Abstract
Transposon insertional mutagenesis is an effective alternative to T-DNA mutagenesis when transformation through tissue culture is inefficient as is the case for many crop species. When used as activation tags, transposons can be exploited to generate novel gain-of-function phenotypes without transformation and are of particular value in the study of polyploid plants where gene knockouts will not have phenotypes. We have developed an in cis-activation-tagging Ac-Ds transposon system in which a T-DNA vector carries a Dissociation (Ds) element containing 4× cauliflower mosaic virus enhancers along with the Activator (Ac) transposase gene. Stable Ds insertions were selected using green fluorescent protein and red fluorescent protein genes driven by promoters that are functional in maize (Zea mays) and rice (Oryza sativa). The system has been tested in rice, where 638 stable Ds insertions were selected from an initial set of 26 primary transformants. By analysis of 311 flanking sequences mapped to the rice genome, we could demonstrate the wide distribution of the elements over the rice chromosomes. Enhanced expression of rice genes adjacent to Ds insertions was detected in the insertion lines using semiquantitative reverse transcription-PCR method. The in cis-two-element vector system requires minimal number of primary transformants and eliminates the need for crossing, while the use of fluorescent markers instead of antibiotic or herbicide resistance increases the applicability to other plants and eliminates problems with escapes. Because Ac-Ds has been shown to transpose widely in the plant kingdom, the activation vector system developed in this study should be of utility more generally to other monocots.
Genetic mutants have always played a central role as tools for functional analysis of plant genes. Many plant genes have been isolated by the strategy of insertional mutagens. In the model plants of Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), large-scale T-DNA and transposon insertion libraries and flanking sequence tag (FST) databases have been generated, which serve the plant biologists worldwide for both forward and reverse genetics studies (Parinov et al., 1999; Tissier et al., 1999; Jeon et al., 2000; Ito et al., 2002; Kuromori et al., 2004; Ito et al., 2005). Transposon insertional mutagenesis is an effective alternative to T-DNA mutagenesis when transformation through tissue culture is inefficient, as is the case for many crop species. The strategy for transposon mutagenesis requires just a limited number of primary transformants, with insertions being generated through propagation.
On the other hand, although the classic insertional mutagenesis strategies play an important role in plant functional genomics studies, a limitation of such methods is the difficulty of identifying genes that are redundant in plant genomes and whose knockouts do not induce phenotypes. Activation tagging is an effective approach for overcoming this limitation. Activation tagging involves introduction of a T-DNA-containing regulatory sequence such as the enhancer of the cauliflower mosaic virus (CaMV) 35S promoter randomly into a plant genome to enhance expression of nearby genes, which potentially resulted in a gain-of-function phenotype (Kardailsky et al., 1999; Borevitz et al., 2000; Weigel et al., 2000; Jeong et al., 2002; Mathews et al., 2003; Jeong et al., 2006; Mori et al., 2007; Hsing et al., 2007).
As for the functional genomics of cereals and monocot plants, maize (Zea mays) transposable elements are useful tools for generating large collection of gene knockouts, because high-throughput T-DNA transformation is not a viable approach for those plant species. A number of maize genes have been isolated from transposon-tagged mutants. Several studies have shown that the maize Ac-Ds transposable elements function in barley (Hordeum vulgare), and mapped Ds launching pads were developed for the study of functional genomics (Koprek et al., 2000; Cooper et al., 2004; Singh et al., 2006; Zhao et al., 2006). Ayliffe et al. (2007) reported a novel Ac-Ds system in which a modified Ds element (UbiDs) carrying two maize ubiquitin 1 promoters (Christensen and Quail, 1996) was utilized for read-through transcription of adjacent flanking sequences.
We have been developing vectors using maize transposable elements for plant functional genomics. In rice, we previously developed an Ac-Ds insertional mutagenesis system in which a GFP fluorescence gene functioned as negative selection marker against the immobilized Ac and a Basta resistance marker worked for selection of transposed Ds elements (Kolesnik et al., 2004). In the development of an En/Spm-tagging system in rice, our transposition selection scheme was improved based on GFP and red fluorescent protein (RFP) double fluorescence markers (Kumar et al., 2005). In this study, we developed an activation tagging Ac-Ds in cis-system in which the advantage of GFP and RFP double fluorescence markers was utilized. The system was shown to be effective for distributing a large number of activation-tagging Ds elements in the rice genome. As the Ac-Ds has been shown to transpose widely in the plant kingdom, we anticipate the applicability of our activation vector system for studies in monocot plants, including other cereals.
RESULTS
Vector Construction and Production of Starter Lines
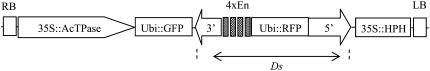
The T-DNA vector pSQ5 carrying a nonautonomous Ds element along with an immobilized Ac element in cis is shown in Figure 1. The immobilized Ac contains the Ac transposase gene under the control of the CaMV 35S promoter (Kolesnik et al., 2004). The Ds element has the 1,785-bp 5′ terminus and 222-bp 3′ terminus of the wild-type Ac element (Sundaresan et al., 1995). A tetramer of the transcriptional enhancer of CaMV 35S promoter and the Discosoma sp. RFP (DsRed) gene were cloned between the 5′ and 3′ Ds termini. The DsRed (or RFP) gene, encoding a RFP (Clontech; Baird et al., 2000), is driven by the maize ubiquitin 1 promoter (Christensen and Quail, 1996). The vector carries a hygromycin phosphotransferase gene for plant transformation selection. A synthetic GFP (sGFP) gene was cloned next to the immobilized Ac transposase source in the T-DNA. The GFP and RFP fluorescence markers make it possible to visually track the Ds element and the immobilized Ac element, respectively, in transgenic progeny.
Figure 1.
T-DNA of the activation-tagging Ac-Ds vector pSQ5. RB and LB, Right and left borders of the T-DNA; Ubi, maize ubiquitin 1 promoter; GFP, GFP gene as a negative selection marker; RFP, RFP (or DsRed) gene as selection marker for the Ds element; 4xEn, a tetramer of CaMV 35S enhancers; HPH, hygromycin phosphotransferase gene as plant transformation selection marker. The T-DNA is in the backbone of pCAMBIA-1300.
The pSQ5 Ac-Ds vector was introduced into rice sp. ‘Nipponbare’ via Agrobacterium tumefaciens-mediated rice transformation. Eighty fertile transformants that were double fluorescent (GFP+ and RFP+) were produced. In the T2 generation, 50 seeds of each transformant were germinated and assayed for GFP and RFP. For GFP segregation, T2 plants of 58 transformants showed a 3:1 ratio and therefore carried a single T-DNA locus in the rice genome. Eight transformants had multiple T-DNA integration loci based on the GFP segregation ratios (>3:1) of T2 plants. The T2 generation of 14 transformants did not have any plants showing GFP or RFP, possibly because of Ds transposition into the GFP gene or due to transgene silencing (see “Discussion”).
High Frequency of Germinal Transposition in T1 and T2 Transgenic Plants
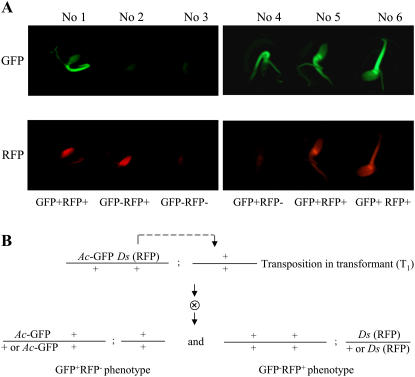
Because the activation-tagging Ds element and the T-DNA-based Ac transposase can be tracked by RFP and GFP, respectively, we determined the GFP and RFP phenotype of each T2 plant from 53 single-T-DNA-locus populations (Table I). An average of 48 T2 plants in each population were assayed for GFP and RFP fluorescence. The phenotype of individual plants was determined as shown in Figure 2A. Four different phenotypes were observed in these T2 populations. Because each T2 population was generated from a single primary transformant (T1) and in the T1 generation the Ds element carrying the RFP gene can transpose, the RFP gene and the T-DNA-anchored GFP gene may have different chromosomal locations and may segregate in the T2 plants (Fig. 2B). Therefore, it is likely that the GFP−RFP+ or GFP+RFP− plants in the T2 generation were derived from transposition events in the T1 generation. In GFP and RFP assays of T2 plants of 53 transformants (Table I), GFP−RFP+ plants were identified from T2 populations of 23 transformants (nos. 19–41). These GFP−RFP+ plants indicated that 43.4% (23/53) of the T1 transformants carried transposed Ds elements that were germinally transmitted to the T2 generation. Among the 53 T2 populations, 12 populations (nos. 42–53) had GFP+RFP− plants but no GFP−RFP+ plants, which suggested that a fraction of excised Ds elements did not reinsert in the rice genome (see “Discussion”).
Table I.
GFP and RFP assays of 53 T2 families
| No. | Transformant | No. of T2 Plants | GFP+RFP+ | GFP−RFP− | GFP+RFP− | GFP−RFP+ |
|---|---|---|---|---|---|---|
| 1 | A3 | 50 | 38 | 12 | 0 | 0 |
| 2 | A5 | 48 | 34 | 14 | 0 | 0 |
| 3 | A14 | 50 | 39 | 11 | 0 | 0 |
| 4 | A15 | 47 | 36 | 11 | 0 | 0 |
| 5 | A28 | 57 | 42 | 15 | 0 | 0 |
| 6 | A30 | 50 | 35 | 15 | 0 | 0 |
| 7 | A39 | 50 | 33 | 17 | 0 | 0 |
| 8 | A51 | 50 | 37 | 13 | 0 | 0 |
| 9 | A52 | 49 | 34 | 15 | 0 | 0 |
| 10 | A72 | 45 | 25 | 20 | 0 | 0 |
| 11 | B4 | 49 | 36 | 13 | 0 | 0 |
| 12 | B16 | 48 | 34 | 14 | 0 | 0 |
| 13 | B30 | 49 | 33 | 16 | 0 | 0 |
| 14 | B46 | 42 | 28 | 14 | 0 | 0 |
| 15 | B49 | 45 | 28 | 17 | 0 | 0 |
| 16 | B50 | 45 | 37 | 8 | 0 | 0 |
| 17 | B52 | 50 | 38 | 12 | 0 | 0 |
| 18 | B53 | 43 | 31 | 12 | 0 | 0 |
| 19 | A35 | 50 | 36 | 11 | 2 | 1 |
| 20 | A56 | 50 | 32 | 12 | 5 | 1 |
| 21 | A57 | 47 | 29 | 11 | 6 | 1 |
| 22 | A66 | 48 | 26 | 3 | 9 | 10 |
| 23 | A68 | 50 | 24 | 9 | 13 | 4 |
| 24 | A70 | 50 | 30 | 2 | 9 | 9 |
| 25 | B9 | 50 | 26 | 11 | 10 | 3 |
| 26 | B15 | 47 | 32 | 3 | 8 | 4 |
| 27 | B17 | 48 | 28 | 9 | 4 | 7 |
| 28 | B18 | 45 | 24 | 8 | 7 | 6 |
| 29 | B23 | 49 | 29 | 12 | 6 | 2 |
| 30 | B28 | 45 | 28 | 13 | 2 | 2 |
| 31 | B31 | 49 | 36 | 12 | 0 | 1 |
| 32 | B45 | 50 | 27 | 10 | 12 | 1 |
| 33 | B47 | 47 | 29 | 7 | 3 | 8 |
| 34 | B54 | 48 | 21 | 5 | 13 | 9 |
| 35 | B62 | 47 | 30 | 10 | 6 | 1 |
| 36 | B65 | 46 | 30 | 5 | 4 | 7 |
| 37 | A13 | 31 | 23 | 1 | 1 | 6 |
| 38 | A36 | 51 | 40 | 3 | 0 | 8 |
| 39 | A37 | 78 | 56 | 9 | 0 | 13 |
| 40 | A50 | 50 | 14 | 15 | 20 | 1 |
| 41 | B36 | 49 | 38 | 5 | 0 | 6 |
| 42 | A8 | 41 | 30 | 10 | 1 | 0 |
| 43 | A34 | 48 | 37 | 10 | 1 | 0 |
| 44 | A74 | 50 | 27 | 13 | 10 | 0 |
| 45 | A83 | 45 | 23 | 11 | 11 | 0 |
| 46 | B10 | 49 | 39 | 9 | 1 | 0 |
| 47 | B12 | 49 | 27 | 15 | 7 | 0 |
| 48 | B13 | 49 | 32 | 14 | 3 | 0 |
| 49 | B24 | 47 | 34 | 12 | 1 | 0 |
| 50 | B41 | 49 | 36 | 9 | 4 | 0 |
| 51 | B48 | 19 | 13 | 5 | 1 | 0 |
| 52 | B69 | 49 | 26 | 13 | 10 | 0 |
| 53 | A55 | 49 | 10 | 9 | 30 | 0 |
Figure 2.
GFP and RFP fluorescence assay of young seedlings (T2) derived from pSQ5 transgenic rice plants. A, GFP and RFP assays and the scheme to determine GFP and RFP phenotype of each T2 plant. B, A diagram showing a transposition event in the primary transformant (T1) and its germinal transmission to T2 population.
To detect transposition in T2 generation, we investigated T3 populations of 37 transformants. For the 37 transformants, 18 transformants (Table I; nos.1–18) did not show transposition in T2 generation, 14 transformants (Table I; nos. 19–21, 30–32, 35, 42–43, 46, 48–51) showed transposition, and T2 families of five transformants (nos. 54–58) were not analyzed. The T3 families of the 37 transformants were obtained by selfing heterozygous T2 plants. As GFP heterozygous T2 seedlings gave a lower level of GFP fluorescence than GFP homozygous seedlings (Kumar et al., 2005), we selected the heterozygous T2 plants based on their lower GFP intensity. In this way we were able to reduce the T2 homozygotes to 8.2% according to the results of GFP assays of T3 generation (data not shown). In each T3 family, 200 to 400 plants were assayed to identify GFP−RFP+ transposant plants.
For the numbers 1 to 18 transformants, at least one GFP−RFP+ plant was identified in T3 families of 12 transformants (Table II; nos. 1–3, 6, 8–12, 14–15, 18). We were able to identify transposition events in the T3 generation but not in their T2 families, because just 42 to 50 plants from each T2 family were analyzed (Table I; nos. 1–3, 6, 8–12, 14–15, 18). For the 14 transformants that showed transposition in the T2 generation, at least one germinal transposition event was identified from T3 families of 13 transformants (Table II; nos. 19–21, 30–32, 35, 42–43, 46, 48–51). In a total of 905 T3 families derived from 37 primary transformants, germinal transposition events were identified in 295 (32.6%) families (Table II). These transposition events in the T3 generation were derived from 30 (81.1%) of the transformants examined. Higher germinal transposition frequencies (20%–83.3%) were detected in the T3 families of 19 single-T-DNA-locus transformants (Table II), which were among one-third (32.7%) of all 58 single-T-DNA-locus transformants.
Table II.
GFP and RFP assays of 905 T3 families
| No. | Transformant | T3 Families Tested | T3 Families with Germinal Transpositiona |
|---|---|---|---|
| 1 | A3 | 18 | 4 (22.2%) |
| 2 | A5 | 20 | 1 (5%) |
| 3 | A14 | 20 | 12 (60%) |
| 4 | A15 | 30 | 0 |
| 5 | A28 | 10 | 0 |
| 6 | A30 | 10 | 3 (30%) |
| 7 | A39 | 20 | 0 |
| 8 | A51 | 20 | 3 (15%) |
| 9 | A52 | 20 | 1 (5%) |
| 10 | A72 | 6 | 5 (83.3%) |
| 11 | B4 | 30 | 10 (33.3%) |
| 12 | B16 | 10 | 2 (20%) |
| 13 | B30 | 20 | 0 |
| 14 | B46 | 10 | 5 (50%) |
| 15 | B49 | 9 | 4 (44.4%) |
| 16 | B50 | 10 | 0 |
| 17 | B52 | 10 | 0 |
| 18 | B53 | 20 | 3 (15%) |
| 19 | A35 | 55 | 27 (49.1%) |
| 20 | A56 | 74 | 49 (66.2%) |
| 21 | A57 | 99 | 36 (36.4%) |
| 30 | B28 | 30 | 5 (16.7%) |
| 31 | B31 | 30 | 13 (43.3%) |
| 32 | B45 | 72 | 42 (58.3%) |
| 35 | B62 | 10 | 5 (50%) |
| 42 | A8 | 34 | 23 (67.6%) |
| 43 | A34 | 20 | 2 (10%) |
| 46 | B10 | 20 | 3 (15%) |
| 48 | B13 | 20 | 3 (15%) |
| 49 | B24 | 20 | 0 |
| 50 | B41 | 24 | 13 (54.1%) |
| 51 | B48 | 10 | 3 (30%) |
| 54 | A29 | 21 | 1 (4.7%) |
| 55 | A62 | 10 | 5 (50%) |
| 56 | A73 | 30 | 10 (33.3%) |
| 57 | B3 | 23 | 1 (4.5%) |
| 58 | B76 | 10 | 1 (10%) |
| Total | 905 | 295 (32.6%) |
Germinal transposition was based on identification of ≥1 of GFP−RFP+ plant. In parentheses is percent of T3 families containing germinal transposition.
Generation of Activation-Tagging Ds Lines of Rice and Isolation of Ds FSTs
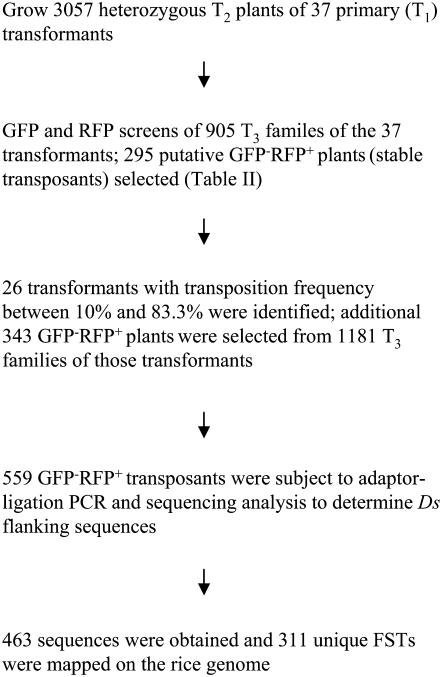
We grew a total of 3,057 T2 plants that were derived from 37 primary transformants. An average of 300 seeds (T3) were produced from each T2 plant. The GFP negative selection marker and the RFP positive selection marker were utilized to screen T3 families to obtain putative stable transposants (GFP−RFP+ plants). Using the scheme as shown in Figure 3, we initially screened 905 T3 families that were derived from the 37 transformants (Table II). In the next screening step, we did not continue with the T3 families derived from transformants for which transposition frequencies in the T3 generation were less than 10%. Instead, we focused on the 1,181 T3 families of the 26 transformants that generated transposition frequencies in the T3 of 10% to 83.3% (Table II). Among the 1,181 T3 families, 343 families were found to have at least one GFP−RFP+ plant per family. Taken together, a total of 2,086 T3 families were screened, and GFP−RFP+ transposants were selected from 638 (30.6%) of those families.
Figure 3.
Selection of stable Ds insertion lines and generation of Ds FSTs.
The putative stable transposants were subjected to adaptor-ligation PCR to determine Ds flanking sequence (Fig. 3). We analyzed 559 transposants that were derived from 26 primary transformants. A total of 463 sequences were obtained after adaptor-ligation PCR, sequencing, and filtration against the T-DNA sequence (to eliminate the Ds donor locus). In a Blast search against the Salk Institute Rice Functional Genomics database (http://signal.salk.edu/cgi-bin/RiceGE), 311 (67.2%) sequences were mapped on the rice chromosomes (Table III). The 152 remaining sequences were 73 (15.8%) redundant FSTs from siblings carrying the same insertions and 79 (17.1%) unmapped sequences due to insertions in rice repeat sequence regions. Our explanation for the redundant FSTs is that a small number of T2 siblings carried the same Ds insertion due to transpositions in the T1 parent (Table I), which was propagated by T2 siblings to the T3 generation (see “Discussion”). In further analysis of the FSTs, The Institute for Genomic Research rice genome database (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml) was searched for gene homology between the FSTs and rice cDNA sequences. There were 186 mapped FSTs with hits to rice cDNA sequences. Such FSTs were 40.2% (186/463) among the FSTs obtained and represented Ds insertions in the genic regions.
Table III.
Interchromosomal distribution of ds insertions derived from 26 transformants
| Transformant | Rice Chromosomes
|
Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| A62 | 9 | 7 | 9 | 4 | 5 | 3 | 3 | 4 | 4 | 3 | 3 | 7 | 61 |
| A73 | 4 | 2 | 6 | 3 | 5 | 3 | 3 | 2 | 1 | 3 | 8 | 2 | 42 |
| A14 | 8 | 3 | 5 | 6 | 3 | 2 | 1 | 1 | 3 | 2 | 2 | 36 | |
| A57 | 6 | 1 | 3 | 1 | 2 | 3 | 2 | 1 | 4 | 2 | 3 | 1 | 29 |
| A35 | 4 | 2 | 1 | 2 | 2 | 3 | 1 | 1 | 1 | 3 | 20 | ||
| B62 | 3 | 3 | 3 | 1 | 1 | 1 | 3 | 2 | 2 | 19 | |||
| B28 | 5 | 1 | 2 | 2 | 3 | 1 | 3 | 1 | 18 | ||||
| A8 | 1 | 2 | 3 | 1 | 6 | 1 | 1 | 1 | 1 | 17 | |||
| B4 | 1 | 3 | 4 | 2 | 1 | 3 | 1 | 15 | |||||
| A56 | 3 | 1 | 1 | 1 | 1 | 1 | 8 | ||||||
| B31 | 2 | 1 | 2 | 1 | 1 | 1 | 8 | ||||||
| B41 | 1 | 1 | 2 | 1 | 1 | 1 | 7 | ||||||
| B49 | 1 | 1 | 1 | 1 | 1 | 5 | |||||||
| B45 | 2 | 1 | 1 | 4 | |||||||||
| A72 | 1 | 1 | 1 | 3 | |||||||||
| B13 | 1 | 1 | 1 | 3 | |||||||||
| B53 | 1 | 1 | 1 | 3 | |||||||||
| A3 | 1 | 1 | 2 | ||||||||||
| A34 | 1 | 1 | 2 | ||||||||||
| B16 | 1 | 1 | 2 | ||||||||||
| B46 | 1 | 1 | 2 | ||||||||||
| A51 | 1 | 1 | |||||||||||
| A52 | 1 | 1 | |||||||||||
| A29 | 1 | 1 | |||||||||||
| A30 | 1 | 1 | |||||||||||
| B10 | 1 | 1 | |||||||||||
| Total | 49 | 27 | 44 | 21 | 35 | 21 | 20 | 19 | 18 | 18 | 19 | 20 | 311 |
| % | 15.8 | 8.7 | 14.1 | 6.8 | 11.3 | 6.8 | 6.4 | 6.1 | 5.8 | 5.8 | 6.1 | 6.4 | |
We analyzed the distribution of Ds insertions among the rice chromosomes. For each single-T-DNA-locus transformant, in the T3 generation, the Ds element was found to be distributed among different rice chromosomes (Table III). When all the 26 transformants are taken in account, the largest chromosomes (1 and 3) carry the largest numbers (49 and 44, respectively) of Ds insertions, and the smallest chromosomes (9 and 10) carry the smallest numbers of Ds insertions (18 and 18). Based on the size of each chromosome, the Ds insertions appear to be evenly distributed in the rice genome.
Enhanced Expression of Rice Genes Adjacent to the Activation-Tagging Ds
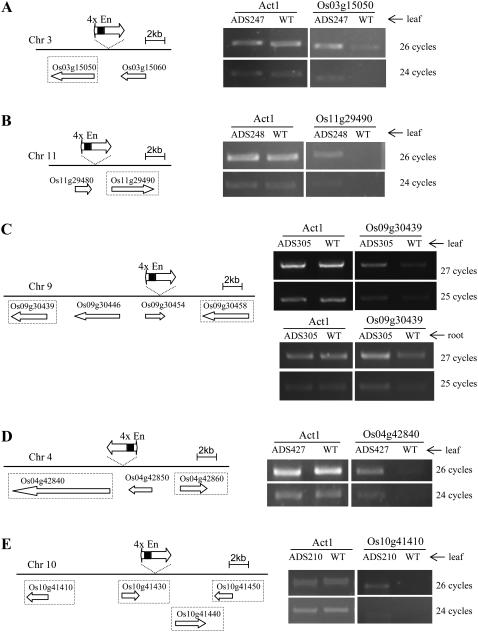
To examine whether expression of the Ds-tagged rice genes was altered, reverse transcriptase (RT)-PCR was performed on transposant lines with Ds insertion adjacent to the rice genes. We randomly selected 24 transposant lines and examined gene transcript levels using the semiquantitative RT-PCR method. Initially, we analyzed the closest rice gene for each Ds insertion, which was at a distance of 1 to 7 kb upstream or downstream of the Ds element. RNA was extracted from the leaves or roots of 60-d-old rice plants. RT-PCR was performed to compare gene expression levels in the 24 candidate lines to those in the wild-type rice ‘Nipponbare’. Two pairs of primers specific to each gene were tested and each RT-PCR experiment was repeated at least three times under the same condition. As shown in Figure 4A, gene expression was enhanced in the line ADS247 whose Ds insertion was 1,609 bp upstream of Os03g15050 as compared to ‘Nipponbare’. In ADS248, whose Ds insertion was 1,880 bp upstream of a plasma membrane-type ATPase gene (Os11g29490), the transcript level was significantly higher than in ‘Nipponbare’, as suggested by the RT-PCR results (Fig. 4B).
Figure 4.
Semiquantitative RT-PCR analysis of five Ds insertion lines. Left, Schematic representation of the Ds insertion and the CaMV 35S enhancers (4xEn) relative to the nearby genes. The map of each chromosomal region was based on the Rice GE genome database RiceGE Functional Genomics Database (http://signal.salk.edu/cgi-bin/RiceGE). Genes that were examined in semiquantitative RT-PCR are marked with dotted-line boxes. Right, RT-PCR analysis of activation-tagged genes. Rice Act1 transcript was amplified as control. WT, Wild-type rice ‘Nipponbare’. RT-PCR using leaf or root RNA was indicated. A, Line ADS247 was tagged by the activation-tagging Ds element 1,609 bp upstream of Os03g15050, which encodes the phosphenolpyruvate carboxykinase. B, Line ADS248 was tagged by a Ds 1,880 bp upstream of Os11g29490, which encodes a plasma membrane-type ATPase. C, Line ADS305 was tagged by a Ds 12.2 kb upstream of Os09g30439, which encodes a heat shock protein. D, Line ADS427 was tagged by a Ds 1,349 bp upstream of Os04g42840, which encodes a HEAT repeat family protein. E, Line ADS210 was tagged by a Ds 11.8 kb upstream of Os10g41410, which encodes the nucleoside diphosphate kinase 1. For each gene, the RT-PCR result was confirmed using two different pairs of specific primers and the result from using one pair of primers was shown. Each PCR experiment was repeated at least three times to confirm the result.
However, the other 22 transposant lines showed the same results as ‘Nipponbare’ in semiquantitative RT-PCR when just one gene was analyzed. We further tested eight of such transposant lines by amplifying other genes in the vicinity of the Ds insertion. Three overexpressed Ds lines were identified through RT-PCR examination of the additional genes (Fig. 4, C–E). In ADS305, whose Ds insertion was 12.2 kb upstream of the gene Os09g30439, the transcript level was slightly higher than in ‘Nipponbare’ as indicated in the results of both leaf and root tissues (Fig. 4C). In ADS427, where the Ds was inserted 1,349 bp upstream of Os04g42840, the transcript level was increased significantly based on the amount of the RT-PCR product in ADS427 and wild-type rice (Fig. 4D). In the RT-PCR analysis of ADS210, we examined expression levels of four genes that are 1.8 to 11.8 kb from the Ds insertion (Fig. 4E). Just for the gene Os10g41410, which is 11.8 kb upstream of the Ds element, transcript level was significantly enhanced in ADS210 as compared to ‘Nipponbare’.
It was observed that the CaMV enhancers in Ds in ADS210 activated the farthest gene Os10g41410 but did not affect expression of the other closer genes (Fig. 4E). In ADS305, as compared to Os09g30439, Os09g30458 is closer to the Ds insertion but did not get activated (Fig. 4C). On the other hand, in ADS427, the CaMV enhancers in Ds affected expression of only Os04g42840, which is closer to the Ds element than the gene Os04g42860 (Fig. 4D). Therefore, for the function of the CaMV enhancers, likelihood of gene activation was not directly related to the distance from the CaMV enhancers to the gene.
DISCUSSION
We have developed an in cis-activation-tagging Ac-Ds system in which a T-DNA vector carries a Ds element containing 4× CaMV enhancers along with the Ac transposase gene. In testing the vector in rice, a total of 638 stable Ds transposant plants were selected from seedlings (T3) of 2,086 T2 plants that were derived from 26 primary transformants. In analysis of 559 transposant lines, 311 of the Ds insertions were mapped on rice chromosomes.
Enrichment of unlinked transpositions is an important step for random insertional mutagenesis using the Ac-Ds transposable elements, as these elements have a feature of preferential transposition to closely linked sites. In the Ac-Ds vector systems we developed in Arabidopsis (Sundaresan et al., 1995) and rice (Kolesnik et al., 2004), a negative-positive selection scheme was successfully utilized for enrichment of unlinked transpositions. The Ac-Ds vectors were designed in such a way that an IaaH or GFP gene functioned as a counter selection (or negative selection) marker against the Ac T-DNA, the Ds-donor T-DNA, and linked Ds transpositions (Sundaresan et al., 1995; Kolesnik et al., 2004). In this study, the activation vector system utilizes an improved negative-positive scheme that is based on dual GFP and RFP fluorescence markers for high-throughput unlinked transposition selection (Kumar et al., 2005). Additionally, our vector system combines the merits of in cis-Ac-Ds elements that do not need genetic crossing and activation/knockout dual function in the Ds element.
The activation-tagging Ds element showed germinal transposition in the primary transformants (T1). In analysis of 53 single-T-DNA-locus transformants, GFP−RFP+ plants were identified in the T2 generation of 23 (43.4%) transformants. These GFP−RFP+ plants represented germinal transposition events that were transmitted from the T1 generation. For 22.6% (12/53) of the transformants, a fraction of excised Ds elements did not reinsert in the rice genome because the T2 populations had GFP+RFP− plants but not the GFP−RFP+ plants. It should be noted that these results were obtained using just 42 to 50 T2 plants per transformant. Because a rice plant produces 200 to 400 seeds, if more T2 progeny were assayed for GFP and RFP fluorescence, the frequency of primary transformants generating germinal transposition events could be higher than 43.4%.
From analysis of the T2 families, we also found the germinal transposition frequency varied depending on transformants. The T3 families of one-third of the single-T-DNA-locus transformants showed high transposition frequencies (20%–83.3%). The result was similar to the result of Kolesnik et al. (2004) that various frequencies were observed in different cross combinations of Ds line and Ac line. Therefore, it was important to select the right starter transformants to establish tagging populations. First, when generating transgenic plants of the activation vector, we did GFP and RFP fluorescence assays of the hygromycin-resistant calluses and plantlets to select transformants with normal GFP and RFP function. Transgenic rice plants carrying multiple or rearranged T-DNA copies may result in transgene silencing (Sallaud et al., 2003). Second, single-T-DNA-locus transformants were identified based on GFP segregation in T2 seedlings, and single-T-DNA-locus T2 plants were selected for generating T3 populations. In screen for putative stable Ds transposants, we initially performed a pilot screen of all T3 families and then focused on families with high germinal transposition frequencies. These strategies together with the GFP and RFP visual method highly facilitated screening of stable transposants.
In the design of a transposon tagging system, it is also essential to prevent a large number of transposant siblings from entering the pipeline. In this study, an important step was to avoid using transformants whose T2 generation exhibited several transposants (GFP−RFP+ and GFP+RFP− plants). For each of the transformants, we performed GFP and RFP assays on a small number of T2 plants to know about the frequency of transposants. In establishing most of the T3 populations, we utilized transformants whose T2 progeny contained no or a low to moderate rate of transposition events. Among the 463 Ds flanking sequences obtained, 73 (15.8%) sequences were redundant FSTs from siblings carrying the same Ds insertions. The number of sibling FSTs could be further reduced if the T2 families for generating T3 populations were more strictly selected.
In testing the activation tag vector system in rice, we obtained a total of 463 FSTs sequences, of which 311 (67.1%) FSTs were from mapped Ds insertions, 79 (17.1%) were from insertions in repeat sequences, and 73 (15.8%) were redundant sibling FSTs as discussed above. Among the 67.1% (311/463) mapped FSTs, 40.2% (186/463) had hits to rice cDNA sequences and represented Ds insertions in the genic regions. For the other 26.9% (125/463) FSTs that showed no homology to rice cDNA sequences, we were not able to make a consistently clear distinction between the FSTs from 5′ or 3′ regulatory sequences of rice genes and the FSTs from Ds insertions in the intergenic regions. These results confirmed the previous reports that the Ac-Ds elements preferentially transpose to genic regions (Enoki et al., 1999; Kolesnik et al., 2004; van Enckevort et al., 2005). For the strategy of activation tagging, Ds insertions in the intergenic regions are desirable as compared to the insertions in genic regions. However, for the transposant lines where the Ds element is inserted in a rice gene, the CaMV enhancers in Ds could still activate other neighboring genes. As indicated in the semiquantitative RT-PCR results, genes that were over 10 kb away from the Ds insertions were activated by the CaMV enhancers in Ds.
We analyzed interchromosomal distribution of the 311 Ds insertions that were mapped. It appeared that the insertions were evenly distributed in the rice genome and the insertion number of each chromosome was proportional to the chromosome size. This is different from the previous report (Kolesnik et al., 2004) in which Ds transposition showed a bias toward chromosome 1. The different results can be explained by the numbers of Ds starter lines that were used in generation of the Ds transposant collections. We used a total of 26 Ds starter lines, while Kolesnik et al. (2004) used just six starter lines. A single Ds launching pad might have preferential transposition on some chromosomes, but utilization of more starter lines might have reduced such bias. We did not analyze intrachromosomal distribution of Ds insertions due to the limited number of Ds insertions on each chromosome. The Ds element in our system may have preferential transposition regions like the hot spot of Ds insertion (Kolesnik et al., 2004) because a similar scheme of enrichment of unlinked transposition was used in this study.
We performed semiquantitative RT-PCR of rice genes in 24 transposant lines and observed enhanced expression of Ds-tagged genes in five lines. The RT-PCR results were confirmed in repeated experiments using the same condition. We never had a result that the RT-PCR product of a transposant line was less than that of wild-type rice. Therefore, the CaMV enhancers in the Ds element were capable of activating rice genes adjacent to the Ds insertion, which was similar to the result of T-DNA-based activation tagging in rice (Jeong et al., 2002, 2006). However, the frequency of activation-tagged lines was 20.8% (5/24) in this study and the frequency is lower than the frequency of 52.7% in the previous report using T-DNA in rice (Jeong et al., 2006). One reason might be that we analyzed just one gene in semiquantitative RT-PCR for most (16 among 24) of the candidate lines. For the other eight lines, two to four rice genes in each line that are close to the Ds insertion were examined in RT-PCR and enhanced expression of Ds-tagged genes was observed in three lines. Therefore, our rate of activation-tagged lines could be increased if more genes in the candidate lines were analyzed. For the function of the CaMV enhancers, it was reported that no good relationship was found between frequency of activation and distance from the CaMV enhancers to the gene and that no correlation was observed between degree of activation and distance (Jeong et al., 2006). We also observed the similar results in RT-PCR analysis of three activation-tagged lines.
Although large T-DNA insertion libraries have been generated in rice, an efficient T-DNA transformation system is available just for the japonica subspecies. For the indica rice subspecies that is more widely grown in rice farming areas around the world, T-DNA transformation is still difficult. Also, most of the T-DNA transformation methods involve a tissue culture step that generates high frequency of somaclonal variation, which disturbs the process of forward mutant screens. Therefore, transposon mutagenesis is an effective approach for plant functional genomics and can serve as an alternative to T-DNA when transformation through tissue culture is inefficient, as is the case for the indica rice species as well as many other crops. Transposable elements can be mobilized or immobilized on demand and the approach requires only a few primary transformants to generate a large collection of independent transposon insertions within the genome. For this reason, generation of insertions in heterologous plants mainly depends on propagation as in the case of transposon mutagenesis in maize.
The Ac-Ds elements have features like small size of cis-required sequence (approximately 600 bp of minimal Ds), large cargo insert capacity, and reasonable high transposition frequency, which have made the Ac-Ds elements particularly amenable for genetic studies. The maize Ac-Ds elements can efficiently transpose in many heterologous plant species. Also, the Ac-Ds showed trans-kingdom transposition in yeast (Saccharomyces cerevisiae) and animal species (Weil and Kunze, 2000; Emelyanov et al., 2006), which suggested that the Ac-Ds elements do not rely on host-specific factors for transposition and that host factors involved in their mobility mechanism are widely conserved. Activation-tagging strategies based on maize transposable elements have been developed in tobacco (Nicotiana tabacum; Suzuki et al., 2001) and Arabidopsis (Marsch-Martinez et al., 2002; Schneider et al., 2005). The activation-tagging Ac-Ds system developed in this study was aimed for rice and other monocot plants. This in cis-two-element system eliminates the need for crossing and utilizes RFP and GFP for tracking Ds element and counter selection against the Ac transposase, respectively, to stabilize transposed Ds elements. The use of fluorescent markers instead of antibiotic or herbicide resistance increases the applicability to other plants and eliminates problems with escapes. As the dual fluorescence markers were under control of the maize ubiquitin 1 promoter (Christensen and Quail, 1996) that is functional in maize, rice, barley, wheat (Triticum aestivum), and many grasses, the principles developed here are applicable to many other monocot plants. We find that the Ds element preferentially transposes into genic regions, which is similar to previous reports about the maize transposable elements (Enoki et al., 1999; Cowperthwaite et al., 2002; Kolesnik et al., 2004; van Enckevort et al., 2005). When utilized as the carrier of the CaMV enhancers, the Ds element offers the advantage of both activation-tagging and knockout mutations. These features make transposon-based activation tagging particularly useful for large genomes with many duplicated genes such as maize, as well as polyploid plant crops. The Ac-Ds-based activation vector system developed in this study is publicly available without intellectual property restrictions and should be applicable for the functional genomics of a range of plants and especially for that of cereals and monocot plants.
MATERIALS AND METHODS
Vector Construction
In construction of the activation-tagging Ac-Ds vector pSQ5, the Ubi-DsRed-Nos cassette was from pSK62 (Kumar et al., 2005), the 1,785-bp 5′Ds and 222-bp 3′Ds were from pWS32 (Sundaresan et al., 1995), and the 4× CaMV 35S enhancers were from the AcREH construct (Suzuki et al., 2001). First, pWS32 was digested with NcoI/EcoRI and ligated to an NcoI/HindIII/EcoRI linker to generate pSQ1. Second, the 4× CaMV 35S enhancers were released as a 1.4-kb HindIII fragment from the AcREH construct and cloned at the pSQ1 HindIII site to make pSQ2. At the third step, multi-DNA ligation was performed using (1) the 1.7-kb 3′Ds/4× enhancers fragment from SacI/EcoRI digested pSQ2; (2) the 1,785-bp 5′Ds from SacI/HindIII digested pWS32; (3) a 3.3-kb Ubi-DsRed-Nos cassette from HindIII/EcoRI digested pSK62. The multiligation products were digested with SacI and 6.8-kb product (i.e. the activation-tagging Ds element) was purified and cloned into a SacI site of pCAMBIA-1300. The activation-tagging Ds element was partially sequenced to confirm junctions of fragments of the multiligation. The Ds element was finally cloned at the SacI site of the pSSZ40 construct (Kolesnik et al., 2004) carrying the CaMV35S-Ac and Ubi-sGFP-Nos cassettes. The backbone of the pSQ5 vector was from pCAMBIA-1300 (GenBank accession no. AF234296). The vector pSQ5 is in the public domain and will be made available by the corresponding author upon request.
Plant Transformation
Mature seeds of rice sp. japonica ‘Nipponbare’ were cultured on NB medium (Li et al., 1993) for 3 to 4 weeks to induce embryogenic callus. The callus was cocultivated with Agrobacterium strain LBA4404 containing the pSQ5 Ac-Ds vector and selected on NB medium containing 50 μg L−1 of hygromycin B (Roche Diagnostics) as described (Yin and Wang, 2000; Sallaud et al., 2003). GFP+RFP+ calluses were transferred to preregeneration medium (Yin and Wang, 2000) and cultured at 25°C and in dark for 10 to 15 d. The GFP+RFP+ calluses were further cultured on regeneration medium (Yin and Wang, 2000) under light for 2 to 3 weeks. GFP+RFP+ transformant plantlets were finally transferred to greenhouse.
GFP/RFP Fluorescence Assays
Transformed rice callus and plantlets were examined under a Zeiss SV11 fluorescence microscope (Zeiss) using appropriate filters of GFP and RFP fluorescence. Mature seeds of transgenic plants were germinated at 25°C or 30°C in dark for 4 to 6 d. The emerging seedlings were screened for GFP fluorescence using the Green Fluorescent Protein Macro Detector set (model GFP-MDS-20BB, BLS). RFP fluorescence visualization of seedlings was under the Zeiss SV11 fluorescence microscope.
Cloning of Ds Flanking Sequences Using Adaptor-Ligation PCR
Rice DNA of stable Ds transposants was isolated using the DNeasy 96 Plant kit (Qiagen). In adaptor-ligation PCR, about 100 ng of rice genomic DNA was added to digestion-ligation reaction mixture, which contained the HindIII and EcoRI adaptors, the enzymes of EcoRI, HindIII, and T4 DNA ligase (Alonso et al., 2003). PCR primers of the first-round PCR were Ds5-1A (5′-acggtcgggaaactagctctacc-3′) and AP1 (5′-gtaatacgactcactatagggc-3′; Alonso et al., 2003). Products of the first round PCR were diluted 10 times with water. The second round PCR was performed using primers Ds5-4 (5′-ctcgggttcgaaatcgatcgggat-3′) and AP2-C (5′-tggtcgacggcccgggctgc-3′). PCR products were sequenced using ABI Biosystems BigDye v3.1 and ABI 3730 × 1 DNA sequencers.
RT-PCR
Total RNA was extracted from leaves or roots of Ds insertion lines and wild-type rice using the Trizol Regent (Invitrogen, catalog no. 15596-018). One microgram of RNA was reverse transcribed in a total volume of 25 μL that contained 0.4 nmol of oligo(dT)18 primer, 5 mm dNTPs, 20 units of SUPERase-in (Ambion, catalog no. AM2694), and 200 units of Moloney murine leukemia virus RT (Promega, catalog no. M1701) in the murine leukemia virus reaction buffer. PCR was performed in 25 μL of solution containing 1× TaKaRa Ex Taq polymerase buffer, 0.2 μm gene-specific primers, 2.5 mm dNTPs, 0.5 unit of TaKaRa Ex Taq polymerase, and 1 μL cDNA reaction product. The reaction condition included denaturing template DNA at 95°C for 5 min, 24 to 27 cycles of PCR (94°C, 30 s; 56°C, 40 s; 72°C, 50 s), and a final incubation step (72°C, 5 min).
Acknowledgments
We thank Patrick Hogan and Asuman Buzkan for their assistance in planting and harvesting rice. We are also grateful to Cameron Johnson for performing sequencing data analysis, to Chellian Santhosh Kumar for discussion, and to Liza Conrad for critical reading of the manuscript.
This work was supported by the National Science Foundation Plant Genome Research Program (award no. 0211924) and by the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service National Research Initiative Program (grant no. 2005–04915).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Venkatesan Sundaresan (sundar@ucdavis.edu).
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Ayliffe MA, Pallotta M, Langridge P, Pryor AJ (2007) A barley activation tagging system. Plant Mol Biol 64 329–347 [DOI] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY (2000) Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA 97 11984–11989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5 213–218 [DOI] [PubMed] [Google Scholar]
- Cooper LD, Marquez-Cedillo L, Singh J, Sturbaum AK, Zhang S, Edwards V, Johnson K, Kleinhofs A, Rangel S, Carollo V, et al (2004) Mapping Ds insertions in barley using a sequence-based approach. Mol Genet Genomics 272 181–193 [DOI] [PubMed] [Google Scholar]
- Cowperthwaite M, Park W, Xu Z, Yan X, Maurais SC, Dooner HK (2002) Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov A, Gao Y, Naqvi NI, Parinov S (2006) Trans-kingdom transposition of the maize dissociation element. Genetics 174 1095–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki H, Izawa T, Kawahara M, Komatsu M, Koh S, Kyozuka J, Shimamoto K (1999) Ac as a tool for the functional genomics of rice. Plant J 19 605–613 [DOI] [PubMed] [Google Scholar]
- Hsing YI, Chern CG, Fan MJ, Lu PC, Chen KT, Lo SF, Sun PK, Ho SL, Lee KW, Wang YC, et al (2007) A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol Biol 63 351–364 [DOI] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Mizukado S, Sakurai T, Kanahara H, Seki M, Shinozaki K (2002) A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol 129 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Noutoshi Y, Seki M, Kamiya A, Mizukado S, Sakurai T, Shinozaki K (2005) A resource of 5814 Dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol 46 1149–1153 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, et al (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22 561–570 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G (2002) T-DNA insertion mutagenesis for activation tagging in rice. Plant Physiol 130 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45 123–132 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, Ramamoorthy R, Cai M, Ma ZG, Sundaresan V, Ramachandran S (2004) Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J 37 301–314 [DOI] [PubMed] [Google Scholar]
- Koprek T, McElroy D, Louwerse J, Williams-Carrier R, Lemaux PG (2000) An efficient method for dispersing Ds elements in the barley genome as a tool for determining gene function. Plant J 24 253–263 [DOI] [PubMed] [Google Scholar]
- Kumar CS, Wing RA, Sundaresan V (2005) Efficient insertional mutagenesis in rice using the maize En/Spm elements. Plant J 44 879–892 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K (2004) A collection of 11,800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J 37 897– 905 [DOI] [PubMed] [Google Scholar]
- Li L, Qu R, Kochko A, Fauquet C, Beachy RN (1993) An improved rice transformation system using the biolistic method. Plant Cell Rep 12 250–255 [DOI] [PubMed] [Google Scholar]
- Marsch-Martinez N, Greco R, Van Arkel G, Herrera-Estrella L, Pereira A (2002) Activation tagging using the En-I maize transposon system in Arabidopsis. Plant Physiol 129 1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, et al (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Tomita C, Sugimoto K, Hasegawa M, Hayashi N, Dubouzet JG, Ochiai H, Sekimoto H, Hirochika H, Kikuchi S (2007) Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol 63 847–860 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from Dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Meynard D, van Boxtel J, Gay C, Bès M, Brizard JP, Larmande P, Ortega D, Raynal M, Portefaix M, et al (2003) Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor Appl Genet 106 1396–1408 [DOI] [PubMed] [Google Scholar]
- Schneider A, Kirch T, Gigolashvili T, Mock HP, Sonnewald U, Simon R, Flugge UI, Werr W (2005) A transposon-based activation-tagging population in Arabidopsis thaliana (TAMARA) and its application in the identification of dominant developmental and metabolic mutations. FEBS Lett 579 4622–4628 [DOI] [PubMed] [Google Scholar]
- Singh J, Zhang S, Chen C, Cooper L, Bregitzer P, Sturbaum A, Hayes PM, Lemaux PG (2006) High-frequency Ds remobilization over multiple generations in barley facilitates gene tagging in large genome cereals. Plant Mol Biol 62 937–950 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9 1797–1810 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Uemura S, Saito Y, Murofushi N, Schmitz G, Theres K, Yamaguchi I (2001) A novel transposon tagging element for obtaining gain-of-function mutants based on a self-stabilizing Ac derivative. Plant Mol Biol 45 123–131 [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JD (1999) Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Enckevort LJ, Droc G, Piffanelli P, Greco R, Gagneur C, Weber C, González VM, Cabot P, Fornara F, Berri S, et al (2005) EU-OSTID: a collection of transposon insertional mutants for functional genomics in rice. Plant Mol Biol 59 99–110 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil CF, Kunze R (2000) Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat Genet 26 187–190 [DOI] [PubMed] [Google Scholar]
- Yin Z, Wang GL (2000) Evidence of multiple complex patterns of T-DNA integration into the rice genome. Theor Appl Genet 100 461–470 [Google Scholar]
- Zhao T, Palotta M, Langridge P, Prasad M, Graner A, Schulze-Lefert P, Koprek T (2006) Mapped Ds/T-DNA launch pads for functional genomics in barley. Plant J 47 811–826 [DOI] [PubMed] [Google Scholar]