee. We thank Jeff Gelles for essential assistance with analytical ultracentrifugation, and Rob Blaustein, Meredith Lemasurier, and Joe Mindell for comments on the manuscript. Sequence data were obtained from TIGR website at http://www.tigr.org. Sequencing of Methanococcus jannaschii and Archaeoglobus fulgidus was accomplished with support from the Department of Energy.
Sequencing of E. coli was accomplished with support from the National Human Genome Research Institute. Sequencing of Mycobacterium tuberculosis was accomplished with support from the Wellcome Trust. Sequencing of Vibrio cholerae was accomplished with support from the National Institute of Allergy and Infectious Diseases. M. Maduke was supported by a grant from the W.M. Keck Institute for Cellular Visualization.
Footnotes
1used in this paper: DDM, dodecylmaltoside; MES, morpholino ethanesulfonic acid; SDS, sodium dodecylsulfate
References
- Abagyan R.A., Batalov S. Do aligned sequences share the same fold? J. Mol. Biol. 1997;273:355–368. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLASTa new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F.R., Plunkett G., Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Brenner S.E., Chothia C., Hubbard T.J. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl. Acad. Sci. USA. 1998;95:6073–6078. doi: 10.1073/pnas.95.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult C.J., White O., Olsen G.J., Zhou L., Fleischmann R.D., Sutton G.G., Blake J.A., FitzGerald L.M., Clayton R.A., Gocayne J.D. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii . Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Canals F. Signal transmission by epidermal growth factor receptorcoincidence of activation and dimerization. Biochemistry. 1992;31:4493–4501. doi: 10.1021/bi00133a016. [DOI] [PubMed] [Google Scholar]
- Chang G., Spencer R.H., Lee A.T., Barclay M.T., Rees D.C. Structure of the MscL homolog from Mycobacterium tuberculosisa gated mechanosensitive ion channel. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- Cohn E.J., Edsall J.T. Density and apparent specific volume of proteins. In: Cohn E.J., Edsall J.T., editors. Proteins, Amino Acids and Peptides. Reinhold Publishing Corp; New York, NY: 1943. pp. 370–381. [Google Scholar]
- Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Craig W.S. Determination of the distribution of sodium and potassium ion activated adenosinetriphosphatase among the various oligomers formed in solutions of nonionic detergents Biochemistry 21 1982. 2667 2674a [DOI] [PubMed] [Google Scholar]
- Craig W.S. Monomer of sodium and potassium ion activated adenosinetriphosphatase displays complete enzymatic function Biochemistry 21 1982. 5707 5717b [DOI] [PubMed] [Google Scholar]
- Creighton, T.E. 1984. Chemical nature of polypeptides. In Proteins: Structure and Molecular Properties. W.H. Freeman and Co., New York, NY. 1–60.
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. The structure of the potassium channelmolecular basis of K+ conduction and selectivity. Science. 1998;280:69–76. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Fahlke C., Knittle T., Gurnett C.A., Campbell K.P., George A.L. Subunit stoichiometry of human muscle chloride channels J. Gen. Physiol. 109 1997. 93 104a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlke C., Durr C., George A.L. Mechanism of ion permeation in skeletal muscle chloride channels J. Gen. Physiol. 110 1997. 551 564b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlke C., Yu H.T., Beck C.L., Rhodes T.H., George A.L. Pore-forming segments in voltage-gated chloride channels Nature. 390 1997. 529 532c [DOI] [PubMed] [Google Scholar]
- Fahlke C., Beck C.L., George A.L. A mutation in autosomal dominant myotonia congenita affects pore properties of the muscle chloride channel Proc. Natl. Acad. Sci. USA. 94 1997. 2729 2734d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming K.G., Ackerman A.L., Engelman D.A. The effect of point mutations on the free energy of transmembrane α-helix dimerization. J. Mol. Biol. 1997;272:266–275. doi: 10.1006/jmbi.1997.1236. [DOI] [PubMed] [Google Scholar]
- Goldberg A.F.X., Miller C. Solubilization and functional reconstitution of a chloride channel from Torpedo californica electroplax. J. Membr. Biol. 1991;124:199–206. doi: 10.1007/BF01994354. [DOI] [PubMed] [Google Scholar]
- Heginbotham L., Odessey E., Miller C. Tetrameric stoichiometry of a prokaryotic K+ channel. Biochemistry. 1997;36:10335–10342. doi: 10.1021/bi970988i. [DOI] [PubMed] [Google Scholar]
- Heginbotham L., Kolmakova-Partensky L., Miller C. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 1998;111:741–750. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Ionic Channels in Excitable Membranes 2nd ed 1992. Sinauer Associates, Inc; Sunderland, MA: pp. 607 [Google Scholar]
- Jentsch T.J., Gunther W., Pusch M., Schwappach B. Properties of voltage-gated chloride channels of the ClC gene family. J. Physiol. 1995;482:19S–25S. doi: 10.1113/jphysiol.1995.sp020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch T.J., Friedrich T., Schriever A., Yamada H. The CLC channel family. Pflügers Arch. 1999;437:783–795. doi: 10.1007/s004240050847. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sasamoto S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi Y., Sawada M., Horikawa H., Haikawa Y., Hino Y., Yamamoto S., Sekine M., Baba S., Kosugi H., Hosoyama A. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- Klenk H.P., Clayton R.A., Tomb J.F., White O., Nelson K.E., Ketchum K.A., Dodson R.J., Gwinn M., Hickey E.K., Peterson J.D. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus [published erratum appears in Nature. 1998. 394(6688):101] Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ludewig U., Pusch M., Jentsch T.J. Two physically distinct pores in the dimeric ClC-0 chloride channel. Nature. 1996;383:340–343. doi: 10.1038/383340a0. [DOI] [PubMed] [Google Scholar]
- Ludewig U., Jentsch T.J., Pusch M. Inward rectification in ClC-0 chloride channels caused by mutations in several protein regions. J. Gen. Physiol. 1997;110:165–171. doi: 10.1085/jgp.110.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon R., Doyle D.A. Prokaryotes offer hope for potassium channel structural studies. Nat. Struct. Biol. 1997;4:877–879. doi: 10.1038/nsb1197-877. [DOI] [PubMed] [Google Scholar]
- McMeekin T.L., Marshall K. Specific volumes of proteins and the relationship to their amino acid contents. Science. 1952;116:142–143. doi: 10.1126/science.116.3006.142. [DOI] [PubMed] [Google Scholar]
- Middleton R.E., Pheasant D.J., Miller C. Reconstitution of detergent-solubilized Cl− channels and analysis by concentrative uptake of 36Cl− and planar lipid bilayers Methods (Orlando). 6 1994. 28 36a [Google Scholar]
- Middleton R.E., Pheasant D.J., Miller C. Purification, reconstitution, and subunit composition of a voltage-gated chloride channel from Torpedo electroplax Biochemistry. 33 1994. 13189 13198b [DOI] [PubMed] [Google Scholar]
- Middleton R.E., Pheasant D.J., Miller C. Homodimeric architecture of a ClC-type chloride ion channel. Nature. 1996;383:337–340. doi: 10.1038/383337a0. [DOI] [PubMed] [Google Scholar]
- Mimms L.T., Zampighi G., Nozaki Y., Tanford C., Reynolds J.A. Phospholipid vesicle formation and transmembrane protein incorporation using octyl glucoside. Biochemistry. 1981;20:833–840. doi: 10.1021/bi00507a028. [DOI] [PubMed] [Google Scholar]
- Molday R.S., MacKenzie D. Monoclonal antibodies to rhodopsincharacterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- Philo J.S. An improved function for fitting sedimentation velocity data for low-molecular-weight solutes. Biophys. J. 1997;72:435–444. doi: 10.1016/S0006-3495(97)78684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabon E.C., Bassilian S., Jakobsen L.J. Glutaraldehyde crosslinking analysis of the C12E8 solubilized H,K-ATPase. Biochim. Biophys. Acta. 1990;1039:277–289. doi: 10.1016/0167-4838(90)90260-m. [DOI] [PubMed] [Google Scholar]
- Rand R.P., Parsegian V.A. Hydration forces between lipid bilayers. Biochim. Biophys. Acta. 1989;988:351–376. [Google Scholar]
- Reynolds J.A., McCaslin D.R. Determination of protein molecular weight in complexes with detergent without knowledge of binding. Methods Enzymology. 1985;117:41–53. doi: 10.1016/s0076-6879(85)17005-9. [DOI] [PubMed] [Google Scholar]
- Reynolds J.A., Tanford C. Determination of molecular weight of the protein moiety in protein-detergent complexes without direct knowledge of detergent binding. Proc. Natl. Acad. Sci. USA. 1976;73:4467–4470. doi: 10.1073/pnas.73.12.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychov G.Y., Pusch M., Roberts M.L., Jentsch T.J., Bretag A.H. Permeation and block of the skeletal muscle chloride channel, ClC-1, by foreign anions. J. Gen. Physiol. 1998;111:653–665. doi: 10.1085/jgp.111.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning, A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schmidt-Rose T., Jentsch T.J. Transmembrane topology of a CLC chloride channel. Proc. Natl. Acad. Sci. USA. 1997;94:7633–7638. doi: 10.1073/pnas.94.14.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene. 1994;151:131–135. doi: 10.1016/0378-1119(94)90643-2. [DOI] [PubMed] [Google Scholar]
- Weiss M.S., Abele U., Weckesser J., Welte W., Schultz E., Schulz G.E. Molecular architecture and electrostatic properties of a bacterial porin. Science. 1991;254:1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- White M.M., Miller C. Probes of the conduction process of a voltage-gated chloride channel from Torpedo electroplax. J. Gen. Physiol. 1981;78:1–19. doi: 10.1085/jgp.78.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J.W. 1972. Molecular homogeneity and its demonstration. In Ultracentrifugation of Macromolecules: Modern Topics. Academic Press, Inc., New York, NY. 99–107.