Abstract
Several legume seed proteins that are potentially allergenic, poorly digested by farm animals, and/or have undesirable functional properties, have been described. One of these is the albumin protein in pea (Pisum sativum) called PA2. A naturally occurring mutant line that lacks PA2 has been exploited in studies to determine the biological function of this nonstorage protein in seed development. The mutant, which has a small seed, a tall plant phenotype, and lacks most of the PA2-encoding genes, has been crossed with a standard cultivar, ‘Birte,’ which contains PA2 to give rise to a recombinant inbred (RI) population. An F3 line carrying the mutation and having a short plant phenotype has been used to generate backcross (BC) lines with ‘Birte.’ Despite having a lower albumin content, seeds from the mutant parent and RI lines lacking PA2 have an equivalent or higher seed nitrogen content. Metabolite profiling of seeds revealed major differences in amino acid composition and polyamine content in the two parent lines. This was investigated further in BC lines, where the effects of differences in seed size and plant height between the two parents were eliminated. Here, differences in polyamine synthesis were maintained as was a difference in total seed protein between the BC line lacking PA2 and ‘Birte.’ Analysis of enzyme activities in the pathways of polyamine synthesis revealed that the differences in spermidine content were attributable to changes in the overall activities of spermidine synthase and arginine decarboxylase. Although the genes encoding spermidine synthase and PA2 both localized to the pea linkage group I, the two loci were shown not to be closely linked and to have recombined in the BC lines. A distinct locus on linkage group III contains a gene that is related to PA2 but expressed predominantly in flowers. The results provide evidence for a role of PA2 in regulating polyamine metabolism, which has important functions in development, metabolism, and stress responses in plants.
Pea (Pisum sativum) albumin 2 (PA2) is a seed protein that has been shown to be resistant to digestion in piglets and chickens and can persist until the end of the digestive tract (Crévieu et al., 1997; Salgado et al., 2003; Le Gall et al., 2007). Studies of the equivalent protein in chickpea (Cicer arietinum) have shown that both chickpea and pea proteins behave as potential allergens in humans (Vioque et al., 1998). Sera from chickpea-sensitive individuals have been shown to give positive reactions against PA2 from both legumes, and both proteins were shown to agglutinate papainized erythrocytes (Vioque et al., 1998). Studies of the physicochemical properties of pea PA2 suggested that a free sulfhydryl group could lead to polymerization, through disulphide interchange, and partial insolubility of protein isolates (Gruen et al., 1987). Taken together, these data suggest that a reduced content of PA2 in pea seeds would lead to a number of improved seed quality characteristics.
The biological relevance of PA2 is unclear. It is not a classic storage protein, lacks a signal peptide, and is not degraded on germination (Higgins et al., 1987). The protein is composed of four copies of a conserved repeat, described as a hemopexin-type repeat, and is thus related structurally to a group of mammalian regulatory proteins that includes vitronectin (Jenne, 1991). It has been shown that chickpea PA2 binds hemin in agreement with this relationship (Pedroche et al., 2005). It is possible that PA2 binds other small ligands, including thiamine (Adamek-Świerczyńska and Kozik, 2002). These data suggest that PA2 may act as a regulatory protein in controlling biological processes dependent on ligand availability.
In this work, a naturally occurring pea mutant that lacks this protein has been identified through germplasm screening. Despite having a lower albumin content, mutant seeds appear to have an elevated nitrogen (N) content under a range of growth conditions. So, provided there are no negative pleiotropic effects, this mutation may have value for crop improvement. The mutation, which is in a wild uncultivated line, was shown to be due to a deletion of most of the structural genes encoding PA2. Genetic crosses involving the mutant line were established and metabolomic approaches employed to identify biochemical changes during seed development that may be linked directly or indirectly to the mutation.
The results indicate that metabolic changes occur in the polyamine pathway with altered amounts of spermidine in the PA2-deficient parent and backcross (BC) mutant lines compared with ‘Birte.’ Polyamines have been implicated in a wide range of biological processes in plants, including growth, development, and stress responses; however, the regulation of their metabolism is still unknown (Kumar et al., 1997). The biosynthetic pathways of spermidine synthesis are well established and involve the synthesis of putrescine via Orn decarboxylase or Arg decarboxylase (ADC) and the conversion of putrescine to spermidine via spermidine synthase (SP), using decarboxylated S-adenosyl-Met (SAM) provided by SAM decarboxylase. Analysis of ADC and SP activities in seeds of PA2-deficient lines provide evidence for a direct biochemical link between PA2 and polyamine metabolism.
RESULTS
Selection and Characterization of PA2 Variant Lines
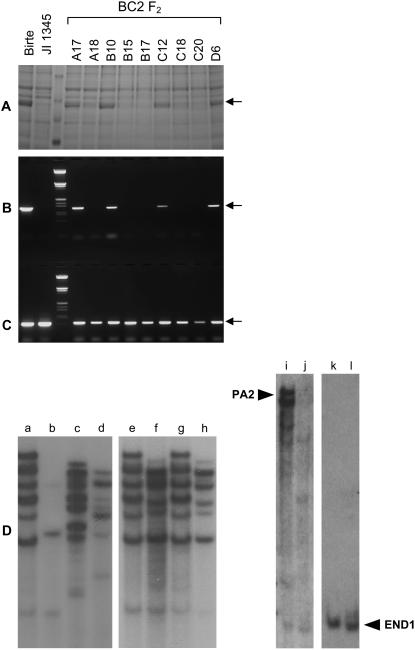
A wide screen of 65 accessions from the pea germplasm collection led to the selection of JI 1345 as a variant, with an elevated total protein content (see below) and a reduced content of the albumin protein, PA2 (Fig. 1A). Genomic-blot analyses showed that the mutant line lacked most of the structural genes for PA2 (Fig. 1D). A recombinant inbred (RI) population of 152 lines was established based on a cross between JI 1345 (Le, long internodes) and a standard pea, ‘Birte’ (le, short internodes). One RI line (RIL; le) was selected to introgress the PA2 mutation into ‘Birte.’ A DNA marker was developed to follow the mutation in RI and BC lines (Fig. 1, A–C). From several combinations of coding and promoter sequence primers that were tested, a promoter region was chosen as a marker, as these primers avoided completely any regions of similarity between PA2 and ENDOTHECIUM1 (END1; see later) genes. Following screening (Fig. 1, A–C), BC mutant lines were selected to grow under controlled environmental conditions for metabolite analysis.
Figure 1.
Analysis of PA2 protein and DNA in variant lines. In A, segregation of PA2 protein (arrow) in BC lines, compared with the parent lines ‘Birte’ and JI 1345. Plantlets from the same seeds were screened by PCR using PA2 promoter (B) or control (C) primers, giving products of 1,234 and 618 bp, respectively. Protein (A) and DNA markers (B and C) separate the parent tracks from the segregants. Southern analysis (D) of: PA2 genes in JI 281 (a), JI 1345 (b), JI 225 (c), ‘Birte’ (d), JI 281 (e), JI 399 (f), two JI 281 × JI 399 RILs (g and h), ‘Birte’ (i), JI 1345 (j), and of END1 genes in ‘Birte’ (k) and JI 1345 (l). Blots were washed at medium (50°C, a–j) or high (65°C, k and l) stringency.
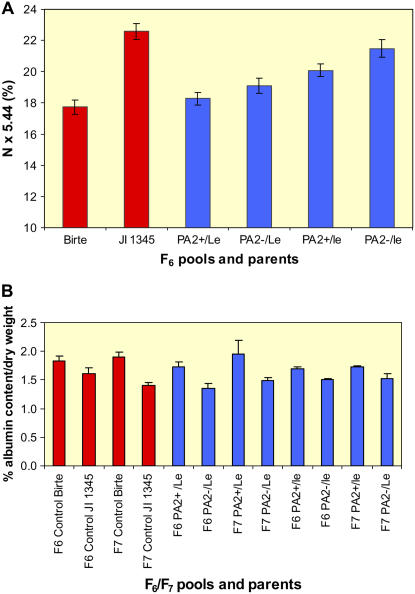
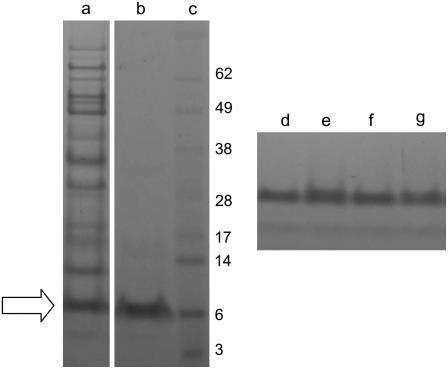
N analyses of the RILs were performed on pooled seeds from progeny lines based on their segregation for le and the PA2 mutation. Analysis of seeds from RIL pools showed that lines lacking PA2 have an elevated N content compared with those that contain PA2 in both Le and le backgrounds (Fig. 2A), suggesting a compensatory increase in other seed proteins in the mutant background. Measurement of the albumin protein fractions from the same RIL pools and parents showed that the PA2 mutation was associated with a significantly lower albumin content (t test, P < 0.001; Fig. 2B). Gel analyses suggested that the mutant parent, JI 1345, contained a higher amount of the pea albumin 1 polypeptide PA1b (approximate Mr 6,000) than ‘Birte’ (Fig. 3). Measurements of the yields of this methanol-soluble protein (Fig. 3, tracks a and b) from RI segregants using a gel scanning and quantification procedure showed that there was no difference between the PA2 segregant classes but a 21% ± 6% higher yield of PA1b from the parent JI 1345 compared with ‘Birte’ (Fig. 3, tracks d–g). These data suggest that the higher N content in the PA2 mutant background is not associated with the albumin fraction.
Figure 2.
Comparison of total seed protein content (N × 5.44) and albumin content in parent lines (red) and RIL segregant classes (blue). RILs have been pooled according to the segregation of internode length (le) and the presence/absence of PA2 protein. Results are the mean ± se (n = 3).
Figure 3.
Analysis of methanol-soluble albumins in parent and segregant classes. The purification from a crude albumin preparation from JI 1345 (a) of the predominant methanol-soluble protein of Mr 6,000 (PA1b, arrow) is shown in b compared with markers (c). The yields of PA1b from parent (d, ‘Birte’; e, JI 1345) and RIL segregant classes (f, Le/PA2+; g, Le/PA2−) were measured by gel scanning. Marker sizes (×10−3) are indicated for track c.
Metabolite Profiling of PA2 Variant Lines
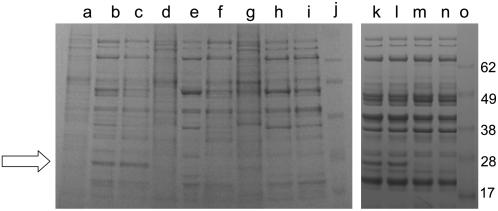
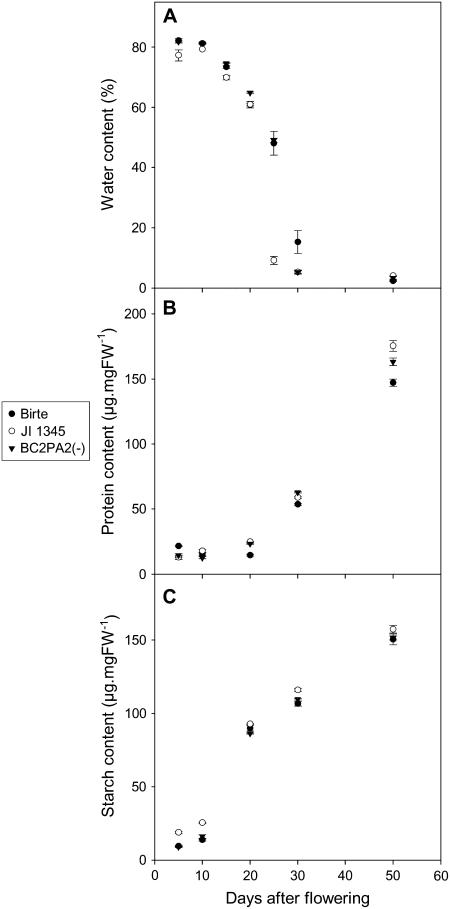
Metabolite profiles were investigated in the mutant line lacking PA2 and in PA2-deficient lines where the PA2 mutation was backcrossed into ‘Birte’ (BC2) to identify biochemical changes during seed development that may be linked directly or indirectly to the PA2 mutation and to seed N content. Because the major part of PA2 accumulation occurs from 20 d after flowering (DAF) onwards (Chandler et al., 1984; Higgins et al., 1987; Fig. 5, tracks a–c), metabolite levels were measured at different stages of seed development. Seed development is characterized by specific changes in the water content of the seeds. As revealed by Figure 4A, there were no substantial changes in the water content of seeds of the mutant and BC lines compared with ‘Birte,’ except at 25 and 30 DAF, where seed-water content decreased. This indicates a slight acceleration of seed maturation in the mutant and BC lines compared with ‘Birte’ at this stage of seed development.
Figure 5.
Comparison of the immature (10 DAF, a, d, and g; 20 DAF, b, e, and h; 30 DAF, c, f, and i) seed protein profiles of ‘Birte’ (a–c), BC2 (PA2-; d–f), and JI 1345 (g–i), and mature seed protein profiles of ‘Birte’ (k and l) and BC2 (PA2-) lines (m and n). An arrow indicates PA2 in ‘Birte.’ The sizes of markers (tracks j and o) are indicated on the right (×10−3).
Figure 4.
Changes in seed composition during development in lines lacking PA2 (JI 1345 and BC) and ‘Birte’: water content calculated by comparing dry weight and fresh weight of the seeds (A), total protein content (B), and starch content (C). Results are the mean ± se (n = 4–7).
Consistent with the elevated total seed N content, the BC and mutant lines lacking PA2 revealed higher total seed protein contents compared with ‘Birte’ at 20, 30, and 50 DAF (the last represents mature seeds; Fig. 4B). The protein profiles for immature and mature BC seeds were very similar to those obtained for ‘Birte,’ apart from the PA2 protein (Fig. 5). In contrast to this, starch content was only slightly elevated in the PA2-deficient mutant, JI 1345, and remained unchanged in the BC (PA2-deficient) line (Fig. 4C).
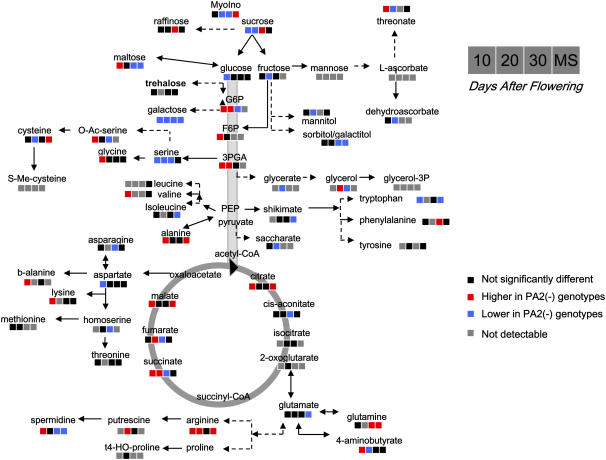
Metabolite profiles were determined for seeds at 10, 20, 30, and 50 DAF and expressed as described in Roessner et al. (2001). Figure 6 presents a schematic overview of the metabolite changes in both PA2-deficient genotypes relative to ‘Birte,’ visualized within a metabolic scheme, whereas Supplemental Figure S1 provides a comprehensive list of all metabolites measured and documents the extent and significance of these changes.
Figure 6.
Changes in metabolite profiles in PA2-deficient lines (JI 1345 mutant and BC lines where the mutation was backcrossed to ‘Birte’) compared with those of ‘Birte’ at different stages of seed development (10, 20, 30, and 50 DAF, the last refers to mature seeds [MS]). Changes in metabolite levels were calculated as the ratio between the level in the PA2-deficient lines and the control (‘Birte’) and are listed in Supplemental Figure S1. To visualize the changes, significant increases or decreases are indicated in red and blue, respectively, within a metabolic scheme (P < 0.05 using the Student's t test of Excel).
Inspection of the data shows that, at 10 to 20 DAF, there was a decrease in all the main sugars (Suc and hexoses) and some minor sugars (such as Gal) in the lines lacking PA2, while glycolytic intermediates (such as hexose phosphates, 3-phosphoglyceric acid), most intermediates of the TCA cycle (such as citrate, succinate, and malate), several amino acids (such as Val, Ala, Lys, and Arg), 4-aminobutyrate, and polyamines (such as spermidine) were increased. Obviously, a deficiency of PA2 led to increased conversion of incoming sugars to amino acids which, however, was not translated to an increase in protein at this early stage.
At later stages of seed development (30–50 DAF) a different picture emerged. A lack of PA2 expression led to unchanged or increased levels in major sugars, while intermediates of glycolysis and TCA cycle were unchanged or showed no consistent trend. However, there were clear changes in several individual amino acids governing the increase in total protein synthesis in the PA2 deficient lines. There was an increase in the levels of Ala, Phe, Cys, Arg, and Gln, while Ile, Tyr, and Glu decreased. Interestingly, lack of PA2 led to a decrease in the level of spermidine, while the level of the precursor of spermidine synthesis, Arg, increased. This indicates that a lack of PA2 leads to a specific block in the pathway of spermidine synthesis involving ADC and SP. Further analysis of mature seeds using an HPLC technique showed that the decrease in spermidine was accompanied by a similar decrease in spermine levels. The levels of spermine were 18.4 ± 0.7, 15.9 ± 0.8, and 13.7 ± 1.3, and those of spermidine 81.2 ± 2.4, 69.6 ± 3.7, and 59.5 ± 6.9 for ‘Birte,’ JI 1345, and BC lines, respectively (expressed as nmol g fresh weight−1, means ± se, n = 3). The values for the mutant lines were significantly different from those of the ‘Birte’ control, according to the Student's t test (P < 0.05).
Analysis of Enzyme Activities in the Pathway of Spermidine Synthesis
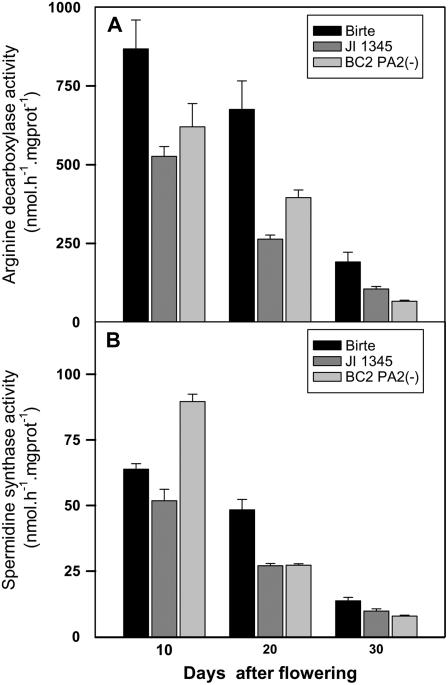
To investigate the link between PA2 and spermidine synthesis more directly, analyses of the overall activities of ADC and SP in developing seeds of PA2-deficient parent and BC mutant lines, in comparison to ‘Birte’ as a control, were performed. The results show that a PA2 deficiency led to a decrease in the specific activity of ADC, especially at 20 and 30 DAF, where ADC activity decreased markedly compared to the control (Fig. 7A). A deficiency in PA2 also led to a decrease in SP activity at 20 and 30 DAF (approximately 2-fold at 20 DAF), while the activity at 10 DAF was not consistently different in the parent and BC mutant lines in comparison to the ‘Birte’ control (Fig. 7B). Irrespective of the genotype, there was a general decrease in ADC- and SP-specific activities during seed development when expressed on a protein basis.
Figure 7.
Changes in the activities of enzymes of spermidine synthesis in PA2-deficient lines (JI 1345 mutant and BC lines where the mutation was backcrossed to ‘Birte’) compared with ‘Birte’ at different stages of seed development. Seeds were frozen under liquid N to analyze the specific activities of ADC (A) and SP (B). Results are the mean ± se (n = 3).
Mapping Polyamine Biosynthetic and PA2-Related Genes
The differences observed in polyamine metabolism between the two parent lines (‘Birte’ and JI 1345) could reflect biochemical differences as a consequence of the PA2 mutation or genetic differences associated with linkage to the PA2 mutation. The latter possibility was investigated by isolating and mapping the relevant genes. The genes encoding PA2 were mapped in two populations (JI 281 × JI 399 and JI 15 × JI 1194, using 65–80 progeny lines) to linkage group I, based on polymorphisms in EcoRI digests of genomic DNA (Fig. 1D, tracks e–h). A gene related to PA2, END1, which is expressed predominantly in flowers (data not shown; Gómez et al., 2004), was found to be present in the PA2 mutant line, JI 1345, by Southern analysis (Fig. 1D, track l) and by PCR (data not shown). It seems likely that END1 represents one of a number of PA2-related genes present in JI 1345 (compare Fig. 1D, tracks b, j, and l). END1 was mapped in the JI 15 × JI 1194 population using PCR primers that amplified a promoter region from the JI 15 but not the JI 1194 gene (data not shown) to linkage group III, close to the position of an anchor morphological marker, st (approximately 9.4 map units from the DNA marker Tps1/262). The gene encoding the polyamine biosynthetic enzyme, SP (spds1), was mapped by single nucleotide polymorphism (SNP) analysis (G/A within intron 3) to linkage group I but at a distance of approximately 47 cM from PA2. This location is in agreement with the genetic location of spds genes on linkage group 5 in Medicago, based on the known synteny between these two linkage groups in the two species (Choi et al., 2004; Kaló et al., 2004). The spds1 allele present in the BC lines was identified as that from the ‘Birte’ parent, thus precluding any effects of linkage drag from the PA2 mutation on SP activity. BLAST analysis of pea ADC (Swissprot accession no. Q43075) indicates corresponding map positions on the Medicago truncatula chromosomes 3 and 4. These correspond to pea linkage groups III and VII, respectively, making any direct effects of genetic linkage on ADC activity in BC lines unlikely.
DISCUSSION
In this work, metabolomic approaches have been adopted to investigate the biological function of the albumin protein PA2 in pea. A mutant lacking PA2 in seeds has been exploited in genetic and metabolite studies. The fact that PA2 is composed of four hemopexin repeats has suggested possible functions for PA2 as a regulator of metabolic processes (Jenne, 1991). Several mammalian proteins, including matrix metalloproteinases, have hemopexin-like motifs adjacent to their catalytic domains (Le et al., 2007). One such protein, vitronectin, has been shown to interact with Ser protease inhibitors and may be important in regulating antiproteolytic activity during cytolysis (Jenne, 1991). In mammals, hemopexins appear to act as scavengers and transporters of toxic plasma heme and have been postulated to play a key role in the homeostasis of nitric oxide (Ascenzi et al., 2007). Hemopexin motifs are likely to be widespread in nature, with prokaryotes having equivalent proteins, predicted to scavenge heme during pathogenicity (Crennell et al., 2000).
Metabolite analyses of the PA2-deficient parent and BC mutant lines showed that a lack of PA2 leads to increased accumulation of seed protein in later stages of seed development (Fig. 4B), which is correlated with changes in the levels of individual amino acids (Fig. 6; Supplemental Fig. S1). A lack of PA2 was accompanied by an increase in the levels of Ala, Phe, Cys, Arg, and Gln. Because accumulation of PA2 contributes approximately 11% to the sulfur-amino acids in pea seeds (Higgins et al., 1987), the increase in free Cys could be the direct consequence of decreased PA2 synthesis. The increase in the N-rich amino acids Arg and Gln and the increase in the Gln to Glu ratio indicate that lack of PA2 leads to a higher N status in the seeds. This is consistent with measurements of total N content in seeds, showing an increase in seeds lacking PA2. It is unlikely that the acceleration in seed desiccation observed at 25 and 30 DAF (Fig. 4A) is responsible for the increased N and total protein content in the PA2-deficient lines. Seed protein was also increased when mature seeds were analyzed, where no significant changes were observed in seed water content between the different genotypes (data not shown).
In addition to the changes in amino acid pattern and protein synthesis, a lack of PA2 led to decreased amounts of the polyamine spermidine during later stages of seed development (see Fig. 6; Supplemental Fig. S1). Our data provide two lines of evidence indicating that this is attributable to a decrease in activity of enzymes of spermidine synthesis. First, the decrease in spermidine was accompanied by an increase in Arg, indicating that conversion of Arg to spermidine involving ADC and SP had been inhibited (Fig. 6). Second, direct measurements of enzyme activities in developing seeds indicated that inhibition of spermidine synthesis is accompanied by a decrease in the activities of ADC and SP (Fig. 7). It seems unlikely that decreased delivery of decarboxylated SAM as a precursor for the conversion of putrescine to spermidine contributed substantially to the decrease in spermidine accumulation, because the decrease in spermidine was not accompanied by an increase in putrescine levels.
The link between PA2 and polyamines is interesting, because the latter have been implicated in a wide range of biological processes, including growth, development, and stress responses (Kumar et al., 1997). Arabidopsis mutants with T-DNA insertions in both of the genes coding for SP (SPDS1 and SPDS2) provide evidence for a critical role of spermidine in embryo development (Imai et al., 2004), whereas Arabidopsis mutants unable to produce spermine exhibited increased water loss and sensitivity to drought stress (Yamaguchi et al., 2006). The decrease in spermidine in lines lacking PA2 could therefore provide a possible explanation for the observed acceleration in seed maturation (compare Figs. 4A and 6).
Apart from direct roles in development and stress responses, polyamines may also be involved in regulating metabolism in developing seeds. The decrease in spermidine and spermine in lines lacking PA2 was accompanied by changes in the levels of sugars, organic acids, and amino acids, indicative of an increase in the organic N content and a decrease in the carbon (C) to N ratio in seeds (Fig. 6). More recently, transgenic tomato (Solanum lycopersicum) lines were used to investigate the effect of increased spermidine and spermine levels on metabolism (Mattoo et al., 2006). Here, changes in spermidine/spermine in the transgenic fruits were accompanied by alterations in the levels of C and N metabolites, supporting a link between spermidine/spermine levels and organic N sensing at the late ripening phase.
It will be interesting to determine why a lack of PA2 leads to a decrease in ADC and SP activities in developing seeds of the mutant and BC lines. Our data show that this is due to biochemical effects rather than genetic linkage. Although the genes encoding SP and PA2 both localized to the pea linkage group I, the two loci were shown not to be closely linked and to have recombined in the generation of the BC lines. We therefore propose that PA2 is biochemically linked to polyamine metabolism in developing seeds by affecting SP and ADC activities. More studies are required to investigate this relationship. The PA2-related gene that is expressed in flowers (Gómez et al., 2004) may give further information on the biological function of this protein family in plants. Targeted mutation of the deduced metal binding sites within END1 is under way to elucidate the function of the active site hemopexin-type domains.
In conclusion, we identified and characterized a natural pea mutant with a reduced content of PA2 in seeds offering a number of potential improved seed quality characteristics, including improved digestibility and reduced allergenic potential. Our results show that seeds lacking PA2 have a higher N content and increased levels of overall seed protein, while starch levels were not substantially changed. Metabolic profiling and enzyme measurements revealed a link between PA2 and spermidine accumulation, providing evidence for a role of PA2 in regulating polyamine metabolism in developing seeds.
MATERIALS AND METHODS
Plant Material
The pea (Pisum sativum) lines (‘Birte’ and JI 1345) were from the John Innes germplasm collection. Sixty-five accessions from the pea germplasm collection were used in the seed protein screen that led to the selection of the PA2 mutant line, JI 1345. A RI population was established from a cross between ‘Birte’ and JI 1345 by single seed descent of 152 progeny lines. Detailed analysis of seed proteins has been carried out at F5, F6, and F7 generations. One RIL (H86 at F3), having both short internodes and the JI 1345-derived PA2 mutation, was used to introgress the mutation into ‘Birte.’ Mutants were identified among the BC1 F2 segregating population and the backcrossing repeated to give BC2 mutant lines. The pea RI mapping populations, derived from the crosses JI 281 × JI 399 and JI 15 × JI 1194, were used to map the loci encoding PA2, a PA2-related protein, and SP.
Plants were grown in a greenhouse with supplementary light and heat in winter months or under controlled environmental conditions for seed development and metabolite analyses. The conditions were: 25°C/d and 20°C/night in a 16-h-day/8-h-night photoperiod at a light intensity of 300 μmol photons m−2 s−1. Emerging flowers were tagged and seed age was expressed in DAF.
Determination of Seed Weights and Water Content
Seed development was followed by measurement of seed weight and water content. Calculations of seed fresh weight, dry weight, and water content were based on weights determined before and after lyophilization of seed samples for 24 h. Mature dry seeds were harvested from the lower nodes of the plants (usually up to node 5), avoiding the smaller late pods.
DNA Screening Methods
Leaves were harvested from young plantlets and used for genomic DNA preparation or leaf imprints were prepared on FTA paper (Whatman) and discs removed for PCR analysis, according to the manufacturer's recommendations.
A DNA-based marker was developed to follow the PA2 mutation in RI and BC lines. PA2 genes were isolated from ‘Birte’ by genome walking using the Advantage Genomic PCR and GenomeWalker systems (CLONTECH), cloned, and sequenced. One gene from a StuI library (PA2stu1) had a promoter of approximately 2.8 kb, whereas two from a DraI library (PA2dra5 and PA2dra8) were shorter, with promoter sequences of 200 bp (EMBL accession nos. AJ831474, AJ833962, and AJ844650, respectively). Forward and reverse primers were designed based on these sequences and tested in various combinations in genomic PCR using DNA from ‘Birte’ and JI 1345. The primers PA2-7 (5′-AACTTTTGCTAACTATTTACC-3′) and PA2-10 (5′-CATTTATCCGTCTAACTAAC-3′) were selected as the optimal forward and reverse primers, respectively, to screen for PA2 gene variants in RI and BC lines.
The sequence of the PA2-related gene, END1 (EMBL accession no. AY324651), was used to design primers based on the promoter region. The primers TB3 (5′-AACCAGTGTCCATATATC-3′) and TB1_rev (5′-AAGGTTATGTTGTGAGC-3′) were used to identify variant END1 genes and to screen RI populations. Southern analysis of PA2 and END1 genes was performed using EcoRI digests of genomic DNA.
The sequences of pea SP cDNAs (EMBL accession nos. AF043108 and AF043109) were compared with corresponding genomic sequence from Medicago truncatula to design primers for isolating and mapping the pea genes. The primers SPDS1-2f (5′-ATTCCAAACCCAAAAAGG-3′) and SPDS1-2r (5′-CRATGTCCTCRATGATRT-3′, where R = A + G to cover interspecific differences) were used to amplify a region of the pea spds1 gene. A SNP was identified within intron 3 in the amplified genomic DNA from JI 281 and JI 399 (G → A, respectively). The primers SPDS1-2f and SPDS2-1r (5′-GGAGCTGCCTTCAAAAATGC-3′) were used as sequencing primers to score and map the intron-based SNP within the JI 281 × JI 399 RI population. The same SNP was evident in comparisons of spds1 in ‘Birte’ and JI 1345 (G → A, respectively), and the same primers were used to check the nature of the allele in the BC lines.
Protein Screening Methods
Mature seeds were screened using meal that was removed with a sharp scalpel blade from seeds before sowing. Immature seeds were ground to a fine meal following lyophilization. Proteins were extracted from seed meal either by heating directly in 5× LDS buffer (Invitrogen) or by shaking in 20 mm ammonium acetate, pH 5.0 (25 mg/mL seed meal) for 1.5 h at 4°C. Ammonium acetate supernatants (albumin fractions) were analyzed on polyacrylamide gels (NuPage BisTris 12% gels in MOPS buffer, Invitrogen), following addition of 5× lithium dodecyl sulfate buffer and 1,4-dithiothreitol to 0.05 m and incubation at 70°C for 10 min. Albumins were fractionated according to methanol solubility following addition of four volumes of 75% methanol and precipitation of insoluble proteins at −80°C for 30 min. Soluble proteins were recovered from supernatants following addition of four volumes of acetone at −20°C. Samples were analyzed on polyacrylamide gels (as above, but using 4%–12% gradient gels in MES buffer) and the relative amounts of methanol-soluble proteins determined by scanning the gels. Protein concentrations were determined using a Bio-Rad protein assay kit and bovine serum albumin in an appropriate buffer as standard.
Total seed N was determined for meals derived from cotyledons following removal of testas. The relative contents of total C and N in dried, powdered meal samples were measured using an elemental analyzer (Vario EL; Elementaranalysensysteme). A minimum of six seeds was analyzed per sample for parent lines; where RILs were pooled according to genotype, the minimum pool size was 46 seeds, with at least two seeds per line. N × 5.44 was used as the N to protein conversion factor for pea (Mossé, 1990). All measurements were performed on triplicate samples.
Metabolite Analyses
Pods were frozen in liquid N and seeds were manually separated from the pods under liquid N2 before being ground to a fine powder using a liquid N mill (Retsch Schwingmühle M200). Ground seeds (50 mg) were extracted according to a method modified from that described previously (Fait et al., 2006) and the relative metabolite contents were determined by gas chromatography-mass spectroscopy as previously described and validated (Roessner-Tunali et al., 2003), using ribitol as internal standard (Roessner et al., 2000). The chromatograms and mass spectra were evaluated using the MassLab program (ThermoQuest). A retention time and mass spectral library for automatic peak quantification of metabolite derivatives was implemented within the MassLab method. Data are normalized to the mean response calculated for the ‘Birte’ samples as described by Roessner et al. (2001).
Free soluble spermine and spermidine were also determined using HPLC (Dionex) after dansylation and quantification with a fluorescence detector according to Smith and Davies (1985).
Protein content was measured according to Eastmond and Rawsthorne (2000). The protein concentration was determined using the dye-binding assay (Bradford, 1976) with bovine serum albumin as the standard. Starch content was measured as described in Geigenberger et al. (1998).
Analysis of Enzyme Activities
Enzyme extracts were prepared from frozen seed material according to Geigenberger and Stitt (1993). ADC (EC 4.1.1.19) was measured as in Pérez-Amador and Carbonell (1995). SP activity (EC 2.5.1.22) was measured by monitoring the production of spermidine from putrescine and decarboxylated SAM by HPLC. Assays were incubated at 37°C. The reaction mixture contained 0.1 m Tris-HCl pH 8.0 buffer, 0.1 mm putrescine, 80 μm decarboxylated SAM, and an aliquot of protein extract. After 30 min, the reaction was stopped by heating (5 min, 95°C). In the control assay, the reaction mixture was placed immediately at 95°C.
Free soluble spermidine was quantified with a fluorescence detector via HPLC (Dionex) after dansylation according to Smith and Davies (1985). The enzyme assays were diluted 1:1 with equal amounts of 0.2 n HClO4 and 1.5 m Na2CO3 before dansylation. A total of 20 μL of sample was injected onto a reverse phase Supelcosil LC-18 column (Supelco) protected by a guard column. Samples were eluted from the column with a solvent gradient (v/v) of water to methanol, changing from 70% to 100% in 15 min at a flow rate of 1 mL/min. The column was washed with 100% methanol for 5 min and reequilibrated at 70% methanol for 5 min before the next sample was injected. Eluates from the column were detected by an attached fluorescence detector with an excitation wavelength of 365 nm and an emission wavelength of 519 nm. Data were analyzed using the Chromeleon (Dionex) software.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ831474, AJ833962, and AJ844650.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Relative metabolite levels in developing seeds of ‘Birte’ (black), JI 1345 (red), and BC2PA2(-) (green).
Supplementary Material
Acknowledgments
We are very grateful to Dr. Hans Weber, Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben (Germany), for carrying out the N analyses in his laboratory, and to Jevneet Kular and Dominic Conquest, John Innes Centre, for assistance with plant screening and assays. We are thankful to Alisdair R. Fernie, Nicolas Schauer, and Maria Ines Zanor, MPI, for help with gas chromatography-mass spectroscopy analyses, and to Karin Koehl for help with plant growth in the Max Planck Institute, Golm.
This work was supported by the European Union (Grain Legumes Integrated Project, a Framework Programme 6 project, grant no. FOOD–CT–2004–506223) and by Defra, United Kingdom (grant nos. AR0105 and AR0711).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Claire Domoney (claire.domoney@bbsrc.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adamek-Świerczyńska S, Kozik A (2002) Multiple thiamine-binding proteins of legume seeds: thiamine-binding vicilin of Vicia faba versus thiamine-binding albumin of Pisum sativum. Plant Physiol Biochem 40 735–741 [Google Scholar]
- Ascenzi P, Bocedi A, Antonini G, Bolognesi M, Fasano M (2007) Reductive nitrosylation and peroxynitrite-mediated oxidation of heme-hemopexin. FEBS J 274 551–562 [DOI] [PubMed] [Google Scholar]
- Bradford M (1976) A rapid and sensitive method for the quantification of microgram quantities of protein using the principle of protein dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Spencer D, Randall PJ, Higgins TJ (1984) Influence of sulfur nutrition on developmental patterns of some major pea seed proteins and their mRNAs. Plant Physiol 75 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Kim D, Uhm T, Limpens E, Lim H, Mun JH, Kalo P, Penmetsa RV, Seres A, Kulikova O, et al (2004) A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M. sativa. Genetics 166 1463–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crennell SJ, Tickler PM, Bowen DJ, ffrench-Constant RH (2000) The predicted structure of photopexin from Photorhabdus shows the first haemopexin-like motif in prokaryotes. FEMS Microbiol Lett 191 139–144 [DOI] [PubMed] [Google Scholar]
- Crévieu I, Carré B, Chagneau AM, Quillien L, Guéguen J, Bérot S (1997) Identification of resistant pea (Pisum sativum L.) proteins in the digestive tract of chickens. J Agric Food Chem 45 1295–1300 [Google Scholar]
- Eastmond PJ, Rawsthorne S (2000) Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryo. Plant Physiol 122 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Hajirezaei M, Geiger M, Deiting U, Sonnewald U, Stitt M (1998) Overexpression of pyrophosphatase leads to increased sucrose degradation and starch synthesis, increased activities of enzymes for sucrose-starch interconversions, and increased levels of nucleotides in growing potato tubers. Planta 205 428–437 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M (1993) Sucrose synthase catalyses a readily reversible reaction in developing potato tubers and other plant tissues. Planta 189 329–339 [DOI] [PubMed] [Google Scholar]
- Gómez MD, Beltrán JP, Cañas LA (2004) The pea END1 promoter drives anther-specific gene expression in different plant species. Planta 219 967–981 [DOI] [PubMed] [Google Scholar]
- Gruen LC, Guthrie RE, Blagrove RJ (1987) Structure of a major pea seed albumin: implication of a free sulphydryl group. J Sci Food Agric 41 167–178 [Google Scholar]
- Higgins TJV, Beach LR, Spencer D, Chandler PM, Randall PJ, Blagrove RJ, Kortt AA, Guthrie RE (1987) cDNA and protein sequence of a major pea seed albumin (PA2: Mr∼26 000). Plant Mol Biol 8 37–45 [DOI] [PubMed] [Google Scholar]
- Imai A, Matsuyama T, Hanzawa Y, Akiyama T, Tamaoki M, Saji H, Shirano Y, Kato T, Hayashi H, Shibata D, et al (2004) Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol 135 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne D (1991) Homology of placental protein 11 and pea seed albumin 2 with vitronectin. Biochem Biophys Res Commun 176 1000–1006 [DOI] [PubMed] [Google Scholar]
- Kaló P, Seres A, Taylor SA, Jakab J, Kevei Z, Kereszt A, Endre G, Ellis THN, Kiss GB (2004) Comparative mapping between Medicago sativa and Pisum sativum. Mol Genet Genomics 272 235–246 [DOI] [PubMed] [Google Scholar]
- Kumar A, Altabella T, Taylor MA, Tiburcio AF (1997) Recent advances in polyamine research. Trends Plant Sci 2 124–130 [Google Scholar]
- Le Gall M, Quillien L, Sève B, Guéguen J, Lallès JP (2007) Weaned piglets display low gastrointestinal digestion of pea (Pisum sativum L.) lectin and albumin pea albumin 2. J Anim Sci 85 2972–2981 [DOI] [PubMed] [Google Scholar]
- Le NTV, Xue M, Castelnoble LA, Jackson CJ (2007) The dual personalities of matrix metalloproteinases in inflammation. Front Biosci 12 1475–1487 [DOI] [PubMed] [Google Scholar]
- Mattoo AK, Sobolev AP, Neelam A, Goyal RK, Handa AK, Segre AL (2006) Nuclear magnetic resonance spectroscopy-based metabolite profiling of transgenic tomato fruit engineered to accumulate spermidine and spermine reveals enhanced anabolic and nitrogen-carbon interactions. Plant Physiol 142 1759–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossé J (1990) Nitrogen to protein conversion factor for ten cereals and six legumes or oilseeds. A reappraisal of its definition and determination. Variation according to species and to seed protein content. J Agric Food Chem 38 18–24 [Google Scholar]
- Pedroche J, Yust MM, Lqari H, Megías C, Girón-Calle J, Alaiz M, Millán F, Vioque J (2005) Chickpea PA2 albumin binds hemin. Plant Sci 168 1109–1114 [Google Scholar]
- Pérez-Amador MA, Carbonell J (1995) Arginine decarboxylase and putrescine oxidase in ovaries of Pisum sativum L. (changes during ovary senescence and early stages of fruit development). Plant Physiol 107 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectroscopy. Plant J 23 131–142 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado P, Freire JPB, Ferreira RB, Teixera A, Bento O, Abreu MC, Toullec R, Lalles JP (2003) Immunodetection of legume proteins resistant to small intestinal digestion in weaned piglets. J Sci Food Agric 83 1571–1580 [Google Scholar]
- Smith MA, Davies PJ (1985) Separation and quantification of polyamines in plant tissue by high performance liquid chromatography of their dansyl derivatives. Plant Physiol 78 89–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vioque J, Clemente A, Sánchez-Vioque R, Pedroche J, Bautista J, Millán F (1998) Comparative study of chickpea and pea PA2 albumins. J Agric Food Chem 46 3609–3613 [Google Scholar]
- Yamaguchi K, Takahashi Y, Berberich T, Imai A, Miyazaki A, Takahashi T, Michael A, Kusano T (2006) The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett 580 6783–6788 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.