Abstract
The β-substituted alanine (Ala) synthase (Bsas) family in the large superfamily of pyridoxal 5′-phosphate-dependent enzymes comprises cysteine (Cys) synthase (CSase) [O-acetyl-serine (thiol) lyase] and β-cyano-Ala synthase (CASase) in plants. Nine genomic sequences encode putative Bsas proteins in Arabidopsis thaliana. The physiological roles of these Bsas isoforms in vivo were investigated by the characterization of T-DNA insertion mutants. Analyses of gene expression, activities of CSase and CASase, and levels of Cys and glutathione in the bsas mutants indicated that cytosolic Bsas1;1, plastidic Bsas2;1, and mitochondrial Bsas2;2 play major roles in Cys biosynthesis. Cytosolic Bsas1;1 has the most dominant contribution both in leaf and root, and mitochondrial Bsas2;2 plays a significant role in root. Mitochondrial Bsas3;1 is a genuine CASase. Nontargeted metabolome analyses of knockout mutants were carried out by a combination of gas chromatography time-of-flight mass spectrometry and capillary electrophoresis time-of-flight mass spectrometry. The level of γ-glutamyl-β-cyano-Ala decreased in the mutant bsas3;1, indicating the crucial role of Bsas3;1 in β-cyano-Ala metabolism in vivo.
Sulfur is an essential macronutrient that is required for the growth of plants. Assimilation of sulfur in plants depends mostly upon Cys formation from the sulfate ion. Cys is the first organic precursor in the formation of sulfur-containing metabolites such as Met and glutathione (GSH; Leustek and Saito, 1999; Saito, 2000, 2004). Cys formation is catalyzed by the enzyme Cys synthase (CSase; EC 4.2.99.8) [O-acetyl-Ser (thiol) lyase] by using O-acetyl-Ser (OAS) and sulfide as substrates. CSase contains a pyridoxal-5′-phosphate (PLP) cofactor and has now been classified within the β-substituted Ala synthase (Bsas) subfamily in the large superfamily of PLP-dependent enzymes (Hatzfeld et al., 2000). The CSase-like protein β-cyano-Ala synthase (CASase; EC 4.4.1.9), which shares high homology at the nucleotide and amino acid level with CSases, belongs to the same enzyme family (Hatzfeld et al., 2000; Jost et al., 2000; Droux, 2004; Wirtz et al., 2004). In cyanide detoxification, CASase catalyzes the formation of the nonprotein amino acid β-cyano-Ala from Cys and cyanide, producing sulfide as a product (Blumenthal et al., 1968). β-Cyano-Ala is further metabolized to Asn in most plants (Blumenthal et al., 1963; Castric et al., 1972) or it can be conjugated to γ-glutamyl-β-cyano-Ala in cyanogenic plants such as Vicia sativa (Ressler et al., 1963).
Nine putative Bsas genes have been identified in whole-genome sequencing of Arabidopsis (Arabidopsis thaliana; Arabidopsis Genome Initiative, 2000). To date, six of nine Bsas isoforms have been well characterized with respect to their enzymatic properties and subcellular localization (Table I). Bsas1;1, Bsas2;1, and Bsas2;2, which are localized in the cytosol, plastids, and mitochondria, respectively, of Arabidopsis cells, are considered the predominant CSases according to kinetic analysis (Hell et al., 1994, 1999; Hesse and Höfgen, 1998; Wirtz et al., 2004). In contrast, Bsas3;1 localized in the mitochondria is supposed to act at in vivo conditions as CASase (Hatzfeld et al., 2000; Yamaguchi et al., 2000). Bsas4;1 and Bsas4;2 localized in the cytosol demonstrate low CSase activity (Hatzfeld et al., 2000; Yamaguchi et al., 2000). The remaining isoforms—Bsas1;2, Bsas4;3, and Bsas5;1—are much less characterized.
Table I.
Arabidopsis Bsas gene family and bsas mutants
| Isoform | AGI Code | Subcellular Localization | In Vitro Activity | References | Mutant ID |
|---|---|---|---|---|---|
| Bsas1;1 | At4g14880 | Cytosol | CSase | Hell et al. (1994); Barroso et al. (1995); Jost et al. (2000) | Salk_072213 |
| Bsas1;2a | At3g22460 | Cytosolb | Not determined | Jost et al. (2000) | |
| Bsas2;1 | At2g43750 | Plastids | CSase | Barroso et al. (1995); Hesse et al. (1999); Jost et al. (2000) | Salk_021183 |
| Bsas2;2 | At3g59760 | Mitochondria | CSase | Hesse et al. (1999); Jost et al. (2000) | Salk_000860 |
| Bsas3;1 | At3g61440 | Mitochondria | CASase | Hatzfeld et al. (2000); Yamaguchi et al. (2000) | Salk_022479 |
| Bsas4;1 | At5g28020 | Cytosol | CSase | Hatzfeld et al. (2000); Yamaguchi et al. (2000) | Salk_097875 |
| Bsas4;2 | At3g04940 | Cytosol | CSase | Hatzfeld et al. (2000); Yamaguchi et al. (2000) | Salk_092696 |
| Bsas4;3 | At5g28030 | Cytosolb | Not determined | Salk_103855 | |
| Bsas5;1 | At3g03630 | Plastidsb | Not determined | Nakamura et al. (1997) | Salk_034133 |
The Bsas1;2 gene is a truncated gene. The gene is transcribed, but not translated, into a functional Bsas protein.
Subcellular localizations were predicted by TargetP (http://www.cbs.dtu.dk/services/TargetP/predictions/pred.html.) and WoLF PSORT (http://wolfpsort.seq.cbrc.jp) analysis.
Although these biochemical characterizations and subcellular localization studies were relatively well conducted, the actual roles of Bsas genes in vivo are not necessarily clear. Questions such as whether each Bsas isoform is redundant or has a specific function in vivo and to what extent each Bsas isoform contributes to the synthesis of Cys and β-cyano-Ala remain unresolved. It is therefore important to conduct in vivo analysis to reveal the actual function of individual Bsas isoforms. The available sequence information of the entire Arabidopsis genome and rich genetic resources make it possible to provide genetic evidence for the roles of Bsas genes in vivo. This article reports the analysis of T-DNA-inserted knockout mutants of Arabidopsis for their gene expression, enzyme activities, and metabolite profiles. Furthermore, nontargeted metabolite profiles of bsas mutants revealed that Arabidopsis can accumulate γ-glutamyl-β-cyano-Ala as a consequence of the CASase action of Bsas3;1. Through the conjunctive study of genetic analysis of knockout mutants and previously reported biochemical analysis of recombinant proteins, the physiological function of these Bsas genes in vivo is clarified.
RESULTS
Expression Analysis Indicates That Four Bsas Genes Are Presumed to Play Major Roles
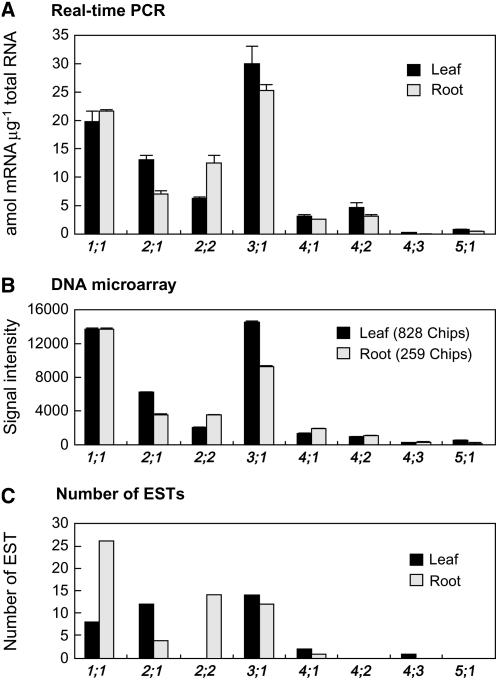
In Arabidopsis, there are nine genes encoding putative Bsas (Arabidopsis Genome Initiative, 2000; Table I). However, Bsas1;2 is unlikely to encode a functional protein because its predicted amino acid sequence is one-half the length of other Bsas isoforms and is probably derived from the N-terminal region-encoding portion of Bsas1;1 (Supplemental Fig. S1; Jost et al., 2000). Therefore, we excluded the Bsas1;2 gene from further study. To accurately determine the expression levels of the eight other Bsas genes, quantitative real-time PCR was performed on the total RNA extracted from leaves and roots of 2-week-old plants (Fig. 1A). A high level of expression for Bsas1;1 and Bsas3;1 and a moderate level of expression for Bsas2;1 and Bsas2;2 were observed and compared with other Bsas genes. With respect to tissue specificity, Bsas1;1 was expressed in similar amounts in leaf and root. Relatively high amounts of Bsas2;1 and Bsas3;1 transcripts were observed in leaf. Bsas2;2 expression was higher in root than in leaf. These tissue specificities were in agreement with the data previously obtained by northern analysis (Hesse et al., 1999; Yamaguchi et al., 2000). Transcripts of the other Bsas genes (Bsas4;1, Bsas4;2, Bsas4;3, and Bsas5;1) could be detected, but at an overall lower expression level. These results for expression levels and tissue specificities were in good agreement with the microarray analysis (Genevestigator; https://www.genevestigator.ethz.ch; Fig. 1B) and the number of ESTs in The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org; Noji et al., 2006; Fig. 1C).
Figure 1.
Gene expression analysis of Bsas genes in the leaf and root. A, Quantitative real-time PCR analysis. Total RNA was extracted from leaves and roots of 2-week-old wild-type plants. Data are the means of triplicate determinations ±sd (bars). B, Expression analysis using the Genevestigator tool based on DNA microarray analysis (https://www.genevestigator.ethz.ch). C, Expression analysis performed using the number of ESTs in the TAIR database (http://www.arabidopsis.org).
Knockout Mutants Showed No Visible Phenotypic Changes
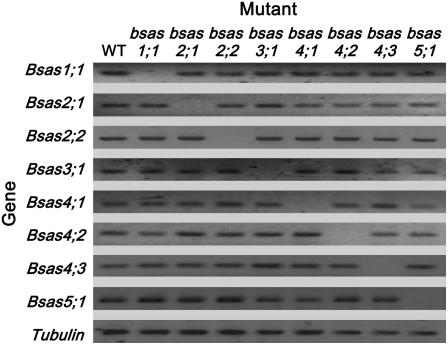
On searching the Salk Institute Insertional Mutation Database, T-DNA insertion mutants (Alonso et al., 2003) were identified for eight Bsas genes. The bsas mutants, all of Columbia (Col-0) background, were identified in a PCR-based screening of the various T-DNA-transformed populations of Arabidopsis for those bearing insertions in the Bsas locus (Supplemental Fig. S2). After selection of the homozygous T-DNA insertion mutants, semiquantitative reverse transcription (RT)-PCR analysis was performed for Bsas transcripts in 2-week-old wild-type plants and bsas mutants (Fig. 2). In all bsas mutants, the expressions of targeted Bsas genes for knockout mutation was repressed, but the expression levels of other Bsas genes besides the knockout Bsas gene were not remarkably changed compared with those of the wild type. The same result was obtained in the analysis of root (data not shown). In addition, no visible phenotypic changes were observed in all bsas mutants compared with the wild type under normal growth conditions. These findings suggest that the knockout of each Bsas gene caused no apparent changes in development and expression of other Bsas genes under normal growth conditions.
Figure 2.
Bsas transcripts in bsas mutants by RT-PCR analysis. Total RNA was extracted from 2-week-old homozygous bsas mutants and the wild type. RT-PCR was performed with specific primers for Bsas genes and a tubulin gene used as the control.
Bsas1;1, Bsas2;1, and Bsas2;2 Predominantly Contribute to Cellular CSase Activity
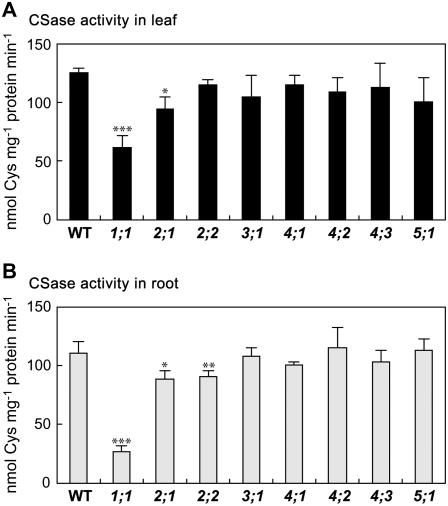
CSase activity was determined in crude protein extracts of the bsas mutants in leaves (Fig. 3A) and in roots (Fig. 3B) and compared with the wild type. Greater reductions in CSase activity were observed in bsas1;1, decreasing to 50% in leaf and 24% in root as compared to that in the wild type. In bsas2;1, CSase activity decreased to 75% in leaf and 80% in root. In bsas2;2, CSase activity did not change in leaf, but decreased to 82% in root. Activities in other bsas mutants did not change significantly as compared with those in the wild type. Cytosolic Bsas1;1, plastidic Bsas2;1, and mitochondrial Bsas2;2, which were considered to be predominant CSases from biochemical analysis (Hell et al., 1994; Hesse and Höfgen, 1998; Hesse et al., 1999; Wirtz et al., 2004), did contribute to CSase activity in vivo in different cellular compartments. The difference in the activity of each Bsas isoform, judging from the CSase activity measurements in mutants, correlated with the difference in Bsas gene expression (Fig. 1). The predominance of three Bsas isoforms (Bsas1;1, Bsas2;1, and Bsas2;2) suggests that these Bsas genes are responsible for the production of Cys used in the different cellular compartments.
Figure 3.
CSase activity in bsas mutants. Total extracts of soluble proteins were prepared from leaves and roots of 2-week-old plants and CSase activity was determined by measuring the formation of Cys in the leaf (A) and root (B). Data represent the mean of four experiments with 15 plants in each (±sd). Differences between the wild type and bsas mutants analyzed using Student's t test were statistically significant (*, P < 0.01; **, P < 0.005; ***, P < 0.001).
Bsas3;1 Plays a Major Role as CASase
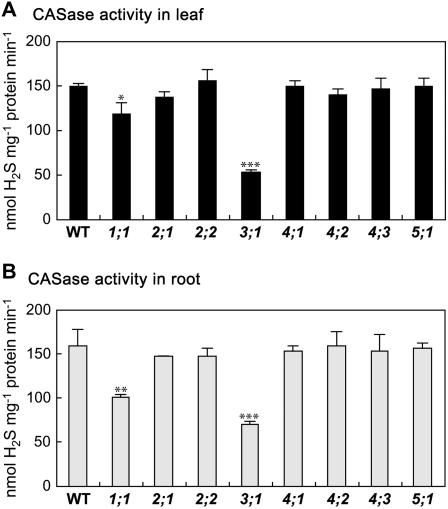
CASase activity was determined in crude protein extracts of the bsas mutants from leaves (Fig. 4A) and from roots (Fig. 4B). Activity in the bsas3;1 mutant decreased dramatically to 36% in leaf and 45% in root as compared to those in the wild type. This result suggests that the mitochondrial Bsas3;1, which was considered to be a genuine CASase judging by the substrate specificity of recombinant protein (Hatzfeld et al., 2000; Warrilow and Hawkesford, 2000; Yamaguchi et al., 2000), played a major role as CASase. Significant reduction of CASase activity was also observed in bsas1;1, decreasing to 80% in leaf and 64% in root. The reduction in CASase activity in bsas1;1 was ascribed to the knockout of Bsas1;1-exhibiting CASase activity as a side reaction apart from CSase. CSase from various plant species displays CASase activity to some extent (Maruyama et al., 1998, 2000, 2001). Although the recombinant proteins of Bsas1;1, Bsas2;1, and Bsas2;2 exhibited bifunctional CSase and CASase activities, all these proteins were determined to be true CSase by kinetic analysis (Jost et al., 2000). Considering all the available information, we conclude that Bsas3;1 acts predominantly as CASase in vivo and Bsas1;1 may have an accessory function for CASase activity.
Figure 4.
CASase activity in bsas mutants. Total extracts of soluble proteins were prepared from leaves and roots of 2-week-old plants and CASase activity was determined by measuring the formation of H2S from Cys in leaf (A) and root (B). Data represent the mean of three experiments with 15 plants in each (±sd). Differences between the wild type and bsas mutants analyzed using Student's t test were statistically significant (*, P < 0.01; **, P < 0.005; ***, P < 0.001).
Cellular Thiol Contents Are Affected by the Knockout of Bsas1;1 and Bsas2;2
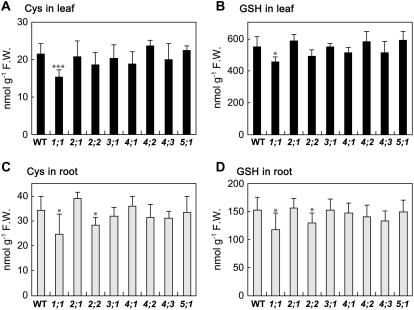
To determine whether Cys and GSH levels were altered in bsas mutants, thiol levels were measured in leaves and roots (Fig. 5). In leaf, thiol levels in bsas1;1 alone were significantly lower compared with the wild type: Cys content was down to 72% and GSH content was down to 83%. In root, significant reductions in thiol levels were observed in bsas1;1 and bsas2;2: Cys and GSH contents were down to 72% and 77% in bsas1;1 and 83% and 85% in bsas2;2, respectively. These results suggest that cytosolic Bsas1;1, the gene expression and the CSase activity of which were high in both leaf and root, was mainly responsible for Cys biosynthesis. The role of mitochondrial Bsas2;2 is presumably more important in the root. The high level of Bsas2;2 expression and reduction of CSase activity in bsas2;2 observed in root (Figs. 1 and 3) support this hypothesis.
Figure 5.
Accumulation of Cys and GSH in bsas mutants. Thiols were extracted from leaves and roots of 2-week-old plants. Cys and GSH in leaf (A and B) and root (C and D) were determined by HPLC analysis. Data represent the mean of five experiments with 15 plants in each (±sd). Differences between the wild type and bsas mutants analyzed using Student's t test were statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.005).
Nontargeted Metabolome Analysis Pinpointed the Lack of Accumulation of γ-Glutamyl-β-Cyano-Ala in the bsas3;1 Mutant
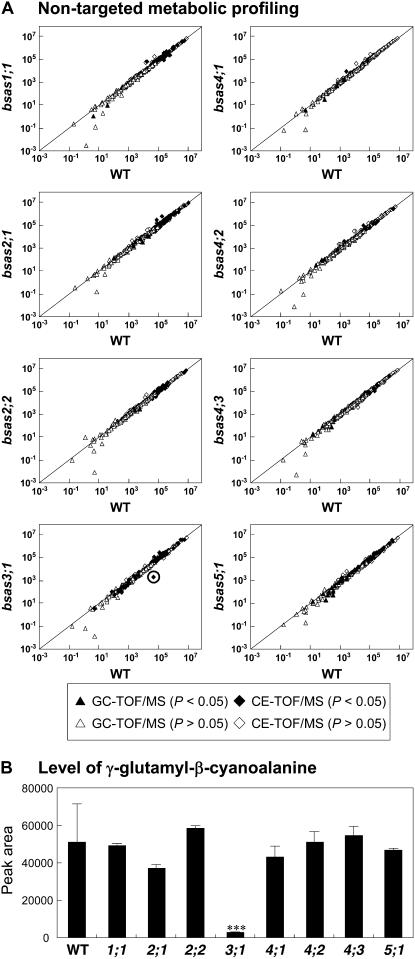
To investigate the whole metabolic change in each bsas mutant, we performed nontargeted gas chromatography time-of-flight mass spectrometry (GC-TOF/MS) and capillary electrophoresis time-of-flight mass spectrometry (CE-TOF/MS) analyses in the leaves of mutant plants. Nontargeted metabolome analysis may lead to the identification of specific metabolites whose levels are altered by knockout genes of unknown functions and may present an overview of the global perturbation of cellular metabolism. Principal component analysis generated from GC-TOF/MS and CE-TOF/MS data matrices showed no obvious differences between the wild-type and all bsas mutant plants (Supplemental Fig. S3). This result indicates that the changes in whole-metabolite accumulation for all bsas mutants were quite small.
To detect differentially accumulated metabolites, metabolite peak areas were compared in the wild type and in each bsas mutant (Fig. 6A). No significant change in β-cyano-Ala contents, which accumulated in trace amounts even in wild-type plants, was observed between the bsas mutants and the wild type despite significant changes in CAS activity in bsas1;1 and bsas3;1. In bsas3;1, however, an unknown peak observed at mass-to-charge ratio (m/z) 244.0924 by CE-TOF/MS was dramatically decreased as compared with that in the wild type (Fig. 6B). This m/z (244.0924) matched the theoretical m/z of γ-glutamyl-β-cyano-Ala (C9H14O3N5; m/z 244.0933). This metabolite was confirmed to be γ-glutamyl-β-cyano-Ala by comparison with the synthetic product by γ-glutamyltranspeptidase (GGT) from β-cyano-Ala and Glu (Braun et al., 1982), using CE-TOF/MS and fluorescence detection on HPLC for amino acids derivatized with O-phthalaldehyde (Supplemental Fig. S4).
Figure 6.
A, Comparison of the metabolite accumulation between the wild type and bsas mutants. The mean peak areas of the wild type and each bsas mutant are shown on the x and y axes, respectively. Data represent the mean of eight experiments for the wild type and three experiments for bsas mutants with 20 plants. Differences between the wild type and bsas mutants analyzed using Welch's t test were statistically significant (black triangles [P < 0.05] and white triangles [P > 0.05] for GC-TOF/MS; black diamonds [P < 0.05] and white diamonds [P > 0.05] for CE-TOF/MS). The black line represents the diagonal line (y = x). The black circle on the plot of the wild type and bsas3;1 indicates γ-glutamyl-β-cyano-Ala. B, Accumulation of γ-glutamyl-β-cyano-Ala in bsas mutants. γ-Glutamyl-β-cyano-Ala contents were measured in leaves by CE-TOF/MS. Data represent the mean of eight experiments for the wild type and three experiments for bsas mutants with 20 plants in each (±sd). Differences between the wild type and bsas mutants analyzed using Welch's t test were statistically significant (***, P < 0.001).
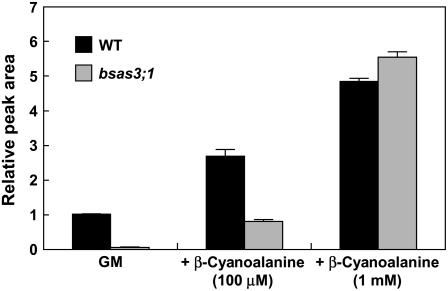
Upon addition of β-cyano-Ala to the growth medium, accumulation of γ-glutamyl-β-cyano-Ala increased in the bsas3;1 mutant plant to levels similar to those found in the wild-type plant undergoing the same treatment (Fig. 7). This result suggests that γ-glutamyl-β-cyano-Ala was a metabolite derived from β-cyano-Ala. These results indicate that Arabidopsis metabolizes cyanide to β-cyano-Ala and eventually to γ-glutamyl-β-cyano-Ala as the presumable storage form, and Bsas3;1 is primarily responsible for the synthesis of β-cyano-Ala and, subsequently, γ-glutamyl-β-cyano-Ala.
Figure 7.
Accumulation of γ-glutamyl-β-cyano-Ala in the leaf of the wild type and bsas3;1 mutant treated with β-cyano-Ala. Plants grown for 3 weeks on GM-agar medium were transferred to GM-agar medium or medium containing β-cyano-Ala (100 μm and 1 mm) and grown for 3 d. γ-Glutamyl-β-cyano-Ala was analyzed by fluorescence HPLC. Data represent the mean of three experiments with 15 plants in each (±sd).
DISCUSSION
Physiological Roles of Bsas Isoforms by in Vivo Experiments
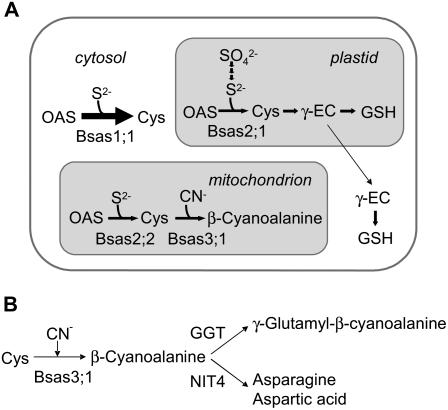
Analyses of Bsas gene expression and characterization of bsas mutants reveals that cytosolic Bsas1;1, plastidic Bsas2;1, and mitochondrial Bsas2;2 are genuine CSases and mitochondrial Bsas3;1 is a true CASase in Arabidopsis (Fig. 8A). The cytosolic Bsas1;1 is primarily responsible for the synthesis of Cys in leaf and root. The role of mitochondrial Bsas2;2 is more important in root than in leaf. Different quantities of mitochondrial Bsas2;2 in photosynthetic leaf and nonphotosynthetic root suggest that these two tissues have different requirements for cellular Cys. In bsas4;1, bsas4;2, bsas4;3, and bsas5;1, no obvious changes were observed in CSase activity, CAS activity, thiol contents, and γ-glutamyl-β-cyano-Ala content as compared with the wild type. These results suggest that Bsas4;1, Bsas4;2, Bsas4;3, and Bsas5;1 participate to a lesser extent in the synthesis of Cys and β-cyano-Ala. Because no specific patterns of expression are seen with those genes in the public transcriptome database Genevestigator (https://www.genevestigator.ethz.ch), these minor Bsas isoforms are presumed to be redundant. However, it is possible that specific functions would be exhibited under particular conditions. It was reported that the CSase of several plant species is also responsible for the synthesis of secondary heterocyclic β-substituted Alas, some of which were physiologically active in a variety of organisms (Ikegami and Murakoshi, 1994). Therefore, rather than function as CSases, some Bsas isoforms may function as enzymes with PLP-dependent β-replacement activity for production of those secondary metabolites, or those genes of lesser CSase function may have potential to evolve to a new gene of more diverse function during the evolutionary process.
Figure 8.
Schematic representation of Cys and β-cyano-Ala metabolism in Arabidopsis. A, Schematic representation of Bsas isoforms involved in Cys metabolism in the cytosol, plastid, and mitochondrion. Solid arrows show enzymatic reactions. γ-EC, γ-Glutamyl-Cys. B, Metabolism of β-cyano-Ala. β-Cyano-Ala is metabolized to γ-glutamyl-β-cyano-Ala by GGT or to Asn and Asp by the gene product of NIT4.
The pioneer study (Riemenschneider et al., 2005) on down-regulation of CSases in antisense potato (Solanum tuberosum) plants describes a predominant role of cytosolic CSase, which is in agreement with this study. However, the antisense potato plants unexpectedly showed slight increases in Cys and GSH levels, with some alterations in several amino acids. This apparent discrepancy in Cys and GSH levels in antisense potato plants compared with Arabidopsis knockout mutants in this study might be explained as follows. First, because the activity of Cys formation is regulated by the ratio of CSase and Ser acetyltransferase (SATase; EC 2.3.1.30) via the CSase/SATase complex (Wirtz and Hell, 2007), this mechanism is more pronounced in CSase antisense plants, where a substantial amount of residual CSase protein remains to form the CSase/SATase complex, than in Arabidopsis knockout mutants. Partial suppression of CSase might cause an increase in the SATase/CSase ratio, resulting in elevated levels of Cys and GSH. In contrast, because no residual CSase isoform protein remains in Arabidopsis T-DNA knockout plants, regulation by the CSase/SATase complex is not effective, thus causing a decrease in Cys and GSH levels as expected. Second, the possible differences in the activity of Cys desulfhydration, the side reaction of CSase, in potato and Arabidopsis may explain the apparent discrepancy. This side activity of Cys degradation when partially suppressed was presumed to cause the unexpected increase in Cys and GSH levels eventually (Riemenschneider et al., 2005). In contrast to what has been believed, if CSase involvement is greater in the degradation of Cys and, consequently, in its recycling, the possible differences in Cys desulfhydrase activity in potato and Arabidopsis enzymes might largely affect the changes in Cys levels. Because it is well known that Cys synthesis is regulated by a number of factors, such as posttranslational regulation, protein complex, feedback regulation, and subcellular compartmentation, further detailed analysis, particularly on the protein levels, is required for better understanding of the system.
A Single Knockout Is Compensated by Other Bsas Genes
The Cys synthetic system comprising CSase and SATase is localized in all cellular compartments where protein synthesis occurs (Lunn et al., 1990; Leustek and Saito, 1999; Saito, 2004). The reason for this is thought to be that transport of Cys between cellular compartments is limited. The knockout mutants of Bsas1;1, Bsas2;1, and Bsas2;2, which are the major CSases in cytosol, plastids, and mitochondria, respectively, were able to grow without any visible phenotypic changes under our normal growth conditions, although some changes have been seen in the enzymatic activity and thiol levels. This observation suggests that Cys or compounds converted from or to Cys were able to transfer across compartments to compensate for the loss of Cys in each compartment of these mutants, or that the reduced activity in each compartment might be compensated in part by other Bsas in different compartments. The problem of transport of Cys and its precursors or derivatives, across compartments, awaits elucidation.
Cytosolic Bsas1;1 Is a Predominant Form for Cys Synthesis in Arabidopsis
Cytosolic Bsas1;1 was most abundant in the leaf and root in Arabidopsis (Figs. 1 and 3). The importance of the cytosolic CSase isoform under cadmium stress has also been reported in Arabidopsis (Dominguez-Solis et al., 2001) and in potato by antisense experiments (Riemenschneider et al., 2005). High CSase activity in plastids may make sense in terms of sulfide supply for net Cys synthesis because sulfide is produced only in plastids (Leustek and Saito, 1999; Saito, 2000, 2004). In terms of OAS supply, high CSase activity in mitochondria is more relevant because high activity of OAS-producing SATase was found in the mitochondria of various plant species, such as Arabidopsis, Spinacea oleracea, and Pisum sativum (Droux, 2003). Nevertheless, the activity of SATase was also reported in three compartments: cytosol, plastids, and mitochondria (Smith, 1972; Ascano and Nicholas, 1977; Brunold and Suter, 1982; Ruffet et al., 1995). This apparent difference of major localization of CSase and SATase may be explained by species-specific differences in the regulation of Cys synthesis. From biochemical and molecular studies, either cytosolic or plastidic SATase was reported to be feedback inhibited by the physiological level of Cys in a plant species-dependent manner (Noji et al., 1998; Noji and Saito, 2002; Droux, 2003). The Cys-sensitive SATase may be important for metabolic regulation in Cys synthesis. In S. oleracea and P. sativum, SATase in plastids—where a highly active CSase is present—was Cys sensitive (Kuske et al., 1996; Noji and Saito, 2002; Droux, 2003). In Arabidopsis, SATase in the cytosol, where CSase activity was highest, was also Cys sensitive (Noji et al., 1998). The cytosol may be the most important compartment for Cys synthesis in Arabidopsis. This observation suggests that sulfur metabolism in the model plant Arabidopsis was not necessarily applicable to other plant species. Free sulfide released from plastids and OAS released from the mitochondria in the cytosol might be incorporated into Cys through cytosolic CSase in Arabidopsis. This idea is supported by the low CSase activity in mitochondria of various plant species despite the abundant OAS supply in the mitochondria (Lunn et al., 1990; Kuske et al., 1996; Droux, 2003). Furthermore, the sulfate reduction pathway may be able to provide sufficient sulfide from plastids to cytosol, and, eventually, OAS may limit Cys synthesis rather than sulfide (Takahashi and Saito, 1996; Woehl et al., 1996; Blaszczyk et al., 1999; Harms et al., 2000; Noji and Saito, 2002; Tsakraklides et al., 2002; Wirtz and Hell, 2003; cited in Wirtz et al., 2004). It might be worthwhile to mention that the OAS levels in the Arabidopsis knockout plants were unaltered in this study.
Isoform-Dependent Differential Action of Bsas as CSase and CASase in Vivo
It was reported that CSase and CASase could carry out both reactions with different substrate affinities and efficiency (Ikegami et al., 1993; Hatzfeld et al., 2000; Jost et al., 2000). Bsas1;1 is the predominant CSase and Bsas3;1 is the true CASase in Arabidopsis. Significant reduction in both CSase and CASase activity was observed in bsas1;1. In contrast, significant reduction in CASase activity alone was observed in bsas3;1 (Figs. 3 and 4). These results suggest that Bsas1;1 probably carries out the CASase reaction, whereas Bsas3;1 probably does not perform the CSase reaction in vivo. This result is consistent with the data from biochemical analysis of recombinant proteins (Hatzfeld et al., 2000; Jost et al., 2000). Furthermore, in bsas3;1, the residual CASase activity and the small amounts of β-cyano-Ala and γ-glutamyl-β-cyano-Ala suggest that Bsas1;1 can carry out the CASase reaction instead of Bsas3;1 to a limited extent.
Detection of γ-Glutamyl-β-Cyano-Ala as a Metabolite of β-Cyano-Ala and Physiological Roles of CASase in Arabidopsis
CASase is presumed to be involved in cyanide detoxification and amino acid metabolism (Blumenthal et al., 1968; Manning, 1988). β-Cyano-Ala produced by CASase is further metabolized to Asns in most plants (Blumenthal et al., 1963; Castric et al., 1972). Arabidopsis nitrilase 4 (NIT4), which metabolizes β-cyano-Ala to Asns and Asps, has been cloned from Arabidopsis (Piotrowski et al., 2001). No remarkable changes in the levels of β-cyano-Ala and other amino acids in bsas3;1 and bsas1;1 were observed despite significant changes in CAS activity. In this study, we discovered that β-cyano-Ala is converted to γ-glutamyl-β-cyano-Ala as the presumable storage form in Arabidopsis (Fig. 8B). The content of γ-glutamyl-β-cyano-Ala in bsas3;1 decreased remarkably when compared with the wild type, indicating the primary involvement of Bsas3;1 in the mitochondria for synthesis of β-cyano-Ala and its presumable storage form γ-glutamyl-β-cyano-Ala. In Arabidopsis, three functional GGT isoforms localized in the apoplast (GGT1 and GGT2) and the vacuole (GGT3) were reported (Ohkama-Ohtsu et al., 2007a, 2007b). If γ-glutamylation could be carried out by the action of these GGTs, β-cyano-Ala needs to be transported from the mitochondria to these sites in the cytosol where NIT4 is present. Further investigation is required to better understand β-cyano-Ala metabolism with respect to its localization.
This knowledge on cyanide metabolism in Arabidopsis raises the question of the significance of this metabolic process. Thus far, CASase has been considered to detoxify cyanide produced during ethylene production. However, its role in cyanide detoxification remains to be clarified (Meyer et al., 2003). Our study showed that the bsas3;1 mutant could grow normally and did not suffer any growth retardation as a result of cyanide treatments (data not shown), although CASase activity and the content of γ-glutamyl-β-cyano-Ala in this mutant were much lower than those in the wild type. This suggests that other enzymes might be involved in cyanide detoxification. It is possible that rhodanese (thiosulfate sulfurtransferase; EC 2.8.1.1) and mercaptopyruvate sulfurtransferase (EC 2.8.1.2), both of which catalyze the formation of thiocyanate from thiosulfate and mercaptopyruvate, respectively, could be responsible for cyanide detoxification. Both enzymes are responsible for cyanide detoxification in mammals (Williams, 1959; Ansell and Lewis, 1970). Two mercaptopyruvate sulfurtransferases have been cloned and characterized from Arabidopsis (Hatzfeld and Saito, 2000; Nakamura et al., 2000; Papenbrock and Schmidt, 2000).
The bsas3;1 mutant could grow similarly to wild-type plants despite its low γ-glutamyl-β-cyano-Ala content under cyanide treatment (data not shown). This suggests that the accumulation of γ-glutamyl-β-cyano-Ala might have another physiological role in plant cells. Both β-cyano-Ala and γ-glutamyl-β-cyano-Ala are known to be the chief neurotoxic principals of V. sativa seed (Harper and Arscott, 1962; Ressler, 1962; Ressler et al., 1963, 1969). Similarly, β-cyano-Ala and γ-glutamyl-β-cyano-Ala in Arabidopsis might act as defense molecules against predators. This possibility needs to be experimentally evaluated by further study.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana ecotype Col-0) plants were used as the wild type in this study. Plants were cultured on germination medium (GM)-agar medium containing 1% Suc (Valvekens et al., 1988) in a growth chamber at 22°C under 16-h-light (approximately 2,500 lux)/8-h-dark cycles for 2 weeks. The leaves and roots of the plants were harvested, immediately frozen with liquid nitrogen, and stored at −80°C until use. Identical plant materials were analyzed for their gene expression, enzyme activities, and metabolite profiles.
Screening of T-DNA Mutants
The T-DNA-inserted mutants of Arabidopsis line 072213 (bsas1;1), line 021183 (bsas2;1), line 000860 (bsas2;2), line 022479 (bsas3;1), line 097875 (bsas4;1), line 092696 (bsas4;2), line 103855 (bsas4;3), and line 034133 (bsas5;1) were obtained from the Salk Institute. Homozygous mutants were identified by following the protocol described at the Salk Insertional Mutant Database by using a PCR method (Alonso et al., 2003). The primer list for the screening of homozygous bsas mutants is provided in detail in Supplemental Table S1. LBa1 primer sequence (5′-TGGTTCACGTAGTGGGCCATCG-3′) is accessible at http://signal.salk.edu/tdna_FAQs.html.
Real-Time PCR and RT-PCR Analysis
Total RNA of the wild type and each bsas mutant was extracted with the RNeasy plant mini kit (Qiagen), and cDNA was synthesized with SuperScript II RNase H− reverse transcriptase (Invitrogen) following the manufacturer's instructions. One hundred nanograms of cDNA were used for quantitative real-time PCR analysis. The primer list for real-time PCR is provided in detail in Supplemental Table S2. SYBR green real-time PCR master mix (Toyobo) was used for amplification according to the protocols provided by the supplier. RT-PCR analysis was standardized based on equal quantities of cDNA samples and the respective plasmid DNA or PCR product of each gene was used to draw a standard curve. Primers were designed for Actin2, as described by Himanen et al. (2002). The primer list for RT-PCR is provided detail in Supplemental Table S3. Primers were designed for α-tubulin, as described by Ludwig et al. (1987). The PCR program for amplification was as follows: 94°C for 3 min, 24 to 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. PCR products were analyzed on 1% (for α-tubulin and Bsas5;1) or 2% (for other Bsas genes) agarose gel.
Assay of CSase and CASase Enzymatic Activity
The enzymatic activity of CSase was determined in the reaction mixture (50 μL) containing 50 mm potassium phosphate (pH 8.0), 5 mm Na2S, and 12.5 mm OAS. The reaction was performed at 30°C for 10 min and terminated by the addition of 10 μL of 7.5% (w/v) TCA. The Cys produced was quantified by spectrophotometry using the acid-ninhydrin method at 560 nm (Gaitonde, 1967). The enzymatic activity of CASase was determined in the reaction mixture (1 mL) containing 107.3 mm Tris base, 5.7 mm Cys-HCl, 1.5 mm lead acetate, and 3.1 mm potassium cyanide. The reaction was performed at 25°C for 1 h with the appearance of a brown precipitate due to the presence of lead sulfide formed by lead cations combining with bisulfide anions and monitored at 550 nm (Warrilow and Hawkesford, 1998).
Determination of Cys and GSH Contents
Quantitative analyses of reduced forms of Cys and GSH were performed by a combination of monobromobimane fluorescent labeling and HPLC (Anderson, 1985; Fahey and Newton, 1987). Rosette leaves and roots were homogenized in 3 volumes of 0.1 m HCl (fresh-weight basis) with mixer mill MM 300 (Qiagen). A mixture of 20 μL of extract and 40 μL of 25 μm N-acetyl-Cys as the internal standard was reacted with 5 μL of 30 mm monobromobimane in acetonitrile and 10 μL of 8.5 mm N-ethylmorpholine for 20 min at 37°C in the dark. The labeling reaction was terminated by the addition of 10 μL of acetic acid and the resulting solution was then subjected to HPLC analysis. HPLC was carried out as described previously (Saito et al., 1994).
Nontargeted GC-TOF/MS and CE-TOF/MS Analysis
Sixteen plants were planted on a single plate separated into fourths to minimize the differences in growth conditions. Four wild-type plants were planted on one-fourth, and four bsas mutant plants were planted on each of the remaining fourths. Five plates were replicated for each bsas mutant. Each sample was extracted with a concentration of 25 mg fresh weight of tissues per microliter of the extraction medium (methanol:chloroform:water [3:1:1; v/v/v]) by using a Retsh mixer mill MM 310 at a frequency of 30 Hz−1 for 3 min at 4°C. After centrifugation for 5 min at 15,100g, 400 μL of the supernatant of each plate were put together in accordance with each section of fourths. Four hundred microliters of the 2-mL supernatant were used for GC-TOF/MS analysis, and another 400 μL were used for CE-TOF/MS analysis.
The analysis of metabolites by GC-TOF/MS, including the derivatization step and the processing of MS data, was performed as described elsewhere (Kusano et al., 2007a, 2007b).
Analysis of metabolites by CE-TOF/MS was performed using an Agilent CE capillary electrophoresis system (Agilent Technologies), an Agilent G3250AA LC/MSD TOF system (Agilent Technologies), an Agilent 1100 series binary HPLC pump, and the Agilent G1603A CE-MS adapter and Agilent G1607A CE-ESI-MS sprayer kit. Agilent G2201AA ChemStation software for CE and Analyst QS software for TOF/MS were used. For cationic compounds, separations were carried out using a fused silica capillary (50 μm i.d. × 100 cm total length) filled with 1 m formic acid as the electrolyte. The sample solutions were injected at 50 mbar for 15 s (15 nL). Prior to each run, the capillary was flushed with electrolyte for 5 min. The applied voltage was set at 30 kV. The capillary temperature was maintained at 20°C, and the sample tray was cooled below 4°C. Fifty percent (v/v) methanol-water containing 0.5 μm reserpine was delivered as the sheath liquid at 10 μL min−1. ESI-TOF/MS was conducted in the positive ion mode and the capillary voltage was set at 4 kV. A flow rate of heated dry nitrogen gas (heater temperature 300°C) was maintained at 10 psig. In TOF/MS, the fragmentor, skimmer, and Oct RFV voltage were set at 110, 50, and 160 V, respectively. In acquiring a fragment ion mass spectrum, the fragmentor voltage was increased to 210 V. Automatic recalibration of each acquired spectrum was performed using reference masses of reference standards. The methanol dimer ion ([2M + H]+; m/z 65.0597) and reserpine ([M + H]+; m/z 609.2806) provided the lock mass for exact mass measurements. Exact mass data were acquired at a rate of 1.5 cycles s−1 over a 50 to 1,000 m/z range. Analysis of anionic compounds and nucleotides was carried out as described previously (Soga et al., 2002a, 2002b).
Statistical Data Analysis
The obtained data matrix (observations: samples, variables: 337 peaks, including peaks with mass spectral tags [137 annotated peaks from GC-TOF/MS plus 200 annotated or stably appearing peaks from CE-TOF/MS analysis; Supplemental Tables S4 and S5]) was used for statistical analysis. For statistical multivariate analysis, principal component analysis was performed with SIMCA-P 11.0 software, using log10-transformed and autoscaled data (Umetrics AB). Simple comparisons of means of obtained peak areas were performed by Welch's t test. A difference of P < 0.05 was considered to be significant. In terms of minimizing the effect of zero substitution, we scaled all normalized peak areas by 10,000 and then added 1 uniformly.
Synthesis of γ-Glutamyl-β-Cyano-Ala by GGT
γ-Glutamyl-β-cyano-Ala was synthesized in the reaction mixture (100 μL) containing 10 mm Tris base, 10 mm β-cyano-Ala, 10 mm Glu, and 1 unit of GGT. GGT was purchased from Sigma (product no. G9270–100UN). The reaction was performed at 25°C for 30 min and terminated by addition of 100 μL of ethanol.
Determination of γ-Glutamyl-β-Cyano-Ala Contents by HPLC
Rosette leaves were extracted twice in 10 volumes (fresh-weight basis) of 80% ethanol at 50°C. The supernatant was evaporated and resuspended in 5 volumes (fresh-weight basis) of water for HPLC analysis. γ-Glutamyl-β-cyano-Ala was detected by fluorescence spectrophotometry after postcolumn reaction with O-phthalaldehyde using HPLC according to Kim et al. (1997).
β-Cyano-Ala Treatment
Wild type and bsas3;1 were grown for 3 weeks on GM-agar medium. Plants were transferred to GM-agar medium or medium containing β-cyano-Ala (100 μm and 1 mm) and cultured for 3 d. Leaves were harvested and γ-glutamyl-β-cyano-Ala contents were measured by HPLC.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_117574 (Bsas1;1), NM_113145 (Bsas1;2), NM_129937 (Bsas2;1), NM_115838 (Bsas2;2), NM_116009 (Bsas3;1), NM_122685 (Bsas4;1), NM_111366 (Bsas4;2), NM_122686 (Bsas4;3), NM_111234 (Bsas5;1), NM_112764 (Actin2), NM_121982 (α-tubulin).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Predicted amino acid sequences of Arabidopsis Bsas genes.
Supplemental Figure S2. Schematic diagram of the insertion for bsas mutants.
Supplemental Figure S3. Principal component analysis score scatter plot of wild type and bsas mutants.
Supplemental Figure S4. Identification of γ-glutamyl-β-cyano-Ala by comparison with the synthetic product of GGT from β-cyano-Ala and Glu.
Supplemental Table S1. Primer design for bsas mutant screening.
Supplemental Table S2. Primer design for real-time PCR analysis.
Supplemental Table S3. Primer design for RT-PCR analysis.
Supplemental Table S4. List of annotated peaks detected by GC-TOF/MS.
Supplemental Table S5. List of annotated peaks detected by CE-TOF/MS.
Supplementary Material
Acknowledgments
We thank Dr. Pär Jonsson (Umeå University, Sweden) and Dr. Thomas Moritz (Umeå Plant Science Centre, Sweden) for providing the scripts for processing of GC-TOF/MS data; Mr. Makoto Kobayashi and Ms. Naomi Hayashi (RIKEN Plant Science Center, Japan) for excellent technical support on GC-TOF/MS analysis; and Mr. Ryo Nakabayashi (Chiba University, Japan) for identification of γ-glutamyl-β-cyano-Ala.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Culture, Sports and Technology, Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kazuki Saito (ksaito@faculty.chiba-u.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson ME (1985) Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol 113 548–555 [DOI] [PubMed] [Google Scholar]
- Ansell M, Lewis FAS (1970) A review of cyanide concentrations found in human organs: a survey of literature concerning cyanide metabolism, “normal,” non-fatal, and fatal body cyanide levels. J Forensic Med 17 148–155 [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Ascano A, Nicholas DJD (1977) Purification and properties of O-acetyl-L-serine sulphydrylase from wheat leaves. Phytochemistry 16 889–893 [Google Scholar]
- Barroso C, Vega JM, Gotor C (1995) A new member of the cytosolic O-acetylserine (thiol) lyase gene family in Arabidopsis thaliana. FEBS Lett 363 1–5 [DOI] [PubMed] [Google Scholar]
- Blaszczyk A, Brodzik R, Sirko A (1999) Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J 20 237–243 [DOI] [PubMed] [Google Scholar]
- Blumenthal SG, Butler GW, Conn EE (1963) Incorporation of hydrocyanic acid labeled with carbon-14 into asparagine in seedlings. Nature 197 718–719 [Google Scholar]
- Blumenthal SG, Hendrickson HR, Abrol YP, Conn EE (1968) Cyanide metabolism in higher plants. 3. The biosynthesis of β-cyanoalanine. J Biol Chem 243 5302–5307 [PubMed] [Google Scholar]
- Braun JP, Bardies J, Thouvenot JP, Benard P, Rico AG (1982) Serum gamma-glutamyltransferase in equids: reference physiologic values. Am J Vet Res 43 339–340 [PubMed] [Google Scholar]
- Brunold C, Suter M (1982) Intracellular localization of serine acetyltransferase in spinach leaves. Planta 155 321–327 [DOI] [PubMed] [Google Scholar]
- Castric PA, Farnden KJF, Conn EE (1972) Cyanide metabolism in higher plants. V. The formation of asparagine from β-cyanoalanine. Arch Biochem Biophys 152 62–69 [DOI] [PubMed] [Google Scholar]
- Dominguez-Solis JR, Gutierrez-Alcala G, Romero LC, Gotor C (2001) The cytosolic O-acetylserine (thiol) lyase gene is regulated by heavy metals and can function in cadmium tolerance. J Biol Chem 276 9297–9302 [DOI] [PubMed] [Google Scholar]
- Droux M (2003) Plant serine acetyltransferase: new insights for regulation of sulphur metabolism in plant cells. Plant Physiol Biochem 41 619–627 [Google Scholar]
- Droux M (2004) Sulfur assimilation and the role of sulfur in plant metabolism: a survey. Photosynth Res 79 331–348 [DOI] [PubMed] [Google Scholar]
- Fahey RC, Newton GL (1987) Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol 143 85–96 [DOI] [PubMed] [Google Scholar]
- Gaitonde MK (1967) A spectrophotometric method for the direct measurement of Cys in the presence of other naturally occurring amino acids. Biochem J 104 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms K, von Ballmoos P, Brunold C, Höfgen R, Hesse H (2000) Expression of a bacterial serine acetyltransferase in transgenic potato plants leads to increased levels of Cys and glutathione. Plant J 22 335–343 [DOI] [PubMed] [Google Scholar]
- Harper JA, Arscott GH (1962) Toxicity of common and hairy vetch seed for poults and chicks. Poult Sci 41 1968–1974 [Google Scholar]
- Hatzfeld Y, Maruyama A, Schmidt A, Noji M, Ishizawa K, Saito K (2000) β-Cyanoalanine synthase is a mitochondrial Cys synthase-like protein in spinach and Arabidopsis. Plant Physiol 123 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld Y, Saito K (2000) Evidence for the existence of rhodanese (thiosulfate: cyanide sulfurtransferase) in plants: preliminary characterization of two rhodanese cDNAs from Arabidopsis thaliana. FEBS Lett 470 147–150 [DOI] [PubMed] [Google Scholar]
- Hell R, Bork C, Bogdanova N, Frolov I, Hauschild R (1994) Isolation and characterization of two cDNAs encoding for compartment specific isoforms of O-acetylserine (thiol) lyase from Arabidopsis thaliana. FEBS Lett 351 257–262 [DOI] [PubMed] [Google Scholar]
- Hesse H, Höfgen R (1998) Isolation of cDNAs encoding cytosolic (accession no. AF044172) and plastidic (accession no. AF044173) Cys synthase isoforms from Solanum tuberosum. Plant Physiol 116 1604 [Google Scholar]
- Hesse H, Lipke J, Altmann T, Höfgen R (1999) Molecular cloning and expression analysis of mitochondrial and plastidic isoforms of Cys synthase (O-acetylserine(thiol)lyase) from Arabidopsis thaliana. Amino Acids 16 113–131 [DOI] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, Engler JD, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami F, Itagaki S, Murakoshi I (1993) Purification and characterization of two forms of Cys synthase from Allium tuberosum. Phytochemistry 32 31–34 [Google Scholar]
- Ikegami F, Murakoshi I (1994) Enzymic synthesis of non-protein β-substituted alanines and some higher homologues in plants. Phytochemistry 35 1089–1104 [Google Scholar]
- Jost R, Berkowitz O, Wirtz M, Hopkins L, Hawkesford MJ, Hell R (2000) Genomic and functional characterization of the oas gene family encoding O-acetylserine (thiol) lyases, enzymes catalyzing the final step in Cys biosynthesis in Arabidopsis thaliana. Gene 253 237–247 [DOI] [PubMed] [Google Scholar]
- Kim H, Awazuhara M, Hayashi H, Chino M, Fujiwara T (1997) Analysis of O-acetyl-L-serine in in vitro cultured soybean cotyledons. In WJ Cram, LJ De Kok, I Stulen, C Brunold, H Rennenberg, eds, Sulphur Metabolism in Higher Plants. Backhuys Publishers, Leiden, The Netherlands, pp 307–309
- Kusano M, Fukushima A, Arita M, Jonsson P, Moritz T, Kobayashi M, Hayashi N, Tohge T, Saito K (2007. a) Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst Biol 1 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Fukushima A, Kobayashi M, Hayashi N, Jonsson P, Moritz T, Ebana K, Saito K (2007. b) Application of a metabolomics method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenotyping of natural variants in rice. J Chromatogr B Analyt Technol Biomed Life Sci 855 71–79 [DOI] [PubMed] [Google Scholar]
- Kuske CR, Hill KK, Guzman E, Jackson PJ (1996) Subcellular location of O-acetylserine sulfhydrylase isoenzymes in cell cultures and plant tissues of Datura innoxia mill. Plant Physiol 112 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Saito K (1999) Sulfate transport and assimilation in plants. Plant Physiol 120 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Oppenheimer DG, Silflow CD, Snustad DP (1987) Characterization of the α-tubulin gene family of Arabidopsis thaliana. Proc Natl Acad Sci USA 84 5833–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Droux M, Martin J, Douce R (1990) Localization of ATP sulfurylase and O-acetylserine (thiol) lyase in spinach leaves. Plant Physiol 94 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K (1988) Detoxification of cyanide by plants and hormone action. In Ciba Foundation, eds, Cyanide Compounds in Biology. John Wiley & Sons, Chichester, UK, pp 92–110 [DOI] [PubMed]
- Maruyama A, Ishizawa K, Takagi T, Esashi Y (1998) Cytosolic β-cyanoalanine synthase activity attributed to Cys synthases in cocklebur seeds. Plant Cell Physiol 39 671–680 [DOI] [PubMed] [Google Scholar]
- Maruyama A, Ishizawa K, Takagi T (2000) Purification and characterization of β-cyanoalanine synthase and Cys synthases from potato tubers: are β-cyanoalanine synthase and mitochondrial Cys synthase same enzyme? Plant Cell Physiol 41 200–208 [DOI] [PubMed] [Google Scholar]
- Maruyama A, Saito K, Ishizawa K (2001) β-Cyanoalanine synthase and Cys synthase from potato: molecular cloning, biochemical characterization, and spatial and hormonal regulation. Plant Mol Biol 46 749–760 [DOI] [PubMed] [Google Scholar]
- Meyer T, Burow M, Bauer M, Papenbrock J (2003) Arabidopsis sulfurtransferases: investigation of their function during senescence and in cyanide detoxification. Planta 217 1–10 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Koizumi N, Sano H (1997) Isolation of a novel cysteine synthase cDNA (AB003041) from Arabidopsis thaliana. Plant Physiol 114 7479235602 [Google Scholar]
- Nakamura T, Yamaguchi Y, Sano H (2000) Plant mercaptopyruvate sulfurtransferases: molecular cloning, subcellular localization and enzymatic activities. Eur J Biochem 267 5621–5630 [DOI] [PubMed] [Google Scholar]
- Noji M, Goulart Kawashima C, Obayashi T, Saito K (2006) In silico assessment of gene function involved in Cys biosynthesis in Arabidopsis: expression analysis of multiple isoforms of serine acetyltransferase. Amino Acids 30 163–171 [DOI] [PubMed] [Google Scholar]
- Noji M, Inoue K, Kimura N, Gouda A, Saito K (1998) Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J Biol Chem 273 32739–32745 [DOI] [PubMed] [Google Scholar]
- Noji M, Saito K (2002) Molecular and biochemical analysis of serine acetyltransferase and Cys synthase towards sulphur metabolic engineering in plants. Amino Acids 22 231–243 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Radwan S, Peterson A, Zhao P, Badr AF, Xiang C, Oliver DJ (2007. a) Characterization of the extracellular γ-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. Plant J 49 865–877 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Zhao P, Xiang C, Oliver DJ (2007. b) Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J 49 878–888 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Schmidt A (2000) Characterization of two sulfurtransferase isozymes from Arabidopsis thaliana. Eur J Biochem 267 5571–5579 [DOI] [PubMed] [Google Scholar]
- Piotrowski M, Schonfelder S, Weiler EW (2001) The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode β-cyano-L-alanine hydratase/nitrilase. J Biol Chem 276 2616–2621 [DOI] [PubMed] [Google Scholar]
- Ressler C (1962) Isolation and identification from common vetch of the neurotoxin β-cyano-L-alanine, a possible factor in neurolathyrism. J Biol Chem 237 733–735 [PubMed] [Google Scholar]
- Ressler C, Giza YH, Nigam SN (1963) Biosynthesis and metabolism in species of vetch and lathyrus of γ-glutamyl-β-cyanoalanine: relation to the biosynthesis of asparagine. J Am Chem Soc 85 2874–2875 [Google Scholar]
- Ressler C, Nigam SN, Giza YH (1969) Toxic principle in vetch. Isolation and identification of γ-L-glutamyl-L-β-cyanoalanine from common vetch seeds. Distribution in some legumes. J Am Chem Soc 91 2758–2765 [DOI] [PubMed] [Google Scholar]
- Riemenschneider A, Riedel K, Hoefgen R, Papenbrock J, Hesse H (2005) Impact of reduced O-acetylserine (thiol) lyase isoform contents on potato plant metabolism. Plant Physiol 137 892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffet ML, Lebrun M, Droux M, Douce R (1995) Subcellular distribution of serine acetyltransferase from Pisum sativum and characterization of an Arabidopsis thaliana putative cytosolic isoform. Eur J Biochem 227 500–509 [DOI] [PubMed] [Google Scholar]
- Saito K (2000) Regulation of sulfate transport and synthesis of sulfur containing amino acids. Curr Opin Plant Biol 3 188–195 [PubMed] [Google Scholar]
- Saito K (2004) Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol 136 2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Kurosawa M, Tatsuguchi K, Takagi Y, Murakoshi I (1994) Modulation of Cys biosynthesis in plastids of transgenic tobacco overexpressing Cys synthase. O-Acetylserine(thiol)-lyase. Plant Physiol 106 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IK (1972) Studies of L-Cys biosynthetic enzymes in Phaseolus vulgaris L. Plant Physiol 50 477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga T, Ueno Y, Naraoka H, Matsuda K, Tomita M, Nishioka T (2002. a) Pressure-assisted capillary electrophoresis electrospray ionization mass spectrometry for analysis multivalent anions. Anal Chem 74 6224–6229 [DOI] [PubMed] [Google Scholar]
- Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T (2002. b) Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathway by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem 74 2233–2239 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Saito K (1996) Subcellular localization of spinach Cys synthase isoforms and regulation of their gene expression by nitrogen and sulfur. Plant Physiol 112 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakraklides G, Martin M, Chalam R, Tarczynski M, Schmidt A, Leustek T (2002) Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant J 32 879–889 [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow AG, Hawkesford MJ (1998) Separation, subcellular location and influence of sulphur nutrition on isoforms of cysteine synthase in spinach. J Exp Bot 49 1625–1636 [Google Scholar]
- Warrilow AG, Hawkesford MJ (2000) Cys synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. J Exp Bot 51 985–993 [DOI] [PubMed] [Google Scholar]
- Williams RT (1959) The metabolism of nitriles. In RT Williams, ed, Detoxification Mechanisms, Ed 2. Chapman and Hall, London, pp 390–409
- Wirtz M, Droux M, Hell R (2004) O-acetylserine (thiol) lyase: an enigmatic enzyme of plant Cys biosynthesis revisited in Arabidopsis thaliana. J Exp Bot 55 1785–1798 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R (2003) Production of Cys for bacterial and plant biotechnology: application of Cys feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 24 195–203 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R (2007) Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. Plant Cell 19 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehl E, Tai C, Dunn M, Cook P (1996) Formation of the α-aminoacrylate immediate limits the overall reaction catalyzed by O-acetylserine sulfhydrylase. Biochemistry 35 4776–4783 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Nakamura T, Kusano T, Sano H (2000) Three Arabidopsis genes encoding proteins with differential activities for Cys synthase and β-cyanoalanine synthase. Plant Cell Physiol 41 465–476 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.