Abstract
Organisms with a high density of transposable elements (TEs) exhibit nesting, with subsequent repeats found inside previously inserted elements. Nesting splits the sequence structure of TEs and makes annotation of repetitive areas challenging. We present TEnest, a repeat identification and display tool made specifically for highly repetitive genomes. TEnest identifies repetitive sequences and reconstructs separated sections to provide full-length repeats and, for long-terminal repeat (LTR) retrotransposons, calculates age since insertion based on LTR divergence. TEnest provides a chronological insertion display to give an accurate visual representation of TE integration history showing timeline, location, and families of each TE identified, thus creating a framework from which evolutionary comparisons can be made among various regions of the genome. A database of repeats has been developed for maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum), and barley (Hordeum vulgare) to illustrate the potential of TEnest software. All currently finished maize bacterial artificial chromosomes totaling 29.3 Mb were analyzed with TEnest to provide a characterization of the repeat insertions. Sixty-seven percent of the maize genome was found to be made up of TEs; of these, 95% are LTR retrotransposons. The rate of solo LTR formation is shown to be dissimilar across retrotransposon families. Phylogenetic analysis of TE families reveals specific events of extreme TE proliferation, which may explain the high quantities of certain TE families found throughout the maize genome. The TEnest software package is available for use on PlantGDB under the tools section (http://www.plantgdb.org/prj/TE_nest/TE_nest.html); the source code is available from http://wiselab.org.
Transposable elements (TEs) are mobile DNA found throughout eukaryotic organisms. Although abundance is extremely high in some organisms, little is known about the processes governing the distribution of TEs across the genome. Each classification level of a TE may exhibit different genetic makeup, different modes of replication, or preference for different genomic habitats. By the nature of their mobility, TEs have the potential to induce change throughout an organism's genome. As a consequence of multiple TE copies, unequal crossover and recombination can occur between chromosome regions. TE insertions can cause gene or regulatory mutations, altering levels of transcripts, or provide new genetic material for novel gene functions to evolve (Kidwell and Lisch, 2000). TE genes may be recruited by the host organism for cellular functions and can serve as transportation systems for genes to new genomic locations (Lal et al., 2003).
Abundance of TEs varies widely across different organisms. Human (Homo sapiens) DNA is composed of 45% (Lander et al., 2001) repetitive sequences, Drosophila melanogaster is 3.9% (Kaminker et al., 2002), and maize (Zea mays) is 67% (Haberer et al., 2005). Even closely related organisms can have vastly different amounts of repetitive elements; for example, rice (Oryza sativa) is 35% repetitive (IRGSP, 2005) compared to 67% for maize. Classes of TEs also vary between organisms; the ratio of long-terminal repeat (LTR) retrotransposon/non-LTR retrotransposon/DNA transposon of human repetitive DNA is 8.3/74.5/2.8, whereas D. melanogaster is 68.6/22.5/8.0 and maize is 94.4/0.1/1.9, respectively. The high amounts and different repetitive makeup of genomes coupled with their potential for inducing evolutionary changes make the annotation of TEs in DNA sequences crucial to decipher genomic processes. Annotation of TEs goes beyond identification of repetitive sequence sections. Many TE sequences are historical artifacts of past replications and have become inert or truncated by evolution. Reconstruction of these past insertion events cannot only show how genes and regulatory regions have altered, but shed light on the evolutionary dynamics of the entire genome. Based on insertion order and calculation of age of insertion, complete annotation of TEs can provide historical chronology of a genomic region. A genome-wide annotation of TEs can also provide insight into repeat biology, including insertion site preferences, family distribution, and differences in repetitive density.
High quantities of TEs, especially the LTRs of retrotransposons, greatly impede sequence assembly as well as genome annotation (Rabinowicz and Bennetzen, 2006). As subsequent TEs integrate into a clustered location, there is a high likelihood the repeat will insert within the boundaries of existing elements. This incident, nesting of one element within another, seen on a small scale in some organisms (Quesneville et al., 2005), is widely observed throughout grass genomes (SanMiguel et al., 1996, 1998). When TEs nest within one another, the existing repeat is fragmented by the sequence of the inserting element. Successive insertion events nested within a cluster will create highly fragmented sequences; correct identification of the repeats in nested groups requires reconstruction of the original sequences.
Current repeat annotation tools have not adequately addressed the issue of nested TEs and are unable to rebuild fragmented elements. Three distinct methods of TE identification have been developed. RepeatMasker (http://www.repeatmasker.org) uses a repeat database to locate sequence matches. This provides correct identification of fragmented TEs in nested repeat clusters, but reconstruction of whole TEs and evolutionary timeline of insertions is not possible. LTR retrotransposon detection software, such as LTR_struct (McCarthy and McDonald, 2003; Kalyanaraman and Aluru, 2005), groups LTR pairs based on sequence alignment identity. With LTR pair locations, one can infer a general retrotransposon insertion order; however, nested repeats are not specifically addressed and an LTR broken from subsequent insertions will not be identified. Furthermore, LTR retrotransposon identification software is unable to identify internal regions of retrotransposons and will not locate non-LTR retrotransposons, DNA transposons, or other TEs. De novo TE identification software, PLIER (Edgar and Myers, 2005), RECON (Bao and Eddy, 2002), and RepeatScout (Price et al., 2005), has the ability to locate and classify previously unknown repeats based on sequence identity of repeated regions. These programs do not have the ability to reconstruct fragmented TEs or provide insight into the genomic evolutionary process.
To fully analyze repeat dense grass genomes, we have developed TEnest. Using a community updated repeat database of LTR retrotransposons, non-LTR retrotransposons, DNA transposons, and other repetitive elements, TEnest will identify all TE insertions in the input sequence. With additional repeat database construction, TEnest will annotate TEs in any organism's genome. For LTR retrotransposons, TEnest will identify the two flanking LTR sequences and calculate the time since insertion based on the rate of mutation accumulation in repetitive sequences of grasses (1.3 × 10−8; Kimura, 1980; Ma and Bennetzen, 2004). TEnest identifies the internal regions of the TE insertion and, for all repetitive element types, will reconstruct sequence fragmentations caused by nesting of TEs. TEnest outputs the coordinate locations of TEs identified as well as a publication-quality graph representing the chronology of the DNA sequence.
Recent evidence suggests that the high percentage of repetitive elements, especially LTR retrotransposons in maize, is due to the replication activities of just a few element families (Meyers et al., 2001). Here, analysis with TEnest shows the retrotransposons Ji, Opie, and Huck of maize each make up 11% to 13% of the total genome sequence. A single TE that rapidly replicates throughout the genome can have significant consequences not only on the evolution of the TE family, but also on the whole genome. TEnest gives the user the ability to reconstruct the ancient TE insertions to their prenested sequence states, allowing phylogenetic analysis upon all the existing members of the TE family throughout the evolutionary history of the organism. A detailed phylogenetic analysis of each high-copy TE family, associated with each element's time since insertion, suggests there have been isolated events of extreme TE proliferation across the genome, allowing Ji, Opie, and Huck to replicate seemingly without restraint. In addition to reconstructing ancient fragmented TEs, TEnest also identifies solo LTRs of retrotransposons formed by unequal recombination. Solo LTRs found throughout the maize genome are shown to be inconsistent across both retrotransposon families, inconsistent with TE length or LTR length.
Throughout this article we follow the TE nomenclature format outlined in Wicker et al. (2007). TEs are hierarchically classified into six divisions from class through subfamily. Classifications most used in this article are class, separated into retrotransposons and DNA transposons; superfamily, including Gypsy and Copia retrotransposons; and family, individual TEs grouped together by sequence similarity. Throughout this article, superfamily and family names are italicized (Wicker et al., 2007).
RESULTS
The nested TE identification software package TEnest has three sections for use in genome sequence analysis: the organism-specific repeat databases; TEnest, a program for identification of TE coordinates; and svg_ltr, a graphical display program for visualization of TE insertions.
TEnest Uses a Repeat Database Kept Up to Date with New Sequences and User Input
The repeat databases are kept up to date by two methods. First, when new genomic contigs are completed, they are entered into PlantGDB (http://www.plantgdb.org; Dong et al., 2004) and a TEnest insertion graph is produced. This triangle insertion graph is examined for unidentified or fragmented nested insertions within TEs. These insertions are compared against a set of potential new TEs to determine whether it has been previously characterized. A similar process will be implemented for wheat (Triticum aestivum) sequence contigs in the TriAnnot wheat annotation pipeline (urgi.versailles.inra.fr/projects/TriAnnot). In maize, several TEs have been identified by this process, including Danelle (GenBank accession no. EF562447), Stella (GenBank accession no. EF621725), Tavish, Tenzig, Klaus, and Hodge.
Second, users of TEnest on PlantGDB can update the repeat databases with newly identified TEs. This submission system requires information about the TE, such as the organism, TE classification, sequence locations of identification, and the proposed name. The new TE is aligned to known TE families in the organism and flagged for manual review; when review is complete, users will be notified of the status of their TE submission. TEnest users can also use this submission system to suggest revisions or repairs to TE database entries.
Annotation of TEs Using TEnest
With use of the plant repeat databases, TEnest identifies all TE insertions in the input sequence, reconstructs fragmented elements, and determines age of insertion for LTR retrotransposons producing a list of coordinates for each TE sequence location. TE insertions are classified as one of four data types: SOLO, corresponding to solo LTR sequences; PAIR, right and left LTRs of a LTR retrotransposon grouped by base pair similarity and the corresponding internal sequences of the TE; NLTR, full-length TEs of classes not containing LTRs (non-LTR retrotransposons and DNA transposons); and FRAG, partial sequences of the NLTR class or internal fragmented regions of LTR retrotransposons.
Identification of Retrotransposon LTR Sequences
Throughout the TEnest process, a two-alignment approach is used to quickly identify exact coordinate locations of TE alignments. First, WU-BLAST blastn (http://blast.wustl.edu) is used to rapidly identify possible TE sequence regions; then FASTA LALIGN (Huang and Miller, 1991) is used to retrieve the exact coordinates of each possible TE type in these regions. The TEnest process begins by identifying LTR sequences within the input sequence. Users can select or deselect specific TEs to include in the analysis. LTR database sequences are aligned to the input sequence with WU-BLAST blastn. If an alignment is found, the coordinates are retrieved, expanded on either side (by the size of the matching LTR), and excised from the input sequence. This set of excised sequences will contain short, incomplete matches to LTRs of multiple TE families. Each excised LTR sequence is sent to the FASTA LALIGN process where a pairwise local alignment between it and the database LTR is performed. If this alignment provides a passing score (default of E value 10−20), this coordinate set is entered into the alignment list, which now contains full-length annotations over the extent of the identified region.
Each pairwise LTR alignment coordinate set is entered into the recombination process where a power set algorithm is used to rejoin separated LTR sections. A power set is the set of all the subsets of a set (Suppes, 1972). For the TEnest algorithm, the original set is the separated LTR sections; the power set is the list of all subsets of the separated LTR set. The power set recombination process is based on coordinates of the matching database LTR (also referred to as the subject of the alignment, whereas the input sequence is the query), and does not allow overlapping sequence regions to result in joined sections containing duplicated sequence regions. For example, three separated LTR sections are entered into the power set recombination process; section A with LTR-based coordinates of 1 to 350, section B with coordinates of 50 to 400, and section C with coordinates of 351 to 500. The power set of the separated LTR set is ({}, {A}, {B}, {C}, {A,B}, {A,C}, {B,C}, {A,B,C}). Sets with overlapping LTR-based coordinates are not allowed; this removes {A,B}, {B,C}, and {A,B,C}. From the remaining five, the set with the largest sequence length is returned; each LTR section must be returned exactly one time. In this case, two sets are returned, sets {A,C} and {B}, with lengths of 500 and 350 bp.
LTR retrotransposons replicate into new locations across the genome by means of reverse transcription and integration. A seven-step process produces an exact DNA intermediate of the retrotransposon with one exception; the two new LTR sequences are reverse transcribed from an intermediate LTR, which itself is a unique LTR sequence formed from the combination of the two original LTRs of the parent retrotransposon. This results in a new integrated retrotransposon with identical LTRs (Boeke and Corces, 1989). Over evolution, these LTRs separately acquire mutations, but over a stretch of sequence the two LTR copies from a retrotransposon insertion will be more alike than LTRs from different insertion events. Thus, sequence similarity can be used to pair LTRs for reconstruction of whole LTR retrotransposons.
For each TE family, each LTR alignment returned from the power set recombination process is excised from the input sequence and joined into a single contiguous sequence. By TE family, the LTR sequences are locally aligned to each other and the base pair substitution rate (BSR) is determined. Any insertion, deletion, or substitution of any length is scored as a single substitution for the alignment. BSR is calculated by the total substitutions divided by the alignment length of the two LTR sequences. LTR sequences are grouped according to the smallest BSR. The two paired LTRs are classified in the PAIR data type; any LTR sequences not paired are assigned to the SOLO class. LTRs can be found in a solo configuration throughout the genome. There are three possibilities where TEnest is unable to assign a LTR to a pair. (1) The second LTR sequence is missing; it is found either off the end of the contig or in a sequence gap. (2) The LTR's true partner was incorrectly paired with another LTR. Although unlikely, if two LTRs from different retrotransposon insertions have evolved to be more similar than those from the same insertion, incorrect LTR pairing can occur and possibly cause solo LTR identification. Any such occurrences of incorrect LTR pairing are resolved by TEnest by the discrepancy function discussed below. (3) The solo LTR is the result of homologous unequal recombination that has caused deletion of the internal retrotransposon region and one, leaving just a single LTR.
Identification of Retrotransposon Internal Regions
The sequence within the boundaries of each paired LTR set is examined for the internal regions of a LTR retrotransposon. LTR pairs are grouped according to nesting sets; each first-level LTR pair found inserted directly into the original, pre-TE insertion DNA sequence is grouped with any subsequently inserted LTR pairs found nested within the first-level LTR pairs. Each LTR pair grouped in each nesting set is examined, the smallest LTR pair coordinate distance first, for internal sequence locations. As in the LTR identification process described above, a similar two-alignment method is performed to quickly and accurately identify the exact coordinates of the LTR retrotransposon internal regions. First, WU-BLAST blastn is used to rapidly identify potential locations; second, LALIGN makes a pairwise alignment of all identified regions to determine exact coordinates; third, the power set recombination process reconstructs separated regions into the final internal middle region.
However, there is one significant difference between the alignment method here and the previously described LTR identification process. First, paired LTRs are arranged by smallest sequence-spanning length first; after each paired LTR is processed, the identified regions are ignored by subsequent alignments. Second, during the alignment process, the sequence database of the initial WU-BLAST alignment contains only one TE sequence, the TE type corresponding to the paired LTRs. These restrictions give TEnest the ability to correctly annotate the entire internal regions of nested LTR retrotransposons. The identified internal regions are added to the PAIR data type signifying classification as whole LTR retrotransposons.
Identification of Non-LTR Retrotransposons and DNA Transposons
At this stage, the TEnest annotations consist of full LTR retrotransposons and solo LTRs. All unidentified regions are examined for potential NLTRs, DNA transposons, or FRAGs of any classification. Unidentified regions may be found anywhere in the sequence not classified as PAIR or SOLO and may be found inserted into the original DNA sequence or found nested within PAIR or SOLO identifications. As described above in the LTR and internal region alignment processes, the two-alignment method is used to identify coordinates of TE annotations. First, WU-BLAST blastn is used to rapidly identify potential locations; then LALIGN is used to determine exact coordinates. As illustrated in Figure 1A, sequence locations are grouped prior to the recombination process so as to allow joined sections to be separated by subsequent TE insertions, but not by recombined sections or PAIR LTR-middle-LTR sets. Recombined sequences are classified by sequence length to the original database sequence; if greater than an 80% match, the joined sections are assigned to the NLTR data type; if less than this value, the recombined group is assigned to FRAG to signify it is a fragmented TE insertion.
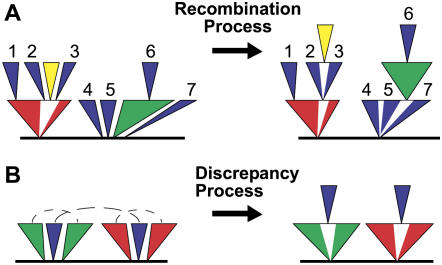
Figure 1.
Recombination of separated TE Sections. A, Recombination rules for non-LTR retrotransposon and fragmented transposon annotations. Separated sections of a single type of TE insertion are shown in blue; other TE insertions are shown as green, red, or yellow. Recombination allows sections separated by subsequent insertions to be rejoined, but does not allow sections to join across previously recombined groups. With correct TE insertion-based coordinates, fragments 2 and 3 will form a group and fragments 4, 5, and 7 will join as a recombined group. Fragments 1 and 6, and the joined sections 2-3 and 4-5-7 cannot join any fragments. B, Coordinate discrepancies of recombined sections. Recombined sections are shown with dotted areas across separated sections. These cases can be caused by local inversions, sequence assembly errors, or incorrect joins from the recombination process. A weighted ratio of number of disagreements, sequence alignment identity, and sequence length of annotations is used to determine which TE-recombined discrepancy is removed.
The power set recombination process of TEnest is useful for joining sections separated by nesting of subsequent TE insertions; however, this process can run into problems. Although uncommon, recombined TE sections are susceptible to coordinate discrepancies, defined as a rejoined sequence set whose grouping configuration disagrees with another rejoined set. This is seen when sections from two or more recombined TE annotations are found in alternating orders across the input sequence (Fig. 1B) as opposed to nested within one another. Whereas a biological process, such as local small inversions, can explain such occurrences, a nested insertion display is unable to represent disagreeing rejoined sections. TEnest, therefore, assumes the discrepancies are caused by either incorrect power set grouping or incorrect LTR pairing. To resolve each recombination discrepancy, each TEnest data type (PAIR, SOLO, NLTR, FRAG) is self checked and each combination of data types is checked for possible coordinate discrepancies by TEnest. Any discrepancies found are scored based on alignment identities, sequence length percentage of the whole TE, and number of discrepancies; the joins of those with the worst discrepancy scores are broken to split combined sections into separate groups.
TEnest Processing Time Is Decreased with Use of Multiprocessors and Clustered Computers
The time required to complete a TEnest run is dependent on the amount of TEs contained within the sequence. As longer plant sequence contigs are produced, the amount of TEs found in the sequence will also increase, and the time required for TEnest runs will grow proportionally. To address increasing sequence lengths, TEnest has been developed with multiprocessor ability and can be run on clustered computers with the use of an included perl script. An average-sized bacterial artificial chromosome (BAC) of 164 kb (GenBank accession no. AC148161) containing 11 PAIRs, one SOLO, five NLTRs, and 30 FRAGs takes 5 min, and the currently largest maize contig of almost 1 Mb (GenBank accession no. EF517601) containing 46 PAIRs, six SOLOs, 36 NLTRs, and 155 FRAGs takes 45 min for a TEnest run using a dual 3.2-GHz desktop PC with 4-Gb RAM. Time-intensive sections of the TEnest algorithm are broken out to different processors for five subroutines: LTR alignment, LTR power set, LTR BSR calculation, PAIR internal sequence detection, and FRAG/NLTR detection.
In addition, to make TEnest a viable resource for chromosome-sized maize pseudomolecules, a TEnest wrapper script, clusterTEnest.pl, has been developed. This script will take a large input sequence and split it into user-defined lengths and send each section to a separate node of a clustered computer to run several instances of multiprocessor TEnest simultaneously. Once each split sequence is complete, the annotation results are regrouped and the identified TEs are removed from the input sequence. A final TEnest is run on the full sequence, ultimately providing the same output as an original TEnest submission. This split function decreases process time for long sequences and decreases incorrect LTR BSR pairing that may be found when analyzing a large number of a retrotransposon type. For example, when the same 1-Mb contig (GenBank accession no. EF517601) was split into 100-kb segments and sent to 10 nodes of a clustered computer (each with dual 3.2 Ghz, 4-Gb RAM), it took 35 min to complete. The benefit of clusterTEnest.pl will increase with longer sequence contigs.
Visualization of Nested TE Structures with svg_ltr
TE insertions identified by TEnest are visualized in a triangle insertion graph with the program svg_ltr (Fig. 2). svg_ltr uses the coordinate table of identified TE locations from TEnest (Supplemental Figs. S1–S4) to produce the main output of TEnest. The nesting display graph represents the original DNA prior to repeat insertions as a black horizontal line and the TE insertions within it as triangles. The horizontal top of a triangle TE insertion corresponds to the length of the TE insertion; the bottom point shows the insertion location. The genome distance between any two points on a triangle graph is determined by the addition of all horizontal lines (including the black DNA representation and TE triangle tops) on all levels between the two points. Spacing and alignment of triangles are adjusted to prevent overlapping triangles; however, TE triangle insertion point locations are preserved to show the true location of a TE. Triangle color corresponds to TE type, shown in the legend at the bottom of the display.
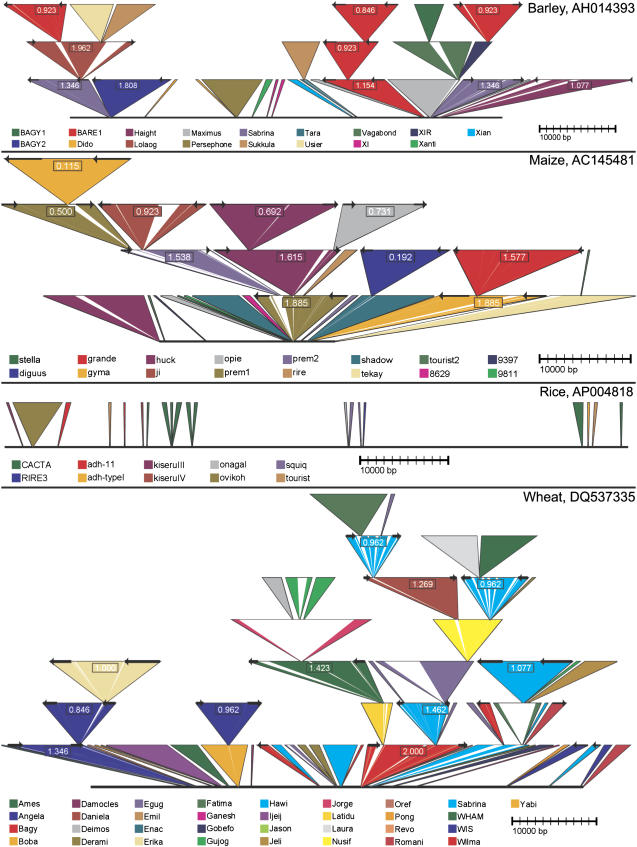
Figure 2.
TEnest graphical display; barley, maize, rice, wheat. TEnest graphical output examples; barley (AH014393; Caldwell et al., 2004), maize (AC145481), rice (AP004818), wheat (DQ537335; Gu et al., 2006). The original, pre-TE insertion DNA sequence is shown as a black horizontal line. TE insertions are shown as colored triangles; names for each type are shown below in the legend. LTRs of LTR retrotransposons are shown as black arrows at the top of the triangle. White areas within triangles are unique or unidentified insertions within the TE. These can possibly correspond to new, unidentified TEs. Insertion age of LTR retrotransposons is shown in Mya inside the retrotransposon triangle. Features, including annotation type, white areas, Mya calculation, and coordinate display can be optimized and selected/deselected by the user.
Several functions are included with svg_ltr to produce graphs containing information needed by the user. Display of data types SOLO, PAIR, NLTR, and FRAG can be toggled on or off. Arrows representing location and direction of LTRs can be shown at the top of the triangle. Either BSR or millions of years ago (Mya) can be displayed inside a box within a LTR retrotransposon triangle; see Figure 3 for calculation of BSR and Mya. Coordinates corresponding to either the TE or the input sequence can be displayed at the top of the TE triangle for each section of the recombined TE group. Insertions found within a TE that do not align to a TE found in the repeat databases can be omitted from the display; this is shown by a white triangle within the TE triangle.
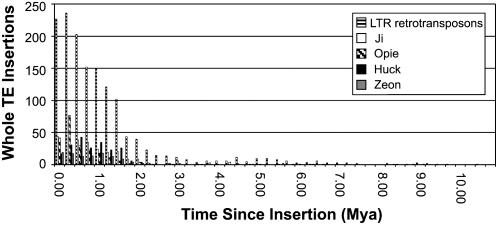
Figure 3.
Chronology of maize whole LTR retrotransposon insertions. Differences in paired LTR sequences are used to calculate the time since insertion in Mya. Mya is calculated by BSR divided by 2 times the substitution rate in repetitive regions of grasses (1.3 × 10−8; Kimura, 1980; SanMiguel et al., 1998; Ma and Bennetzen, 2004). BSR is determined by the amount of mutations between the pairwise alignments of the left and right LTRs divided by the length of the LTR. Mutations are scored by counting each incidence of an insertion, deletion, or substitution between the two LTRs. Insertions or deletions of single or multiple bases are scored as one mutation event. Of 1,456 maize LTR retrotransposon insertions, 50% are <0.875 Mya, 75% are <1.5 Mya.
In addition, several functions are included with svg_ltr that make this a stand-alone program for display of sequence region annotations. svg_ltr can also display two user-inputted data types: GENE, corresponding to gene annotations, and PSDO, corresponding to pseudogene annotations. Both are displayed along with the TE annotations in the svg_ltr display; GENE and PSDO are shown as rectangular regions with direction-indicating arrows, and can show separated sections, such as multiple exon genes. A script, checkTE.pl, included with TEnest, is provided to assist users in adding this additional information to the svg_ltr input file by converting to and from both the TEnest coordinate table output and a generic feature format (GFF3) table.
Computational Validation of TEnest Repeat Annotations
To assess the repetitive element identification capability of TEnest, three verification experiments were conducted. (1) The TEnest outputs of high-density repetitive regions of maize and rice were compared to GenBank-submitted TE annotations. (2) Permutations of TE annotations were evaluated to exclude confounding TE annotations. (3) Simulated genomic maize sequences were made with TE insertions and examined with TEnest.
Comparisons of Maize and Rice to Submitted GenBank Annotations
To evaluate the accuracy of TEnest, curated maize and rice GenBank sequence contigs were compared to their TEnest outputs. Sequenced contigs were chosen that contained repeat region annotations and had five or less sequence gaps. Rice was selected due to its phylogenetic similarities, yet differences, in abundance of repetitive sequences and its complete sequence and annotation. Rice and maize have vastly different TE class proportions; rice retrotransposons make up 19.3% and DNA transposons make up 13.0% of the genome (IRGSP, 2005), whereas maize is composed of 63.3% retrotransposons and 1.3% DNA transposons. However, structures between element classes are similar and some TE families found between the organisms are closely related. Eight maize (Tikhonov et al., 1999; Song et al., 2001; Fu and Dooner, 2002; Fu et al., 2002; Brunner et al., 2005) and two rice (Nagano et al., 2002) annotations were graphed with svg_ltr and visually compared to the TEnest annotations (Table I).
Table I.
Verification: TEnest results compared to GenBank annotated maize BACs
Summary of verification results of TEnest compared to curator-annotated GenBank submitted sequence contigs. Tpn, Transposon.
| Submitted Contiga | Gaps | Total Annotated GenBank
|
Total Annotated TEnest
|
Missing GenBankb | Missing TEnestc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Retro Tpn | LTR Solo | DNA Tpn | Fragments | Retro Tpn | LTR Solo | DNA Tpn | Fragments | ||||
| Maize | |||||||||||
| AF391808 | 0 | 9 | 0 | 6 | 0 | 12 | 0 | 11 | 8 | 3,0,5,8 | 0,0,0,0 |
| AF391808 (P) | 0 | 9 | 0 | 6 | 0 | 12 | 0 | 11 | 8 | 3,0,5,8 | 0,0,0,0 |
| AF123535 | 0 | 11 | 3 | 0 | 2 | 11 | 2 | 3 | 2 | 0,0,3,0 | 0,0,0,0 |
| AF123535 (P) | 0 | 11 | 3 | 0 | 2 | 11 | 2 | 3 | 2 | 0,0,3,0 | 0,0,0,0 |
| AF448416 | 0 | 4 | 0 | 0 | 2 | 4 | 0 | 12 | 4 | 0,0,12,0 | 0,0,0,0 |
| AF448416 (P) | 0 | 4 | 0 | 0 | 2 | 4 | 0 | 12 | 4 | 0,0,12,0 | 0,0,0,0 |
| AF488416 | 0 | 6 | 0 | 2 | 0 | 7 | 0 | 6 | 2 | 0,0,4,2 | 0,0,0,0 |
| AF488416 (P) | 0 | 6 | 0 | 2 | 0 | 7 | 0 | 6 | 2 | 0,0,4,2 | 0,0,0,0 |
| AF090447 | 0 | 3 | 0 | 0 | 0 | 12 | 2 | 7 | 6 | 9,2,7,6 | 0,0,0,0 |
| AF090447 (P) | 0 | 3 | 0 | 0 | 0 | 12 | 2 | 7 | 6 | 9,2,7,6 | 0,0,0,0 |
| AY664414 | 5 | 21 | 1 | 0 | 1 | 25 | 0 | 5 | 8 | 4,0,5,7 | 0,1,0,0 |
| AY664414 (P) | 5 | 21 | 1 | 0 | 1 | 25 | 0 | 5 | 8 | 4,0,5,7 | 0,1,0,0 |
| AY664416 | 4 | 2 | 0 | 0 | 0 | 7 | 1 | 8 | 9 | 5,1,8,9 | 0,0,0,0 |
| AY664416 (P) | 4 | 2 | 0 | 0 | 0 | 7 | 1 | 8 | 9 | 5,1,8,9 | 0,0,0,0 |
| AY691949 | 0 | 6 | 0 | 0 | 0 | 8 | 2 | 2 | 2 | 2,2,2,2 | 0,0,0,0 |
| AY691949 (P) | 0 | 6 | 0 | 0 | 0 | 8 | 2 | 2 | 2 | 2,2,2,2 | 0,0,0,0 |
| Rice | |||||||||||
| AP000559 | 0 | 0 | 0 | 36 | 9 | 0 | 0 | 42 | 7 | 0,0,6,0 | 0,0,2,0 |
| AP002542 | 0 | 0 | 0 | 66 | 21 | 0 | 0 | 76 | 15 | 0,0,10,0 | 0,0,6,0 |
Submitted BAC sequences compared to TEnest results using the general and permutated (P) maize and rice repeat databases. In the permutated database, consensus TE sequences were made from multiple alignments excluding TE sequences found on the same branch or cluster of the TE's phylogenetic analysis. This permutation removed any influence biologically similar TE sequences could have on TEnest annotations. In each case, the general and permutated database gave the same result, showing TEnest results are not artificially enhanced by related TE sequences in the repeat database.
Summary of TE annotations found by TEnest, but not by GenBank-submitted annotations (retrotransposons, solo LTRs, DNA transposons, fragment TEs).
Summary of TE annotations found by GenBank-submitted annotations, but not by TEnest (retrotransposons, solo LTRs, DNA transposons, fragment TEs). AY664414 contains one solo LTR identified in the GenBank submission not identified by TEnest; however, this solo LTR was grouped into a whole LTR retrotransposon by TEnest. The initial run of TEnest missed several annotations in both rice contigs; these were obtained in a second run with parameter alterations to allow for smaller TE sequences.
For maize, TEnest identified every annotation formerly found by the original curators. In addition, TEnest identified many LTR retrotransposons, solo LTRs, fragmented repeats, DNA transposons, and other repeats not found in the original annotations. For rice, TEnest found all but seven miniature inverted-repeat TE (MITE) insertions originally identified by the initial annotations. These missing MITEs were truncated and below the default cutoff values in TEnest; additional runs with altered parameters to allow smaller sequence alignments identified all the missing insertions. In addition, TEnest located 11 additional DNA transposons not found in the original analysis. From the analyzed rice BACs, no TEs were seen in nested configuration, compared with 70% of maize TEs found nested. In additional rice BACs analyzed, nested TEs were seen at a rate of approximately 1 per 75 to 100 kb. This rate of nested TEs is due to the low amounts of repeats in rice as well as the small average lengths of rice TE insertions, caused by the high number of MITE insertions in the rice genome.
Permutations of the Maize Repeat Database
TEnest uses a TE database of representative and consensus sequences to identify repeat insertions. Whereas alignment to this database is much quicker than to the set of all TEs identified for each family, this method can potentially introduce a notable flaw. A TE insertion with extremely similar sequence to the representative TE in the repeat database would be identified by TEnest with little to no difficulty; however, a divergent TE insertion would not match as well and therefore obtaining a complete alignment of the region would be more difficult. To ensure that divergent insertions are identified correctly and exact TE annotations within the repeat database do not artificially influence TEnest results, sample sequences were run with TEnest using permutated repeat databases. The permutated repeat databases were used to analyze the same eight maize GenBank-submitted sequences shown in the previous TEnest verification section. For each of the eight contigs, each TE insertion found in the sequence was removed from the TE family prior to the phylogenetic consensus calculation, thus removing its influence from the database. In addition, the entire branch in the phylogenetic tree of the TE family containing this element was removed from the consensus calculation to prevent biologically similar TE insertions from influencing the TEnest results. As before, TEnest found every annotation identified by the original annotations and so did not show differences in identification between the original and permutated repeat database (Table I, with P notations).
Construction and Analysis of Simulated Maize Genomic Sequences
Simulated maize genome sequences were constructed and analyzed with TEnest to determine correctness of repeat annotations. The percentage of repeat sequence and proportion of repeat class were randomly chosen from frequencies observed from 165 maize sequence contigs. Based on observed individual TE frequencies, repeat families were randomly picked for each repeat class and family and randomly assigned an insertion time estimated from observed age of insertion frequencies. A list of TE insertions and associated insertion time was produced. An original DNA sequence with equal base proportions was made and each TE sequence was inserted into the DNA sequence. The insertion location was randomly determined over the entire length of the DNA sequence, continually updated by previous TE insertions. With this process, single- and nested-repeat insertions were obtained. At each time point, random sequence mutations were made on the whole sequence length. Mutations were made based on the rate of mutation (1.3 × 10−8; Ma and Bennetzen, 2004), with a probability of an insertion, a deletion, or a substitution. Insertion and deletion mutations were either single base or a sequence length based on an insertion/deletion length frequency. At each time point, there was a chance of a LTR retrotransposon insertion reverting to a solo LTR; this probability depended on the amount of insertions within that retrotransposon and the divergence of the two LTR copies. For LTR retrotransposons, the LTR sequence was base pair mutated before insertion and used as both the left and right LTR. Thus, the retrotransposon contained exact LTR sequence copies different from other LTRs of the same TE family; random mutations provided differences between the LTR pair to accurately model divergence for insertion age calculations. Completed simulation sequences were run with TEnest and the output was compared with expected results. Out of 100 simulation sequences that were analyzed, six incorrect annotations were observed, 0.35% of total TE insertions. Each incorrect annotation was repaired in a second TEnest run with parameter alterations allowing for more overlap between reconstructed sections.
A Case Study: Repeat Analysis of the Maize Genome with TEnest
TEnest presents a unique ability to observe TE family distributions across plant genomes in relation to age of insertion, sequence similarity, and sequence retention. At the time of submission, the GenBank sequence database contained 165 ordered and oriented genomic maize sequence contigs greater than 100 kb. This included 56 finished contigs and 109 gapped-sequence submissions with sections presented in correct order and orientation relative to the genome sequence. In total, these BACs equal 29.3 Mb or about 1% of the maize genome. This dataset contains BAC clones sequenced with intentions of gene discovery, as well as BACs randomly selected to survey the entire maize genome. Therefore, this set of sequence contigs may be slightly higher in gene content and lower in repetitive amounts than will be observed across the entire maize genome. These 165 contigs were evaluated with TEnest to provide a broad picture of TE clusters in the maize genome.
Distribution of TE Insertions Is Unequal across Families
A summary of results of TE identification by TEnest are displayed in Table II, with columns for each TE general class: LTR retrotransposons, Ty1/Copia, Ty3/Gypsy, other; solo LTRs, Ty1/Copia, Ty3/Gypsy, other; DNA transposons; and unknown. The number of copies and percent of total analyzed sequence for each class are shown. The sequence percentage for each type shows the total length of TEs divided by the total length of the contig; the last column shows the entire repetitive percentage of the contig. The average TE content for the 165 contigs analyzed is 66.95%, similar to the 65.97% reported by Haberer et al. (2005) in their analysis of 100 randomly selected BACs. The full table, showing all TE insertions annotated by TEnest in each of the 165 contigs is found in Supplemental Table S1.
Table II.
TEnest annotations identified across 29.3 Mb of maize sequence
Sequence length and count of all maize TE annotations found across the 165 submitted sequence contigs by TE class (or superfamily) and TEnest classification.
| TE Categorization (Total [Copia, Gypsy, Other]) | TE Amount Identifieda | TE Length Identified | Percentage of Sequence Length |
|---|---|---|---|
| kb | |||
| LTR retrotransposon | |||
| Whole (total [c, g, o]) | 1,186 [536, 424, 226] | 11,095.91 [4,749.53, 4,471.18, 1,872.27] | 37.87 [16.21, 15.26, 6.39] |
| Partial (total [c, g, o]) | 2,278 [804, 686, 788] | 6,240.90 [1,760.93, 2,566.68, 1,913.29] | 21.30 [6.01, 8.76, 6.53] |
| Total LTR retro (total [c, g, o]) | 17,333.88 [6,510.46, 7,034.93, 3,788.49] | 59.16 [22.22, 24.01, 12.93] | |
| Solo LTR | |||
| Whole (total [c, g, o]) | 80 [33, 16, 31] | 146.50 [49.81, 14.65, 82.04] | 0.50 [0.17, 0.05, 0.28] |
| Partial (total [c, g, o]) | 706 [166, 111, 429] | 272.49 [82.04, 26.37, 164.08] | 0.93 [0.28, 0.09, 0.56] |
| Total LTR solo (total [c, g, o]) | 418.99 [128.92, 41.02, 246.12] | 1.43 [0.44, 0.14, 0.84] | |
| DNA transposon | |||
| Whole | 137 | 111.34 | 0.38 |
| Partial | 295 | 228.54 | 0.78 |
| Total DNA transposon | 336.95 | 1.13 | |
| Unknown | |||
| Whole | 433 | 665.11 | 2.27 |
| Partial | 1,478 | 861.42 | 2.94 |
| Total unknown | 1,526.53 | 5.21 | |
| Total TEs | |||
| 29.3 Mb total sequence | 19,616.35 (19.62 Mb) | 66.95 |
Partial TEs identified may contain more than one member from a single original TE insertion. Total TE counts, the sum of whole and partial TE amounts, are not shown due to the possible inflated value seen from unreconstructed partial TE fragments.
LTR retrotransposons make up 60.59% of the total maize sequence analyzed. This is divided into 37.87% whole LTR retrotransposons, 22.23% partial or fragmented LTR retrotransposons, and 0.50% solo LTR sequences. Whole LTR retrotransposons are defined as containing both flanking LTR sequences and >90% of the internal region based on the TE consensus sequence. Partial LTR retrotransposons are incomplete insertions resulting from deletions or transpositions or gaps in the sequence assembly. Partial LTR retrotransposon annotations include any amount of the internal regions of TE sequences that may be reconstructed sections from later insertions and may also include LTR sequences. In terms of sequence length of the identified TEs, partial LTR retrotransposons cover 6.2 Mb of the BAC sequence in this analysis; whole LTR retrotransposons cover 11.1 Mb; this ratio of partial to whole is 1:1.8., showing that, even in fragmented TE remnants, sequence structure is moderately reconstructable.
Solo LTRs are defined as annotations >50% of the LTR length and not connected to internal regions of the TE. Identified solo LTR regions that equaled <50% of the solo LTR length once reconstructed were not analyzed here and were classified as fragmented LTR retrotransposons. Solo LTR-to-whole LTR ratio varies by TE family (Table III), ranging from Gyma, with 2.3 whole TEs per solo LTR, to Zeon, with 121 whole TEs per solo. The majority of solo-to-whole LTR retrotransposon ratios lie between one solo LTR to seven to 15 whole elements, which includes members of both Copia and Gypsy superfamilies. A much higher recombination frequency (one solo LTR to two to three whole elements) is seen with three retrotransposons, Gyma, Ruda, and Danelle, again in both Copia and Gypsy superfamilies. Huck and Zeon show very infrequent solo LTR formation, with one solo to 70 or 121 whole elements. In addition, many TEs have no corresponding solo and are not shown in Table III. Sequence structure may play a role in solo LTR formation; two families of retrotransposons with similar sequence have almost exact unequal recombination rates: Ji and Opie, with 64.2% sequence identity, have 14.9 and 15.1 solo-to-whole ratios; Danelle and Gyma, with 55.7% sequence identity, have 3.3 and 2.3 solo-to-whole ratios. These are considered closely related for between-family comparisons; within TE families, members may have <60% identity to each other over their entire lengths. Solo LTRs are found on average one per 156 kb across the analyzed sequences; however, sequence AC148093, located in a near-centromeric region of chromosome 4, contains one solo LTR per 16.3 kb. This region contains only one solo LTR from the high-rate solo LTR families (zero Danelle, zero Gyma, one Ruda), rather than the high amount of solo LTRs from a variety of lower rate solo-forming LTR retrotransposon families, and suggests a high level of unequal recombination in this region.
Table III.
Rates of solo LTR formation
Solo LTRs of LTR retrotransposons found in the 165 maize contigs in this analysis. Solo LTR formation rates are inconsistent across types of retrotransposons.
| TE Type | TE Class | Whole TE Amount | Solo Amount | Ratioa | LTR Size | TE Size |
|---|---|---|---|---|---|---|
| bp | bp | |||||
| Gyma | Gypsy | 42 | 18 | 2.3 | 4,198 | 12,067 |
| Ruda | Copia | 20 | 7 | 2.9 | 1,409 | 6,384 |
| Danelle | Gypsy | 53 | 16 | 3.3 | 4,602 | 15,397 |
| Klaus | Unknown | 22 | 6 | 3.7 | 1,075 | 7,040 |
| Prem | Copia | 80 | 11 | 7.3 | 3,246 | 6,039 |
| Milt | Gypsy | 32 | 4 | 8.0 | 565 | 9,189 |
| Xilon | Gypsy | 54 | 4 | 13.5 | 2,950 | 12,973 |
| Ji | Copia | 268 | 18 | 14.9 | 1,306 | 9,030 |
| Opie | Copia | 226 | 15 | 15.1 | 1,251 | 8,906 |
| Huck | Gypsy | 212 | 3 | 70.7 | 1,713 | 14,283 |
| Zeon | Gypsy | 121 | 1 | 121.0 | 698 | 7,412 |
Ratio of full-length LTR retrotransposon to solo LTR is determined by whole TE amount divided by solo amount.
TEnest calculates time since insertion (Mya) for each whole LTR retrotransposon using sequence identity from the paired LTRs. Fourteen-hundred fifty-seven LTR pairs were identified in this set of 165 maize contigs. Fifty percent of all LTR retrotransposon insertions occurred <0.875 Mya, and 75% of all LTR retrotransposon insertions are <1.5 Mya (Fig. 3). As shown in Figure 3, the age of insertion across the four most abundant whole LTR retrotransposons; Ji 268 copies, Opie 226 copies, Huck 212 copies, Zeon 121 copies, remains constant following this distribution of insertion times. Less represented LTR retrotransposon insertions may be younger or older than this general distribution; however, too few copies are present in the analyzed dataset to accurately calculate as individual families.
Evolution of TEs across the Maize Genome Shows Clusters of Insertion Ages
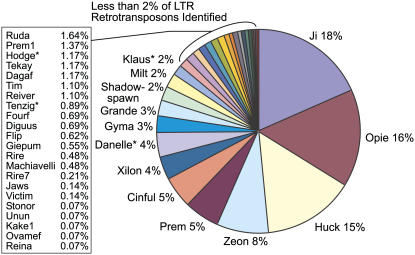
Forty-seven families of LTR retrotransposons were identified, nine Ty1/Copia, 11 Ty3/Gypsy, and 27 others. The three most abundant families Ji, Opie, Huck, each have 15% to 18% of the total amount of LTR retrotransposons identified, comprising more than one-half of the LTR retrotransposons found. As illustrated in Figure 4, there is a considerable drop in TE abundance in the rest of the identified LTR retrotransposon families ranging from 8% to <1%.
Figure 4.
Maize LTR retrotransposon quantification. Quantity of LTR retrotransposons identified across the 165 analyzed maize sequence contigs. Three retrotransposon types, Ji, Opie, and Huck, make up almost 50% (18%, 16%, 15%) of all LTR retrotransposons identified. The three most abundant types represent both Copia and Gypsy classes and exhibit rapid proliferation in phylogenetic analysis. LTR retrotransposons identified in this study are noted with an asterisk (*).
Analysis of the sequence relationship between individual elements within each TE family may give insight into the evolution and expansion of TEs across the genome and may possibly explain the unequal amounts of retrotransposon families. Using the output table of TEnest, a multiple alignment of every element insertion for each family of LTR retrotransposons was made with ClustalW (Thompson et al., 1994). A neighbor-joining phylogenetic tree was constructed with PHYLIP (Felsenstein, 2005); each member was overlaid with the calculated age since insertion (Mya). LTR pairs incomplete due to sequence gaps or other large deletions were not included in this analysis. The retrotransposon family Ji (Fig. 5A) shows distinct clustering of insertion ages in each clade of the phylogenetic tree. In addition, branching order of the tree follows the pattern of insertion age: Closely related clades have similar insertion ages. Other high-copy LTR retrotransposons (Huck, Opie) also follow this phylogenetic pattern (Supplemental Figs. S5 and S6).
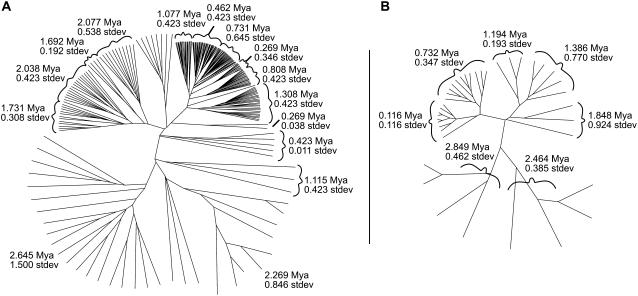
Figure 5.
Phylogenetic analysis of the maize Ji and Grande retrotransposons. Full-length maize LTR retrotransposon insertions for each family were identified with TEnest, excised, aligned with ClustalW, and a neighbor-joining tree was made with PHYLIP. Time since insertion in Mya calculated with TEnest was overlaid for each element. A, The Ji LTR retrotransposon shows that clades of the phylogenetic tree contain elements with similar times insertion ages, shown with sds. We hypothesize this is caused by a number of rapid LTR retrotransposon proliferations of the Ji element. B, Low copy number Grande LTR retrotransposon also shows the proliferation pattern, although with fewer TE insertions this analysis is less certain.
A tree with clustered insertion ages does not follow the expected phylogenetic result of continuously replicating LTR retrotransposons, with each clade representing a distinct family subset replicating individually and concurrently. Here, one expects a tree with a similar range of insertion ages on each phylogenetic branch. Instead, LTR proliferation is observed, where, at specific times throughout the genome evolution, the Ji retrotransposon family has undergone cycles of rapid expansion. This suggests multiple instances of extreme proliferation events by one or a few related members propagating many similar insertions in a small time frame. The Grande retrotransposon family (Fig. 5B) also seems to follow the proposed proliferation process, although with only 39 members in this analysis the clusters of insertion ages are less obvious. This initial evidence from low-copy families excludes the proliferation process as the only explanation for the extremely high copy number of Ji, Opie, and Huck retrotransposons.
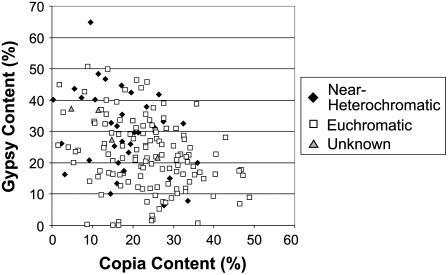
Relative amounts of LTR retrotransposon superfamilies Ty1/Copia and Ty3/Gypsy are similar, 22.66% and 24.16%, respectively, whereas the other class is much less abundant with 13.77% of the sequence content. However, the similar amount of Copia and Gypsy elements does not mean they are found equally across the genome; instead, they correspond to genome locations. In general, Copia and Gypsy sequence quantities per location are inversely proportional (Fig. 6). With the maize WebFPC July 19, 2005 release (Coe et al., 2002; Wei et al., 2007), general designations of near-heterochromatic or euchromatic were given to each BAC in the analysis based on proximity to centromeric and telomeric marker locations in the WebFPC BAC assembly. In this analysis, the term near heterochromatic simply designates a BAC within the FPC contig most near the centromere or telomere; in reality, few of these are truly heterochromatic BACs. No significant differences are seen between Gypsy and Copia superfamilies across near-heterochromatic or euchromatic chromosomal locations. Near-heterochromatic regions do tend to have a higher concentration of Gypsy retrotransposons, but a large percentage of both Copia and Gypsy retrotransposons is found in euchromatic and near-heterochromatic locations of the genome. In addition, chromosome location does not have an impact on total TE content found in a BAC. In general, low-repeat areas are found in euchromatic regions, but highly repetitive BACs can be found throughout the genome. These results show repeat oceans are as important as centromeric regions in attracting or retaining TE insertions and presumably repeat oceans may mimic structures associated with heterochromatin and attract Gypsy elements in a similar fashion. Alternatively, this can suggest a local proliferation process, where retrotransposons replicate to nearby locations inflate quantities of separate types of TEs for specific regions.
Figure 6.
Maize LTR retrotransposon class by BAC. Sequence content of LTR retrotransposon classes of 165 maize sequence contigs. In general, contigs located in near-heterochromatic regions of the genome have a higher Gypsy-to-Copia ratio. In contrast, euchromatic sequences show just a slight preference for Copia retrotransposons. However, high concentrations of Gypsy retrotransposons are found throughout the genome, suggesting large oceans of retrotransposons are as important as heterochromatic regions for attracting Gypsy-type elements. Near-heterochromatic BACs are defined as residing at the contig ends and centromere locations of the maize WebFPC chromosomes (Coe et al., 2002).
DISCUSSION
TEnest: An Efficient Algorithm for Nested TE Annotation
TEnest was initially designed for annotation of maize BAC contigs (e.g. EF517601, EF517601); the repeat database has since been expanded to include rice, wheat, and barley (Hordeum vulgare), and potentially could include other sequenced grasses such as sorghum (Sorghum bicolor) and Brachypodium (Brachypodium distachyon). TEnest can be extended further for use in a variety of organisms; however, the main advantage of TEnest over other repeat identification software is the ability to annotate nested TE insertions, primarily seen in the densely repetitive grass genomes. To evaluate other organisms, users can create a custom repeat database. This custom database can be used with the downloadable version of TEnest or uploaded onto the online version. In addition, users can submit and suggest edits of TE database entries; both of these systems are in place to keep TEnest up to date as more sequence is produced and more TEs are identified.
There are several important steps of the TEnest system that give it the ability to accurately annotate nested TEs and make it a viable resource for genome repeat analysis. The two-alignment method, first using a quick BLAST search to locate general regions of interest, then using a pairwise local alignment to accurately identify the complete sequence alignment, greatly increases speed and precision when using sequences of similar identity, such as repeat databases. The power set reconstruction method builds TE segments separated by nesting, using coordinates based on the TE insertion, giving TEnest the ability to recognize and correctly resolve TE families nested within themselves (a whole Ji retrotransposon nested within another Ji) or to identify a duplicated region within a TE (an extra portion of a Ji found within a Ji element). TEnest pairs the left and right LTRs of retrotransposons based on their divergence, identifying the TE family and the sequence ends of the insertion and allowing TEnest to quickly build the internal region with more relaxed criteria, thus obtaining the complete annotation.
Joining separated sections, both the power set reconstruction method and the LTR pairing method introduce a possibility of join discrepancies, where two or more joined regions disagree (Fig. 1B). If the reconstruction or pairing processes suggest combinations that could not have occurred by TE nesting, but would require a local inversion or translocation, TEnest uses the discrepancy process to separate the most likely incorrect join.
TEnest provides the user with three output formats; an annotation table of TE insertions with insertion ages of LTR retrotransposons, a repeat masked sequence file, and a vector format graphical display of the chronology of TE insertions. Use of these output files can assist with identification of genes and other functionally important locations and can also answer questions regarding the sequence makeup of the genome. When used to analyze a BAC or a single sequence region, TEnest gives information about sequence structure, content, rearrangement, and evolutionary dynamics of the area. Expanding this analysis to multiple regions across the genome, such as the analysis of the currently finished maize BAC contigs within this article, gives a more in-depth example of the capabilities of TEnest. With more sequence information, comparisons between TE families and classes and evolutionary analysis of single TE families can begin to answer questions about TE evolution, replication, and their effect on the genome. The included cluster submission script that splits input sequence and runs TEnest versions on each node of a cluster gives users the ability to evaluate long sequences in relatively short time frames.
Three Verification Tests of TEnest Ensure Accurate Results
Three analyses were used to validate the output from TEnest. The first verification examined TEnest outputs to curated submitted maize and rice contigs. The important information from this analysis is that TEnest was able to identify all known TE insertions. TEnest did identify extra insertions; however, the goals of the original curators were varied and may have intentionally not included all TEs. In addition, at the time of original annotation, community repeat databases were less complete.
TEnest uses a repeat database to identify TE insertions; this repeat database is made of consensus or representative sequences. This process could allow easy identification of similar TEs within the family, whereas not accurately identifying distantly related TEs within the family. In the permutation verification, the TE insertion and similar TEs from the sequence contig were removed from database construction to show that TEnest correctly identified the distantly related TEs within a family. In each case, TEnest was able to correctly identify the permutated TEs showing that, regardless of individual TEs used in the database construction, TEnest gives unbiased repeat annotation. These results are possible because of the unique processes of TEnest, specifically initial identification of paired LTRs and relaxation of internal region alignment parameters.
The final validation highlights a further resource of the TEnest outputs. Hypothetical ancestral sequences, prior to TE insertions, can be constructed by removing whole TE annotations, which we believe are more accurate representations than simple masking of all repeat matching sequences. These ancestral predictions can be used for comparative genome analysis to give a cleaner assessment of the shared sequence regions between genomes or sequence regions. Time-point sequences can be made by removing TEs inserted after a certain age.
Phylogeny of LTR Retrotransposons across the Maize Genome
TEs cause sequence rearrangement and recombination by insertion and translocation of their own and other sequences throughout the genome. Additionally, by their seemingly unbridled expansion of genome size, LTR retrotransposons are significant drivers of sequence evolution. TEnest was designed to quickly and accurately analyze completely sequenced genomes and to explore how TEs affect whole-genome evolution. As with smaller sequence scale analyses, TEnest can provide TE insertion locations, distributions, and insertion preferences to show the current structure of the whole genome. But, on a larger scale, it can give the whole view of each TE family evolution, the sequence divergence history of each type from a common ancestor, along with its age since insertion and its genome location. Combination of TE insertion age, sequence relationships, and location in the genome can be used to investigate fundamental questions about TEs, such as their rates of proliferation across the genome, their paths of replication over time, and ultimately their effects on genome evolution.
Based on data presented here, different TE families experience unequal recombination that results in solo LTRs at different frequencies. The rate of solo LTR formation does not seem to be influenced by length of either the LTR or the whole retrotransposon. Only those LTR retrotransposons with at least one observed solo LTR were included in this analysis; many retrotransposons had no solo LTRs, most likely due to the limited amount of maize genome sequence in this study, as well as the low rate of recombination within the retrotransposon family. Gyma and Danelle and Ji and Opie both share similar rates of solo formation, as well as relatively similar sequence identities, and suggest that TE structure or sequence is an important factor for unequal recombination.
Phylogenetic analysis of LTR retrotransposon families gives similar age of insertion clustered in clades of the tree. We hypothesize that this is caused by proliferation of LTR retrotransposons, where at specific time points in evolution a single or related group of elements has rapidly expanded across the genome. These rapid TE expansions could correspond to times of relaxed mutation standards, such as genome duplication events or environmental stress conditions where mutations caused by TE insertions are less detrimental to the organism. Alternatively, these TE proliferations could be caused by advantageous mutations in the TE sequence, allowing a TE copy to replicate across the genome. Similar proliferation-style phylogenetic trees are observed across many LTR retrotransposon families and therefore the process is not TE specific and cannot explain differences in TE amounts. The causes behind the abundance of certain TE families are due to selective processes not yet understood.
Two other hypotheses for LTR retrotransposon replication do not explain the observed trees. Continual copying of TEs in a family until mutation prevents replication of an individual will give a tree with TEs from any clades of the family the ability to replicate. In this scenario, the phylogenetic tree has clades containing a variety of insertion ages. Alternatively, genome duplication could immediately double the amount of TEs within the genome. A tree following a genome duplication event will contain 2 times as many TEs with every clade, each with the same insertion age, but each clade still contains a variety of insertion ages and those TEs still able to replicate will continue to increase the age ranges. However, genome duplication could play a role in the proliferation hypothesis by allowing proliferation to increase with a decreased chance of harming the genome.
MATERIALS AND METHODS
Construction of Consensus Repeat Databases for TEnest
Maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum), and barley (Hordeum vulgare) repeat databases were constructed from the following sources: GIRI RepBase (Jurka et al., 2005); The Institute for Genomic Research (maize.tigr.org); the Messing lab (Messing et al., 2004); the Wessler lab (daffodil.plantbio.uga.edu/wesslerlab); ISU Maize Genome Assembly (Emrich et al., 2004); and the Triticeae Repeat Sequence Database (http://wheat.pw.usda.gov/ITMI/Repeats/index.shtml; Wicker et al., 2002). These databases each consist of multiple FASTA file formatted repeat sequences. For each organism, each multiple FASTA file was combined and exact duplicate sequence entries were removed. Using a cutoff E value of 10−20, each entry in the combined database was aligned with WU-BLAST blastn. Sequences that passed the cutoff value were removed from the combined database and multiply aligned with ClustalW; a consensus sequence from the multiple alignment was made and added into the combined database. Consensus sequence bases are calculated as >60% of each location in the multiple alignment; any base <60% gives an N. Sequences completely encompassed within the consensus sequence were removed from the database; those still containing unique regions were trimmed, the aligning part removed, and the unique sections added back to the database. This process of clustering and making consensus sequences was repeated while raising the cutoff value until E value reached 10−50. Most trimmed unique repeat sequences aligned to longer TEs clustered in this process were removed from the database; any remaining were classified as potential repeats with single representatives.
Each final set of clustered repeat entries was aligned with ClustalW. Neighbor-joining trees were made using the PHYLIP package. The resulting phylogenetic trees were examined for well-defined separations into subgroups, such as a tree with only two distant clades. If present, these clustered tree sections were split into subgroups of the original repeat set. Consensus sequences were made from each repeat set or each subgroup within a set. Many repeat groups contained high diversity between elements; if >10% of the consensus sequence was Ns or if the sequence had stretches of 90% Ns for more than 100 bases, a consensus sequence was not used. Instead, a representative repeat entry was selected for use in the repeat database from a central branch off the phylogenetic analysis.
Consensus sequences were checked against the GenBank maize database (Benson et al., 2006) and the combined repeat databases; those entries previously characterized as TE families with at least partial sequences were updated with the original nomenclature. Each consensus repeat terminus was examined for LTR sequences; if found, these LTRs were added to a separate LTR database.
User Customizable Parameters of TEnest
Customization of TEnest runs is accomplished using the many available parameter settings. All of these parameter settings are explained in further detail in the TEnest README file found with the TEnest Web service or bundled with a downloaded version. Similar parameter settings are available for each TEnest identification process; LTRs, internal retrotransposon regions, fragmented, and non-LTR retrotransposon regions. Users can alter the number of pairwise alignments reported (default 7), the gap open penalty (default 30 for LTRs, 75 for others), the gap extension penalty (default 15 for LTRs, 75 for others), and the pairwise alignment E-value cutoff score (default 10−20). The amount of base pairs to allow as overlapping when joining sections is also customizable, for pairwise alignments (default 25), or when reconstructing separated sections in the power set process (default 30). The smallest returned reconstructed LTR (default 25) can be raised to limit unnecessary annotations; the maximum distance between power set reconstructed sections can also be altered (default 100 kb). The LTR pairing process can be customized with gap open (default 12) and gap extension (default 4) penalties, and amount of LTR pairs to consider (default 0.1).
TE makeup across organisms is different; some TEnest settings have proved more useful when attempting annotation on other species. Rice has a high number of small MITE insertions, TEnest has better success identifying these elements when ignoring long-spanning TEs (decreasing the power set reconstruction maximum) and allowing for smaller TE alignments (decreasing the E-value cutoff for pairwise alignments and decreasing the size of reported sections). Some success has been seen with TEnest on nonplant species. In Drosophila melanogaster, the LTRs of LTR retrotransposons are very small in relation to the grass species. We have achieved TE annotations with TEnest on D. melanogaster sequences when lowering the LTR overlap lengths, pairwise alignment cutoffs, and LTR size cutoff.
Required Software for Using TEnest
The TEnest software package is available for use on PlantGDB under the tools section (http://www.plantgdb.org/prj/TE_nest/TE_nest.html), the source code, along with maize, rice, wheat, and barley repeat databases, is available from http://wiselab.org. To install a local version of TEnest, Perl (http://www.perl.org), WU-BLAST version 2.0 (http://blast.wustl.edu), and FASTA2 (ftp://ftp.virginia.edu/pub/fasta) are required. To display TEnest annotations svg_ltr uses the scalable vector graphics format (http://www.w3.org/Graphics/SVG), displayable in Mozilla Firefox (www.mozilla.com/firefox) version 2 or later.
GenBank sequence submissions submitted with this manuscript: LTR retrotransposons Danelle, EF562447 and Stella, EF621725.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. TEnest coordinate output of barley contig AH014393.
Supplemental Figure S2. TEnest coordinate output of maize contig AC145481.
Supplemental Figure S3. TEnest coordinate output of rice contig AP004818.
Supplemental Figure S4. TEnest coordinate output of wheat contig DQ537335.
Supplemental Figure S5. Huck LTR retrotransposon phylogenetic analysis.
Supplemental Figure S6. Opie LTR retrotransposon phylogenetic analysis.
Supplemental Table S1. Summary of TEs identified across 165 maize BAC contigs.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Peterson and Dr. Dan Voytas and for critical review of the manuscript. This article is a joint contribution of The Iowa Agriculture and Home Economics Experiment Station and the Corn Insects and Crop Genetics Research Unit, U.S. Department of Agriculture-Agricultural Research Service. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
This work was supported by the U.S. Department of Agriculture-National Research Initiative (grant no. 2002–35301–12064).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Roger P. Wise (rpwise@iastate.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bao Z, Eddy SR (2002) Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res 12 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2006) GenBank. Nucleic Acids Res 34 D16–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Corces VG (1989) Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol 43 403–434 [DOI] [PubMed] [Google Scholar]
- Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A (2005) Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17 343–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell KS, Langridge P, Powell W (2004) Comparative sequence analysis of the region harboring the hardness locus in barley and its collinear region in rice. Plant Physiol 136 3177–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E, Cone K, McMullen M, Chen SS, Davis G, Gardiner J, Liscum E, Polacco M, Paterson A, Sanchez-Villeda H, et al (2002) Access to the maize genome: an integrated physical and genetic map. Plant Physiol 128 9–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Schlueter SD, Brendel V (2004) PlantGDB, plant genome database and analysis tools. Nucleic Acids Res 32 D354–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Myers EW (2005) PILER: identification and classification of genomic repeats. Bioinformatics (Suppl 1) 21 i152–i158 [DOI] [PubMed] [Google Scholar]
- Emrich SJ, Aluru S, Fu Y, Wen TJ, Narayanan M, Guo L, Ashlock DA, Schnable PS (2004) A strategy for assembling the maize (Zea mays L.) genome. Bioinformatics 20 140–147 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) Version 3.6. Department of Genome Sciences, University of Washington, Seattle
- Fu H, Dooner HK (2002) Intraspecific violation of genetic collinearity and its implications in maize. Proc Natl Acad Sci USA 99 9573–9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Zheng Z, Dooner HK (2002) Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc Natl Acad Sci USA 99 1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Salse J, Coleman-Derr D, Dupin A, Crossman C, Lazo GR, Huo N, Belcram H, Ravel C, Charmet G, et al (2006) Types and rates of sequence evolution at the high-molecular-weight glutenin locus in hexaploid wheat and its ancestral genomes. Genetics 174 1493–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Young S, Bharti AK, Gundlach H, Raymond C, Fuks G, Butler E, Wing RA, Rounsley S, Birren B, et al (2005) Structure and architecture of the maize genome. Plant Physiol 139 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Miller W (1991) A time-efficient, linear-space local similarity algorithm. Adv Appl Math 12 337–357 [Google Scholar]
- IRGSP (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110 462–467 [DOI] [PubMed] [Google Scholar]
- Kalyanaraman A, Aluru S (2005) Efficient algorithms and software for detection of full-length LTR retrotransposons. J Bioinform Comput Biol 4 197–216 [DOI] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al (2002) The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol 3 RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR (2000) Transposable elements and host genome evolution. Trends Ecol Evol 15 95–99 [DOI] [PubMed] [Google Scholar]
- Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16 111–120 [DOI] [PubMed] [Google Scholar]
- Lal SK, Giroux MJ, Brendel V, Vallejos CE, Hannah LC (2003) The maize genome contains a helitron insertion. Plant Cell 15 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al (2001) Initial sequencing and analysis of the human genome. Nature 409 860–921 [DOI] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EM, McDonald JF (2003) LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19 362–367 [DOI] [PubMed] [Google Scholar]
- Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, Wei F, Fuks G, Soderlund CA, Mayer KF, et al (2004) Sequence composition and genome organization of maize. Proc Natl Acad Sci USA 101 14349–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11 1660–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano H, Kunii M, Azuma T, Kishima Y, Sano Y (2002) Characterization of the repetitive sequences in a 200-kb region around the rice waxy locus: diversity of transposable elements and presence of veiled repetitive sequences. Genes Genet Syst 77 69–79 [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA (2005) De novo identification of repeat families in large genomes. Bioinformatics (Suppl 1) 21 i351–i358 [DOI] [PubMed] [Google Scholar]
- Quesneville H, Bergman CM, Andrieu O, Autard D, Nouaud D, Ashburner M, Anxolabehere D (2005) Combined evidence annotation of transposable elements in genome sequences. PLoS Comput Biol 1 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Bennetzen JL (2006) The maize genome as a model for efficient sequence analysis of large plant genomes. Curr Opin Plant Biol 9 149–156 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20 43–45 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274 765–768 [DOI] [PubMed] [Google Scholar]
- Song R, Llaca V, Linton E, Messing J (2001) Sequence, regulation, and evolution of the maize 22-kD alpha zein gene family. Genome Res 11 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppes P (1972) Axiomatic Set Theory. Dover, New York, pp 46–49
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov AP, SanMiguel PJ, Nakajima Y, Gorenstein NM, Bennetzen JL, Avramova Z (1999) Collinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc Natl Acad Sci USA 96 7409–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S, et al (2007) Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Matthews DE, Keller B (2002) TREP: a database for Triticeae repetitive elements. Trends Plant Sci 7 561–562 [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leory P, Morgante M, Panaud O, et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8 973–982 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.