Abstract
As part of the endosomal sorting complex required for transport (ESCRT) machinery, Tsg101 is essential for endosomal sorting, membrane receptor degradation and the final stages of cytokinesis. Depletion or overproduction of the protein can cause disruption of these vital processes and results in severe consequences for the cell. Tsg101 expression is thus controlled posttranslationally within a narrow range and this autoregulation has been mapped to the C-terminus of the protein. Here we elucidate further the mechanisms of this regulation and describe a novel function of Tsg101-associated ligase (Tal) in mediating this control. We show that Tal polyubiquitinates lysine residues in the C-terminus of uncomplexed Tsg101, resulting in proteasomal degradation. However, accessibility to these lysines is prevented by the presence of the other ESCRT-I proteins. We show that VPS28 is a limiting factor, and consequently Tsg101 expression surplus to ESCRT-I function is vulnerable to degradation. The role of Tal in the regulation of Tsg101 steady-state control is highlighted when Tsg101 is overexpressed; however, our data also suggest that additional ligases regulate Tsg101 expression under normal conditions. Lastly, we demonstrate that while the C-terminal lysines are targets for polyubiquitination, they are not required for any additional function necessary for ESCRT activity.
INTRODUCTION
Tumor susceptibility gene 101 (Tsg101) forms part of the endosomal sorting complex required for transport (ESCRT-I), which together with ESCRT-0, -II, and -III is involved in ubiquitinated cargo degradation and multivesicular body (MVB) biogenesis, an essential mechanism for down-regulation of signaling by activated growth factors (Babst et al., 2000; Katzmann et al., 2001; Katzmann et al., 2002; Williams and Urbe, 2007). The ESCRT machinery assembles on late endosomal membranes where it sorts and concentrates cargo, such as ubiquitinated epidermal growth factor receptor (EGFR), into invaginating vesicles. Once late endosomes mature into MVBs and the ESCRT complexes are recycled by the AAA-ATPase VPS4 (Babst et al., 1998), the limiting (outer) membrane fuses with the lysosome, exposing the content of the internal vesicles to degradation by lysosomal hydrolases.
Tsg101 and the ESCRT machinery is also recruited by some enveloped viruses to facilitate budding at the plasma membrane, a process topologically equivalent to vesicle budding at the limiting membrane of the MVB (Garrus et al., 2001; Martin-Serrano et al., 2001; VerPlank et al., 2001; Demirov et al., 2002; Bieniasz, 2006). Retroviruses and other enveloped viruses encode short peptide motifs, the so-called late budding domains (L-domains), which recruit the ESCRT machinery via interaction with alternative adaptor proteins in the class E vacuolar protein sorting (VPS) pathway (Martin-Serrano et al., 2003a; Strack et al., 2003; von Schwedler et al., 2003). A well-characterized PTAP motif in HIV-1 and Ebola virus functions as a L-domain by recruitment of the ESCRT machinery through binding to the ubiquitin E2 variant (UEV) domain in Tsg101.
The ESCRT proteins were originally identified in Saccharomyces cerevisiae as class E VPS gene products (Raymond et al., 1992; Katzmann et al., 2002). Yeast ESCRT-I has recently been characterized as a heterotetrameic complex consisting of Vps23 (the yeast homologue of Tsg101), Vps28, Vps37 (Katzmann et al., 2001), and a newly identified fourth subunit, Mvb12 (Chu et al., 2006; Curtiss et al., 2007; Gill et al., 2007; Oestreich et al., 2007). Structural studies of the yeast heterotetramer revealed an elongated complex in which a globular headpiece comprising the Vps23:Vps28:Vps37 core interactions (Kostelansky et al., 2006; Teo et al., 2006) is attached to an extended stalk formed by α helices from Vps23, Vps37, and Mvb12 (Kostelansky et al., 2007). The N-terminal domains of Vps23 and Vps37 are attached to the stalk by a flexible linker region. Likewise, the C-terminal portion of Vps28 extends out from the headpiece and in yeast links to the ESCRT-II complex (Pineda-Molina et al., 2006; Gill et al., 2007). The human heterotetrameric ESCRT-I consists of Tsg101, VPS28, one of four isotypes of VPS37 (A–D; Babst et al., 2000; Bishop and Woodman, 2001; Bache et al., 2004; Stuchell et al., 2004; Eastman et al., 2005), and one of two newly identified proteins, MVB12A or B (Morita et al., 2007). As with the yeast complex, the C-terminal domain of Tsg101 is thought to provide the core stability by binding all other proteins in the complex.
Tsg101 was originally isolated in a random screen for potential tumor suppressors (Li and Cohen, 1996), where its inactivation or overexpression was found to cause metastatic growth of murine fibroblasts, but the role of Tsg101 in tumorigenesis remains unclear. However, an important role of Tsg101 in cell viability is illustrated by the cell death and embryonic lethality phenotype observed in cells and mice that lack Tsg101 (Ruland et al., 2001; Krempler et al., 2002; Wagner et al., 2003). Conversely, increased expression of Tsg101 also leads to inhibition of cell growth (Oh et al., 2002). A role of Tsg101 and the ESCRT machinery in a membrane fission event required for the last step of cell division, namely abscission, has been recently established (Carlton and Martin-Serrano, 2007). This study also showed that either reduced or increased expression of Tsg101 inhibits abscission and results in marked defects in cytokinesis. From these findings it is clear that cellular Tsg101 expression levels must be controlled within a narrow range. Indeed, overexpression of exogenous Tsg101 causes a posttranslational reduction in the endogenous protein, thus maintaining constant total Tsg101 expression. This “auto-regulatory” activity was mapped to the C-terminal “steadiness box” (SB) domain in Tsg101 (Feng et al., 2000).
Work by Amit et al. (2004) has identified the interaction of Tsg101 with a conserved RING E3 ubiquitin ligase, namely Tsg101-associated ligase (Tal). A recycling model has been proposed whereby multiple mono-ubiquitination of the C-terminal region of Tsg101 by Tal inactivates Tsg101 by shuttling it between a membrane-bound active form and an inactive soluble form. We have characterized this interaction further and found an additional action of Tal in Tsg101 protein steady-state control. Our data are consistent with a model in which Tal, and possibly other ubiquitin ligases, target excess Tsg101 for proteasomal degradation by polyubiquitinating lysines in the C-terminal region that overlaps with the previously described steadiness box. In contrast, when Tsg101 is bound to other ESCRT-I subunits, polyubiquitination of Tsg101 would be inhibited either by masking of the target lysines in Tsg101, or alternatively, the binding of these ESCRT-I subunits would sterically prevent the formation of a productive Tal/Tsg101/Ub complex.
MATERIALS AND METHODS
Plasmid Construction
All yeast two-hybrid, yellow fluorescent protein (YFP), Cherry, and glutathione S-transferase (GST)-tagged full-length Tsg101, YFP, cyan fluorescent protein (CFP), or Myc-VPS28, YFP-Cep55, and hemagglutinin (HA)-tagged ubiquitin plasmids have been described elsewhere (Martin-Serrano et al., 2003a,b, 2004; Carlton and Martin-Serrano, 2007). Deletion and substitution mutants of Tsg101 were derived using PCR-based methods, and RNA interference (RNAi)-resistant constructs were created by methods previously described (Carlton and Martin-Serrano, 2007). Tal and Mahogunin coding sequences were amplified by PCR from IMAGE clones 2988664 and 5755971, respectively, using primers directed to the 5′ and 3′ ends of the coding sequence and containing EcoRI (Tal) or EcoRI/NotI (Mahogunin) restriction sites. The PCR products were inserted into pCR3.1/YFP, sequenced, and subcloned into pGBKT7 (Clontech, Palo Alto, CA) and pVP16 (Martin-Serrano et al., 2001) for yeast two-hybrid assays and pCR3.1/MYC and/or pCAGGS/GST for protein expression experiments. The Tal deletion constructs were generated by PCR; Tal1-674 lacks the RING domain and Tal1–648 lacks the double PT/SAP and RING domain, and both were cloned using a NotI restriction site at the 3′ end. TalδPTAP was cloned with a two-step PCR method using a HindIII site either side of the PTAP-PSAP motif (Δ649–664) and a 3′ NotI site. The pCMS28/YFP/ENX vector used for generating stable cell lines was a modified gift from Prof. Mike Malim (King's College London) and has been described previously (Carlton and Martin-Serrano, 2007).
Yeast Two-Hybrid Assay
Yeast cells (Y190) were transformed with pGBKT7 and pVP16 fusion protein expression plasmids (1 μg of each), and selected on SD–tryptophan–leucine agar for 3 d at 30°C. Protein–protein interactions were measured by β-galactosidase activity as described previously (Martin-Serrano et al., 2001). Expression of VP16-Tal constructs in yeast lysates was determined by harvesting yeast cells and boiling in SDS sample buffer before analyzing by Western blot with α-HA mAb directed to an N-terminal HA tag on the protein.
Transient Transfection
HEK 293T cells were transfected using polyethylinimine (PEI; Polysciences, Warrington, PA). Plasmid DNA, 1 μg, was incubated with 4 μg PEI in 50 μl serum-free DMEM for 10 min before addition to cells. HeLa cells were transfected using Lipofectamine 2000 (Life Technologies, Rockville, MD) according to the manufacturer's instructions.
Tsg101 Ubiquitination Assays
GST-Tsg101 constructs and HA-tagged ubiquitin were expressed in the presence or absence of YFP-tagged full-length or mutated Tal or YFP-Mahogunin in 293T cells. Forty-eight hours after transfection the cells were harvested, incubated with glutathione-Sepharose beads (Amersham Biosciences, Piscataway, NJ) as described previously (Agromayor and Martin-Serrano, 2006), and washed in the presence of a protease inhibitor cocktail (Roche, Indianapolis, IN) and 5 mM N-ethylmaleimide (de-ubiquitinating enzymes inhibitor), (Sigma, St. Louis, MO). Precipitated GST-Tsg101 and ubiquitinated conjugates were eluted by boiling in 100 μl SDS sample buffer and analyzed by Western blotting with α-Tsg101, α-HA, and α-poly-ubiquitin antibodies.
Microscopy
HeLa cells were seeded onto glass coverslips and transfected with plasmids encoding YFP-Tal and Cherry-Tsg101, with CFP-VPS28 or CFP vector control. Twenty-four hours after transfection cells were fixed with 4% paraformaldehyde for 15 min, washed with PBS, and mounted in Mowiol. Images were taken with a TCS SP2 AOBS confocal microscope (Leica, Deerfield, MA; HCX PL APO CS 63.0×, 1.4 NA oil objective), employing the AOTF to collect relevant narrow emission-λ windows for each fluorophore.
Generation of Stable Cell Lines
Stable cell lines were produced using a puromycin resistance bicistronic retroviral packaging vector (a modified gift from Prof. Mike Malim) encoding N-terminal YFP-Tsg101 fusion proteins. pCMS28/YFP-Tsg101 constructs, 200 ng, were transfected with 100 ng of a vesicular stomatitis virus-G envelope protein expression plasmid and 650 ng of MLV gag/pol in 293T cells with PEI. Forty-eight hours later the viral supernatants were harvested, clarified, and used to transduce HeLa cells. Twenty-four hours after infection stably transduced cells were selected with 2 μg/ml puromycin dihydrochloride (Sigma). Total cell death was achieved in uninfected control at 24 h after adding the antibiotic. At 30 h, infected cells were lysed or split for lysis at future time points (see Figure 5A) or maintained under continual selection.
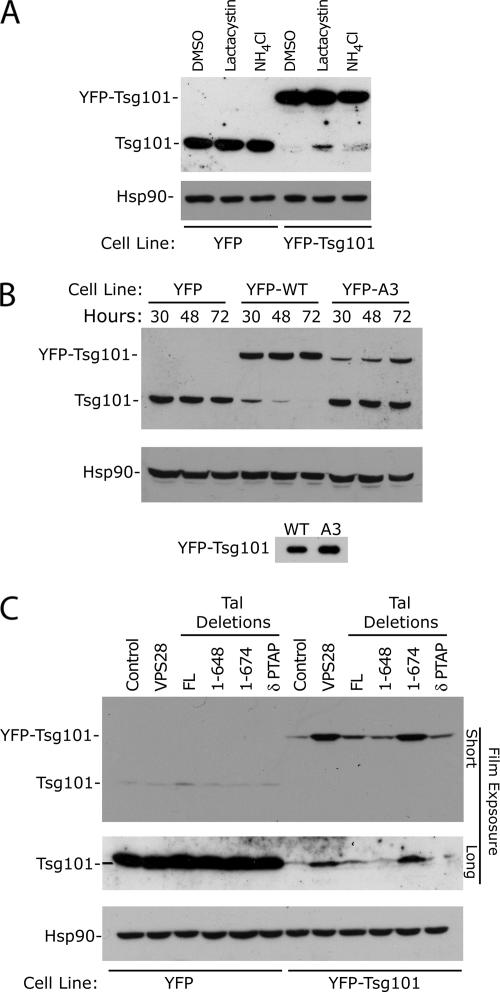
Figure 5.
Investigating cellular mechanisms controlling excess Tsg101. (A) HeLa cells stably expressing YFP or YFP-Tsg101 were treated for 6 h with DMSO (control), lactacystin (proteasome inhibitor), or NH4Cl (lysosome inhibitor) as indicated. Cells were lysed and analyzed by Western blot with α-Tsg101 and α-Hsp90 antibodies. (B) Relative expression of endogenous and exogenous Tsg101 after transduction with retroviral vectors expressing YFP, YFP-Tsg101, or YFP-Tsg101A3 (VPS28 binding site mutant). HeLa cells were lysed at 30, 48, or 72 h after selection and analyzed by Western blot with α-Tsg101 and α-Hsp90 antibodies. The bottom panel shows the relative expression of YFP-Tsg101 and YFP-Tsg101A3 in transiently transfected cells (C) Influence of VPS28 and Tal deletions on Tsg101 expression in stable cell lines. HeLa cells stably expressing YFP or YFP-Tsg101 were transfected with plasmids encoding YFP empty, YFP-VPS28, YFP-Tal, or YFP-Tal deletions as indicated. Cells were lysed 48 h after transfection and analyzed by Western blot using α-Tsg101 and α-Hsp90 antibodies. Exogenous and endogenous Tsg101 expression is detected by short or longer film exposure, respectively.
Proteasome/Lysosome Inhibition Assay
PCMS28/YFP or pCMS28/YFP-Tsg101 expressing stable cell lines were plated, and 12 h later were treated with 1%DMSO media control, 10 mM ammonium chloride (Sigma), or 10 μM clasto-lactacystin β lactone in 1% dimethylsulfoxide (DMSO), 10% fetal calf serum, and DMEM. Six hours after treatment cells were lysed and analyzed by Western blotting with α-Tsg101 and α-Hsp90 antibodies.
Infectivity Assay
Two hours after plating, pCMS28/YFP control or small interfering RNA (siRNA)-resistant Tsg101 mutant cell lines were transfected with siRNA targeting either control (Luciferase) or Tsg101 as described previously (Carlton and Martin-Serrano, 2007). Forty-eight hours later, cells were reseeded and transfected again with siRNA and 300 ng HIV-1 pNL-HXB using Lipofectamine 2000. Forty-eight hours after transfection viral supernatants were clarified and used to infect indicator HeLa-TZM-bl cells (Derdeyn et al., 2000). β-Galactosidase activity in cell lysates was measured 48 h after infection using chemiluminescent detection reagents (Galactostar, Applied Biosystems, Foster City, CA).
Western Blot Analysis
Cell lysates or bead elutates were separated on 7, 10, or 12% acrylamide gels and transferred to nitrocellulose membranes. The blots were probed with mouse monoclonal antibodies α-green fluorescent protein (GFP; Roche), α-HA (HA.11, Covance Laboratories, Madison, WI), α-Tsg101 (4A10, Abcam, Cambridge, MA), α-poly-ubiquitin (FK1, Biomol International, Plymouth Meeting, PA) or α-p24 Gag (183-H12–5C), or rabbit polyclonal antibodies α-Tal (CS-15 or FQ-17, Sigma) or α-Hsp90 (H-114, Santa Cruz Biotechnology, Santa Cruz, CA), followed by a peroxidase-conjugated secondary antibody (α-Mouse AP308, Chemicon, Temecula, CA; or α-Rabbit, Pierce, Rockford, IL). The blots were developed with chemiluminescent substrate reagents (Pierce).
RESULTS
The Main Determinant of Tal-Tsg101 Binding Is the PTAP-UEV Domain Interaction
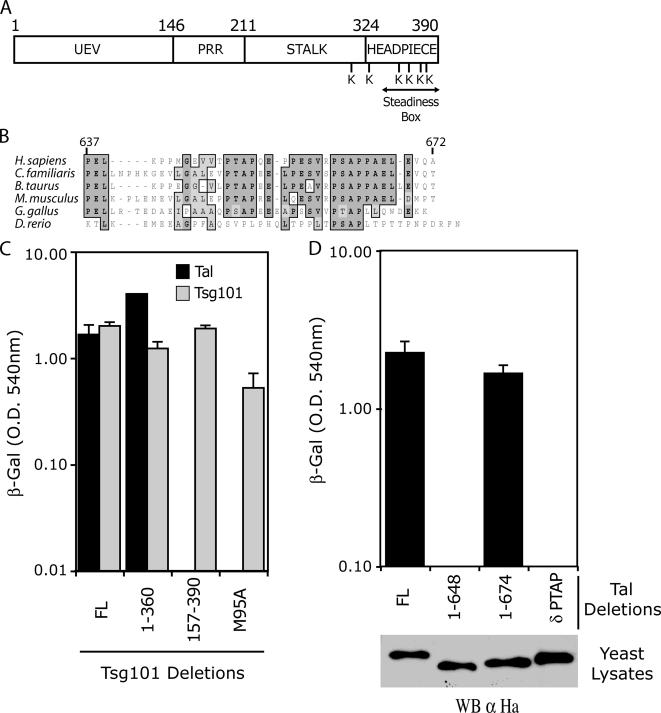
The domain organization of Tsg101 is illustrated in Figure 1A. Tsg101 contains an N-terminal UEV domain that binds P(T/S)AP motifs present in various interacting proteins, such as HIV-1 Gag (Garrus et al., 2001; Martin-Serrano et al., 2001; VerPlank et al., 2001; Demirov et al., 2002; Pornillos et al., 2002a,b), hepatocyte growth factor–regulated tyrosine kinase substrate (Hrs; Bache et al., 2003; Katzmann et al., 2003; Pornillos et al., 2003), VPS37B (Stuchell et al., 2004), ALG-2–interacting protein 1 or ALG-2–interacting protein X (AIP1/Alix; Martin-Serrano et al., 2003a; von Schwedler et al., 2003), and Tal (Amit et al., 2004). Adjacent to the UEV domain, a proline-rich region (PRR) contains the 55-kDa centrosomal protein (Cep55) binding site (Carlton and Martin-Serrano, 2007), a protein required for cytokinesis. This region is linked to a coiled-coil domain, which forms part of the stalk of ESCRT-I. The C-terminal domain binds all other ESCRT-I proteins (Martin-Serrano et al., 2003b; Stuchell et al., 2004; Eastman et al., 2005; Morita et al., 2007), and together they form the headpiece of the complex. Residues 348–390 have been termed the SB because of the autoregulatory activity of this region and its role in maintaining steady-state expression (Feng et al., 2000).
Figure 1.
The main determinant of Tsg101-Tal binding is via a UEV-PTAP interaction. (A) Domain organization of human Tsg101, showing the amino-terminal UEV domain, the proline-rich region, stalk, and headpiece. The C-terminal lysines, and region described as the steadiness box are also indicated. (B) Sequence alignment of the conserved PTAP-PSAP motif in Tal homologues from different species. (C) Yeast two-hybrid assay of Tal–Tsg101 binding, with Tsg101 multimerization as a control. Y190 cells were transformed with GAL4-Tal or GAL-4-Tsg101 expression plasmids along with a plasmid expressing a full-length or deletion mutant VP16-Tsg101 as indicated. The mean level of β-galactosidase expression (±SD) in three pools of transformants is shown. (D) Yeast two-hybrid analysis to determine Tal domains required for Tsg101 binding. Y190 cells were transformed with GAL-4-Tsg101 expression plasmid along with a plasmid expressing full-length or deletion mutant VP16-Tal as indicated. The mean level of β-galactosidase expression and SD of three pools of transformants is shown. Expression of N-terminal HA-tagged VP16-Tal or VP16-Tal deletion mutants in yeast cell lysates was determined by Western blot using α-HA antibodies.
The predicted domain organization of Tal has been described previously (Amit et al., 2004). Functionally defined regions include a C-terminal double PTAP-PSAP motif, which remains evolutionary conserved among vertebrates (Figure 1B), followed by a E3 ligase catalytic RING domain. Previous work describes a bimodal interaction between Tal and Tsg101, in which the UEV domain of Tsg101 interacts with the PTAP-PSAP motif in Tal and the SB interacts with a coiled coil region of Tal (Amit et al., 2004). In this study, we confirmed the Tsg101-Tal interaction, (Figure 1C), and deletion analysis by yeast two-hybrid also showed that binding was lost upon deletion of Tsg101's UEV domain. More specifically, a point mutation in the PTAP binding site (M95A) of Tsg101 (Pornillos et al., 2002a,b) abrogated binding to Tal. The integrity of the different Tsg101 constructs was confirmed by determining their ability to multimerize with the full-length protein (Martin-Serrano et al., 2003b; Figure 1C). In agreement with these results, binding was also undetectable between a Tal mutant lacking the PTAP-PSAP motif (TalδPTAP) and full-length Tsg101 (Figure 1D). Importantly, the different Tal constructs were expressed equivalently as determined by Western blot of the yeast cell lysates. These data suggest that the main determinant of the Tsg101-Tal interaction is the UEV-PTAP binding, and in comparison, the coiled-coil region in Tal and SB in Tsg101 provide a weaker interaction, perhaps important to determine the ubiquitination target in Tsg101.
Tal Specifically Polyubiquitinates and Degrades Tsg101
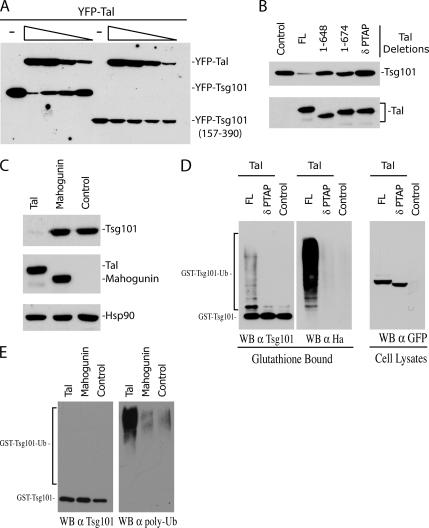
To study the regulation of Tsg101 by Tal, we performed cotransfection experiments in 293T cells. As shown in Figure 2A, coexpression of comparable levels of YFP-Tsg101 and YFP-Tal resulted in a dose-dependent decrease of the steady-state expression of YFP-Tsg101. This effect was not observed with a Tsg101 mutant that lacks the UEV domain and consequently does not bind to Tal (Tsg101157-390). Conversely, a Tal mutant lacking the PT/SAP motifs failed to decrease the steady-state level of Tsg101, confirming the requirement for the PTAP-UEV domain interaction to regulate Tsg101 expression (Figure 2B). Moreover, removal of the RING domain (1-648) also inactivated the degradative action of Tal, indicating that the ubiquitin ligase catalytic activity of Tal is also required to regulate the steady-state expression of Tsg101. Importantly, this phenotype was shown to be a specific function of Tal, as coexpression of Mahogunin, another C3HC4-type E3 ubiquitin ligase that has recently been shown to interact with Tsg101 (Kim et al., 2007), does not result in changes in Tsg101 steady-state expression (Figure 2C).
Figure 2.
Tal specifically degrades Tsg101 via conjugation of polyubiquitin chains. (A) YFP-Tsg101 or YFP-Tsg101157-390 fusion proteins were coexpressed in 293T cells with decreasing amounts of YFP-Tal. Samples were lysed 24 h after transfection and analyzed by immunoblotting with an α-GFP mAb. (B) Coexpression of YFP-Tsg101 fusion protein with GST, GST-fused full-length Tal, or Tal deletions as indicated. Tal1-648 lacks the double PTAP-PSAP motif and RING domain, Tal1-674 lacks the RING domain only and TalδPTAP lacks the PTAP-PSAP motif. Twenty-four hours after transfection cells were lysed and analyzed by Western blot. YFP-Tsg101 expression was detected by immunoblotting with an α-GFP mAb and GST-Tal expression was determined using an α-Tal antibody raised against the N-terminus of Tal. (C) Comparison of GST-Tsg101 expression in the presence of YFP-Tal, YFP-Mahogunin, or empty vector control. Transfected cells were lysed 24 h after transfection and analyzed by Western blot. GST-Tsg101 or YFP- fusion protein expression was detected using α-Tsg101 and α-GFP antibodies, respectively. Lysates were probed with α-Hsp90 as a loading control. (D) Coprecipitation experiment to determine relative ubiquitination of Tsg101 by full-length or mutant Tal. 293T cells were cotransfected with plasmids encoding YFP-Tal, YFP-TalδPTAP, or empty vector with GST-Tsg101 and HA-ubiquitin. Forty-eight hours after transfection cell lysates were precipitated with glutathione-Sepharose beads, and the bound fractions were analyzed by immunoblotting with α-Tsg101 antibody. Samples were normalized for equal Tsg101 expression and probed with α-Tsg101 and α-HA antibodies. YFP-Tal and YFP-TalδPTAP expression in cell lysates was determined using α-GFP antibody. (E) Comparison of GST-Tsg101 polyubiquitination in the presence of YFP-Tal, YFP-Mahogunin or empty vector control. Cell lysates were subjected to precipitation with glutathione-Sepharose beads as before, and analyzed by Western blot. Samples were normalized for equal GST-Tsg101 expression before rerunning and probing with α-Tsg101 and α-polyubiquitin antibodies.
Ubiquitin can be ligated to the target protein in a number of ways. Modification with a single ubiquitin moiety (mono- or multi-monoubiquitination) can alter the substrates location or signal for interaction with other proteins. Addition of a polymeric chain (polyubiquitination), however, is usually a signal for proteasomal degradation (Hicke and Dunn, 2003). To determine if Tal induces polyubiquitinatation of Tsg101, the effect of Tal expression on the relative ubiquitination of Tsg101 was studied by expressing GST-Tsg101 fusions and HA-tagged ubiquitin in 293T cells. A suboptimal dose of Tal was transfected in these experiments to prevent degradation of Tsg101, and the glutathione-bound fractions were normalized for Tsg101 expression (Figure 2D, left panel). In agreement with previous work (Amit et al., 2004) the presence of Tal significantly increased overall ubiquitination of Tsg101 in a PTAP-dependent manner, as determined by anti-HA immunoblot. Detection of Tsg101 with Tsg101-specific antibodies (Figure 2D) reveals a discrete higher molecular weight band, consistent with the addition of a single ubiquitin moiety, but a ubiquitination pattern consistent with polyubiquitination was also observed. The induction of polyubiquitinated Tsg101 by Tal was confirmed by probing the glutathione-bound fraction with an antibody specific for polyubiquitin chains (Figure 2E). As an important control, the polyubiquitination of Tsg101 was not induced by Mahogunin, which has been reported to induce multi-monoubiquitination of Tsg101. These data demonstrate that, although some monoubiquitin modification may also occur, Tal specifically enhances polyubiquitination of Tsg101.
Tal Targets the C-terminus of Tsg101 for Ubiquitination—A Role for VPS28 in Tsg101 Stability
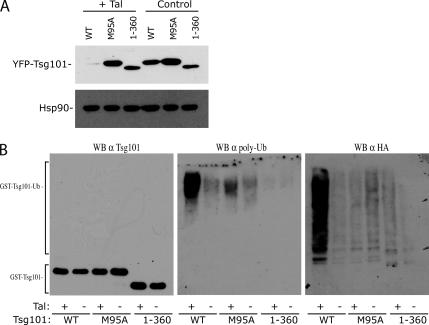
To characterize the regulation of Tsg101 expression by Tal in more detail, we expressed Tsg101 mutants in the presence or absence of Tal. As expected from the binding results obtained in Figure 1, Tsg101M95A was resistant to degradation by Tal. The phenotype observed with this mutant can be explained by the lack of binding to Tsg101, but a C-terminally deleted construct (1-360) that binds Tal was also resistant to degradation (Figure 3A), suggesting that a region at the C-terminus of Tsg101 that overlaps with the steadiness box might be the target of action by Tal. In agreement with these results, Tal was found to increase the polyubiquitination of wild-type, but not C-terminally deleted Tsg101 (Figure 3B).
Figure 3.
Mapping the determinants for Tal-mediated polyubiquitination of Tsg101. (A) Coexpression of wild-type YFP-Tsg101, YFP-Tsg101M95A, or YFP-Tsg1011-360 in the presence of GST-Tal or GST empty vector in 293T cells. Cell lysates were harvested 24 h after transfection and Tsg101 expression was determined by Western blot using α-GFP antibody. Lysates were also probed with α-Hsp90 as a loading control. (B) Pulldown assay to measure relative ubiquitination of wild-type and mutant/truncated GST-Tsg101 by YFP-Tal. GST-Tsg101 constructs were transfected in the presence or absence of YFP-Tal, with HA-ubiquitin in 293T cells. Forty-eight hours after transfection, cell lysates were precipitated with glutathione-Sepharose beads, and the bound fractions were analyzed by immunoblotting with α-Tsg101 antibody. Samples were normalized for equal Tsg101 expression and reprobed with α-Tsg101 (left), α-poly-ubiquitin (middle), and α-HA (right) antibodies.
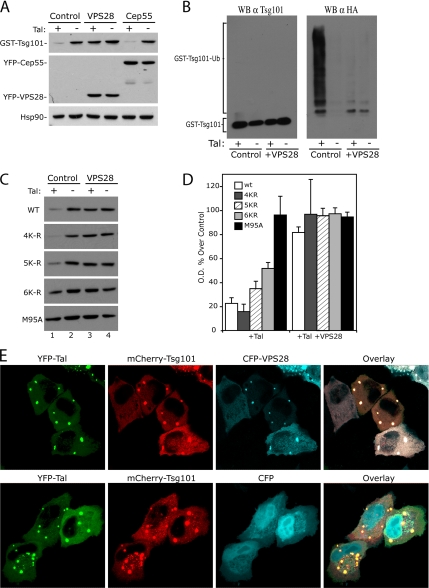
Human VPS28 has also been previously shown to bind to the C-terminal headpiece of Tsg101 (Martin-Serrano et al., 2003b; Stuchell et al., 2004). To determine if VPS28 binding could interfere with the action of Tal on this region, we coexpressed a myc-tagged VPS28 with YFP-Tsg101 and GST-Tal in 293T cells. Interestingly, VPS28 was found to prevent the degradation of Tsg101 by Tal (Figure 4A), and this result correlated with a lack of ubiquitination in the presence of VPS28 (Figure 4B). Similar results were seen upon coexpression of VPS37A–D, which also bind to the C-terminal domain of Tsg101 (data not shown). We show that this is a specific function of the C-terminal binding ESCRT-I proteins, as another Tsg101-binding protein, Cep55, does not prevent degradation of Tsg101 by Tal (Figure 4A). Previous work has shown that VPS28 expression induces a shift in the localization of overexpressed Tsg101 from highly punctate to a diffuse localization (Martin-Serrano et al., 2003b). Using confocal microscopy, we observe that Tsg101 and Tal colocalize in the absence and in the presence of VPS28. Indeed, we observe that all three proteins can localize together in discrete puncta, showing that VPS28 does not prevent the interaction of Tsg101 with Tal. These results suggest that the mechanism of protection by VPS28 is not due to a relocalization of Tsg101 away from Tal (Figure 4E). These findings suggest that other ESCRT-I proteins might mask target lysine residues in the headpiece of Tsg101 that would otherwise be exposed to Tal. On the basis of this hypothesis, we reasoned that mutations in lysine residues in the C-terminal region of Tsg101 (Figure 1A) might alter Tsg101's regulation by Tal. In agreement with this model, mutation of all the lysine residues in the headpiece of Tsg101 to arginines (Tsg101 5K-R: K326R, K361R, K369R, K379R, K382R) resulted in a modest relative resistance to degradation as compared with wild-type Tsg101 (Figure 4C). The additional mutation of an adjacent lysine residue in the stalk region of Tsg101, (Tsg101 6K-R: K304R, K326R, K361R, K369R, K379R, K382R) resulted in a more pronounced resistance to degradation by Tal, suggesting that this region may also be targeted for regulation of Tsg101 expression.
Figure 4.
VPS28 protects C-terminal lysine targets of Tal-mediated polyubiquitination. (A) Coexpression experiment to compare the ability of Tsg101-binding proteins VSP28 and Cep55 to prevent Tsg101 degradation by Tal. Plasmids encoding GST-Tsg101 with GST-Tal or GST-empty vector and empty control vector, YFP-VPS28 or YFP-Cep55, were cotransfected in 293T cells. Cells were lysed 24 h after transfection and analyzed by immunoblotting with α-Tsg101, α-GFP, and α-Hsp90 antibodies. (B) Coprecipitation experiment demonstrating that VPS28 blocks Tsg101 ubiquitination. GST-Tsg101 and HA-ubiquitin expressed in 293T cells in the presence or absence of YFP-Tal and Myc-VPS28 as indicated. Cell lysates were subjected to precipitation with glutathione-Sepharose beads and analyzed by Western blot, with α-Tsg101 to normalize for Tsg101 expression before reprobing with α-Tsg101 and α-HA monoclonal antibodies. (C) Coexpression of wild-type YFP-Tsg101, YFP-Tsg101 lysine to arginine mutants (progressively mutating lysines 3′ to 5′ from C-terminus), or YFP-Tsg101M95A, with GST-Tal or GST-empty and Myc-empty or Myc-VPS28 as indicated. Cells were lysed 24 h after transfection and analyzed by Western blot using α-GFP antibody. (D) Densitometry of Western blot shown in C. Expression of Tsg101 in the presence of Tal, with or without VPS28 is shown as a percentage of the control (lane 2) for each construct (n = 3 ± SD). (E) Confocal microscopy images of HeLa cells expressing YFP-Tal and Cherry-Tsg101 with CFP-VPS28 (top panels), or CFP-empty vector (bottom panels) as indicated.
The Role of Tal in Posttranslational Control of Tsg101 Expression
To investigate the steady-state control of Tsg101, we generated cell lines expressing a YFP-Tsg101 fusion protein with a retroviral vector. As reported previously (Feng et al., 2000), the expression of an exogenous Tsg101 resulted in the degradation of the endogenous protein, thus showing that the overall level of Tsg101 protein is tightly regulated in cells. A role of the proteasome in the posttranslational regulation of Tsg101 in cell lines overexpressing Tsg101 is suggested by results in Figure 5A, showing the reemergence of the endogenous Tsg101 in cells treated with a proteasome inhibitor (lactacystin) compared with either the control or the presence of a lysosome inhibitor (NH4Cl). The fact that we were unable to observe an accumulation of Tsg101 in the lactacystin-treated control HeLa cells, or a complete rescue of the endogenous Tsg101 expression in the YFP-Tsg101 cell line, could be explained by technical limitations of the assay. As the replacement of Tsg101 by the exogenous protein is relatively slow (Figure 5B, described below), we would expect to see a greater accumulation of Tsg101 with longer inhibitor treatment; however, because of the toxicity of lactacystin, we were unable to treat the cells for more than 6 h. An alternative explanation is that Tsg101 expression is also regulated at the transcriptional and/or translational levels to prevent deleterious accumulations of the protein.
Time-course experiments showed that in cell lines stably expressing a YFP-Tsg101 fusion protein, endogenous Tsg101 is replaced over time (30–72 h after selection) compared with cells transduced with YFP. In contrast, a mutation of the VPS28 binding site, termed A3, (Martin-Serrano et al., 2003b) rendered YFP-Tsg101 unable to replace the endogenous Tsg101, thus suggesting that binding to VPS28 is required to compete with the endogenous protein for “stabilizing” factors such as VPS28 itself. Importantly, YFP-Tsg101 and YFP-Tsg101A3 are expressed at similar levels in transiently transfected cells (Figure 5B), suggesting that the difference in protein levels of these Tsg101 forms in stably transduced cell lines is due to differences in VPS28-dependent stability rather than to differences in expression. Altogether, these results suggest a model whereby a limiting amount of VPS28 in cells would protect Tsg101 from degradation by Tal, and in situations where there is an excess of Tsg101, the uncomplexed protein would expose the lysine residues at the steadiness box for polyubiquitination by Tal. This model is supported by results in Figure 5C showing that overexpression of VPS28 resulted in a marked increase in YFP-Tsg101 steady-state levels in cells overexpressing Tsg101, leading also to the appearance of the endogenous protein.
RNAi knockdown of Tal was performed in these cell lines, but despite a significant knockdown, we saw no effect on Tsg101 expression (data not shown). However, we cannot exclude the possibility that the residual Tal protein observed by Western blot in these experiments is sufficient to maintain Tal function. Nonetheless, a role of Tal in regulating the expression of Tsg101 is supported by the increased YFP-Tsg101 expression and rescue of the endogenous Tsg101 in cells expressing a dominant-negative form of Tal (1-674) that lacks the RING domain. We demonstrate that PT/SAP-UEV binding is required to compete with endogenous Tal as nonbinding Tal constructs do not lead to increased Tsg101 expression. In contrast, overexpression of VPS28 or Tal1-674 does not influence Tsg101 levels in normal HeLa cells, suggesting that, in this case, Tsg101 is not freely available in the uncomplexed form. These findings suggest that in cells overexpressing Tsg101, as is the case for some tumor cells (Liu et al., 2002; Zhu et al., 2003; Oh et al., 2007; Young et al., 2007), Tal may become an important factor in controlling Tsg101 expression. However, in other cellular contexts, additional mechanisms may also be used to control Tsg101 expression levels.
Ubiquitination of the Tsg101 Headpiece Is Not Required for ESCRT-I Function
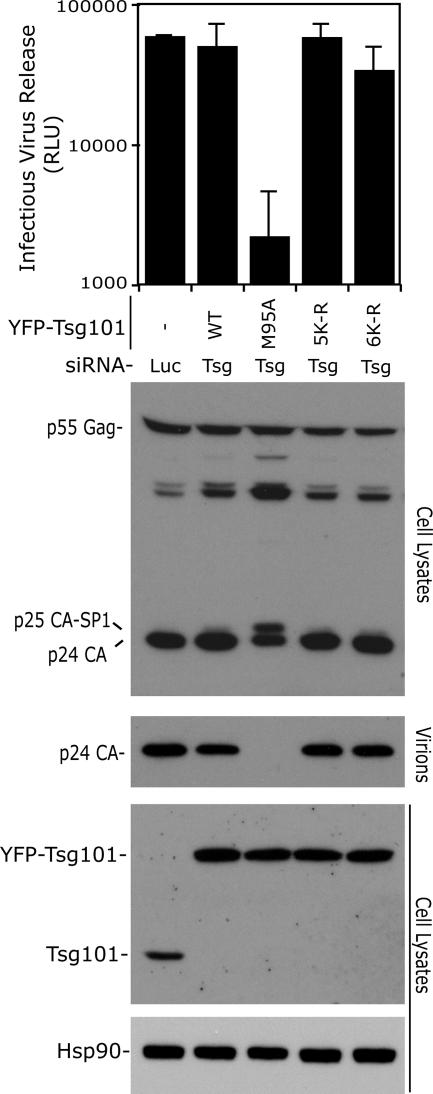
Lastly, a model has been proposed in which ubiquitination of Tsg101 would be essential for ESCRT-I activity by enabling dissociation from the cargo and assembly of the complex (Amit et al., 2004). To address whether the C-terminal region of Tsg101 serves that purpose, we tested the ability of lysine mutants to support HIV-1 budding. An experimental system described previously (Carlton and Martin-Serrano, 2007) in which endogenous Tsg101 is replaced by an RNAi resistant form (YFP fusion protein) was used. In addition, residual endogenous protein was removed by siRNA. We found that YFP-Tsg1015K-R or YFP-Tsg1016K-R activity was comparable to wild-type and YFP-expressing control cells (Figure 6). As a control, we did see the characteristic L-domain gag processing defect (accumulation of p25) and a substantial decrease in virion release in cells expressing YFP-Tsg101M95A. Notably, no increase in Tsg101M95A expression was seen in this cell line, implying that the reduction in virus release is due to a lack of HIV-1 Gag p6 binding (Pornillos et al., 2002b) and not due to a dominant negative effect of Tsg101 overexpression (Goila-Gaur et al., 2003; Martin-Serrano et al., 2003a). These data suggest that ubiquitination of the lysines at the C-terminus is not required for ESCRT function; thus, it is likely that the only action of Tal on these lysines is to mediate polyubiquitin conjugation and degradation of Tsg101.
Figure 6.
Ubiquitination of the C-terminal lysine residues of Tsg101 is not required for ESCRT-I function. HeLa cells stably transduced with retroviral vectors expressing YFP, or the indicated siRNA-resistant YFP-Tsg101 plasmids were treated with Luciferase or Tsg101 siRNA as indicated and transfected with a pNL/HXB HIV-1 proviral plasmid. Infectious virus release was measured with a chemiluminescent assay after infection of TZM-bl reporter cells with supernatants harvested from the transfected HeLa cells and is expressed as relative luminescence units (RLU; n = 3 ± SD). Cell lysates were examined by immunoblotting with α-Gag, α-Tsg101, and α-Hsp90 antibodies, and virion release was analyzed with α-Gag antisera.
DISCUSSION
This study describes a novel role of Tsg101 associated ligase (Tal) in the regulation of Tsg101 steady-state control and links this function to the regulation of Tsg101 expression by the C-terminal SB. Our results demonstrate the specificity of Tal-mediated control of Tsg101 expression as compared with Mahogunin, another E3 ubiquitin ligase that binds Tsg101. Although both ligases bind Tsg101 primarily via a UEV-PT/SAP contact and through a secondary interaction with the C-terminus, we show that only Tal enhances polyubiquitination of Tsg101. The type of ubiquitin modification is important and results in very different consequences for Tsg101. Whereas multi-monoubiquitination by Mahogunin may modulate Tsg101 function and is required for proper endosome-lysosome trafficking (Kim et al., 2007), our data show that polyubiquitination by Tal targets Tsg101 for degradation. Recent work demonstrates that it is possible for a particular ubiquitin ligase to mediate mono- or polyubiquitination of a specific substrate under different circumstances (Carter et al., 2007), hence our results do not exclude the possibility of additional multi-monoubiquitination by Tal. We show that Tal is an important regulator of Tsg101 expression in circumstances where excess Tsg101 is produced. In this context, expression of a mutant lacking the ubiquitin ligase catalytic domain (Tal1-674) leads to an increase in Tsg101 protein steady-state level, presumably by interference with the control mediated by endogenous Tal. However, several lines of evidence suggest that Tal is not the only mechanism to regulate Tsg101 expression. First, in normal HeLa cells overexpression of VPS28 does not stabilize additional Tsg101, suggesting that the uncomplexed form must be a minor fraction in this cellular context. In agreement with this notion, Tal1-674 does not increase Tsg101 expression in this situation. Additionally, and in contrast to the predicted result, no increase in Tsg101 expression is seen on stable expression of Tsg101M95A, indicating that additional ligases, not dependent on binding to the UEV domain, may also target the uncomplexed Tsg101 for degradation. Tal is evolutionally conserved in vertebrates, but it is not found in invertebrate species outside the chordate phylum. Thus compared with Tsg101, which is conserved from yeast to humans, Tal is a relatively new gene. As Tsg101 is crucial for cell viability, it seems likely that more than one mechanism for regulating this protein could have evolved. One such E3 ligase, MDM2, has been reported to target Tsg101 for proteasomal degradation as part of a feedback loop regulating both proteins (Li et al., 2001; Cheng and Cohen, 2007).
Our deletion analysis shows that conjugation of polyubiquitin chains and regulation of Tsg101 expression by Tal, maps to the last 30 amino acids of Tsg101, a region containing VPS28- and VPS37-binding sites. Moreover, the finding that VPS28 expression prevented Tal mediated ubiquitination and degradation of Tsg101 suggests that binding of other ESCRT-I subunits, namely VPS28 or VPD37A-D, may mask the C-terminal target site of Tal. This model is consistent with recent yeast structural studies, describing the relative positions of the three proteins. The C-terminus of the yeast homologue of Tsg101 forms an antiparallel helical hairpin that is sandwiched between two similar hairpins of Vps28 and Vps37 (Kostelansky et al., 2006, 2007; Teo et al., 2006). Extensive interactions between the helices stabilize the region, leaving little accessible surface of Vps23. The mutation analysis is in agreement with this model as mutation of the six terminal lysine residues confers a significant resistance to degradation of Tsg101 by Tal. These mutations do not render Tsg101 completely resistant to regulation, suggesting that lysine residues in the stalk region may be alternative targets for polyubiquitination.
The emerging model from these transient transfection experiments suggests a mechanism for the specific degradation of uncomplexed Tsg101, through polyubiquitination of C-terminal lysine residues. This hypothesis is consistent with previous work, which attributed the tight control of Tsg101 expression to an unidentified property of the C-terminus, termed the SB (Feng et al., 2000). Our experiments show that binding to VPS28, specifically, is the key to this autoregulation. We confirm that overexpression of wild-type YFP-Tsg101 replaces endogenous Tsg101, thus maintaining total Tsg101 concentration. However, an exogenously expressed mutant that does not bind to VPS28 (A3) fails to induce degradation of the endogenous protein. We propose that the steadiness box effect is due to competition between exogenously and endogenously expressed Tsg101 for VPS28 protection of target lysines. In agreement with this model, overexpression of the wild-type protein (but not the A3 mutant) would sequester VPS28 away from the endogenous protein, leaving it vulnerable to degradation.
In summary, we have identified the C-terminal lysines of Tsg101 as targets of polyubiquitination, which are exposed when Tsg101 expression exceeds the level required for ESCRT function. We also describe a novel function of Tal in targeting excess Tsg101 for proteasomal degradation and our data suggest that other ubiquitin ligases are likely to be used for this critical purpose. This regulation of Tsg101 protein level might provide a crucial mechanism to prevent the severe consequences associated with overexpression of Tsg101, such as disruption of endosomal sorting, inhibition of cell growth and failure of cytokinesis.
ACKNOWLEDGMENTS
We thank M. Agromayor and J. Carlton for critical reading of the manuscript. This work was supported by Career Establishment Grant G0400207 from the Medical Research Council UK.
Abbreviations used:
- Tsg101
tumor susceptibility gene 101
- Tal
Tsg101-associated ligase
- ESCRT
endosomal sorting complex required for transport
- UEV
ubiquitin E2 variant
- VPS
vacuolar protein sorting
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0957) on December 12, 2007.
REFERENCES
- Agromayor M., Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 2006;281:23083–23091. doi: 10.1074/jbc.M513803200. [DOI] [PubMed] [Google Scholar]
- Amit I., et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18:1737–1752. doi: 10.1101/gad.294904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Odorizzi G., Estepa E. J., Emr S. D. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K. G., Brech A., Mehlum A., Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K. G., Slagsvold T., Cabezas A., Rosendal K. R., Raiborg C., Stenmark H. The growth-regulatory protein HCRP1/hVps37A is a subunit of mammalian ESCRT-I and mediates receptor down-regulation. Mol. Biol. Cell. 2004;15:4337–4346. doi: 10.1091/mbc.E04-03-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz P. D. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Bishop N., Woodman P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 2001;276:11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- Carlton J. G., Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Carter S., Bischof O., Dejean A., Vousden K. H. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- Cheng T. H., Cohen S. N. Human MDM2 isoforms translated differentially on constitutive versus p53-regulated transcripts have distinct functions in the p53/MDM2 and TSG101/MDM2 feedback control loops. Mol. Cell. Biol. 2007;27:111–119. doi: 10.1128/MCB.00235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu T., Sun J., Saksena S., Emr S. D. New component of ESCRT-I regulates endosomal sorting complex assembly. J. Cell Biol. 2006;175:815–823. doi: 10.1083/jcb.200608053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss M., Jones C., Babst M. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol. Biol. Cell. 2007;18:636–645. doi: 10.1091/mbc.E06-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov D. G., Ono A., Orenstein J. M., Freed E. O. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn C. A., Decker J. M., Sfakianos J. N., Wu X., O'Brien W. A., Ratner L., Kappes J. C., Shaw G. M., Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman S. W., Martin-Serrano J., Chung W., Zang T., Bieniasz P. D. Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding. J. Biol. Chem. 2005;280:628–636. doi: 10.1074/jbc.M410384200. [DOI] [PubMed] [Google Scholar]
- Feng G. H., Lih C. J., Cohen S. N. TSG101 protein steady-state level is regulated posttranslationally by an evolutionarily conserved COOH-terminal sequence. Cancer Res. 2000;60:1736–1741. [PubMed] [Google Scholar]
- Garrus J. E., et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gill D. J., Teo H., Sun J., Perisic O., Veprintsev D. B., Emr S. D., Williams R. L. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. EMBO J. 2007;26:600–612. doi: 10.1038/sj.emboj.7601501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R., Demirov D. G., Orenstein J. M., Ono A., Freed E. O. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol. 2003;77:6507–6519. doi: 10.1128/JVI.77.11.6507-6519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Babst M., Emr S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Stefan C. J., Babst M., Emr S. D. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. Y., Olzmann J. A., Barsh G. S., Chin L. S., Li L. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol. Biol. Cell. 2007;18:1129–1142. doi: 10.1091/mbc.E06-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky M. S., Schluter C., Tam Y. Y., Lee S., Ghirlando R., Beach B., Conibear E., Hurley J. H. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–498. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky M. S., Sun J., Lee S., Kim J., Ghirlando R., Hierro A., Emr S. D., Hurley J. H. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 2006;125:113–126. doi: 10.1016/j.cell.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempler A., Henry M. D., Triplett A. A., Wagner K. U. Targeted deletion of the Tsg101 gene results in cell cycle arrest at G1/S and p53-independent cell death. J. Biol. Chem. 2002;277:43216–43223. doi: 10.1074/jbc.M207662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Cohen S. N. Tsg101, a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- Li L., Liao J., Ruland J., Mak T. W., Cohen S. N. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc. Natl. Acad. Sci. USA. 2001;98:1619–1624. doi: 10.1073/pnas.98.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. T., Huang C. C., You H. L., Chou F. F., Hu C. C., Chao F. P., Chen C. M., Cheng J. T. Overexpression of tumor susceptibility gene TSG101 in human papillary thyroid carcinomas. Oncogene. 2002;21:4830–4837. doi: 10.1038/sj.onc.1205612. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Perez-Caballero D., Bieniasz P. D. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 2004;78:5554–5563. doi: 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J., Yarovoy A., Perez-Caballero D., Bieniasz P. D. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA. 2003a;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P. D. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P. D. Role of ESCRT-I in retroviral budding. J. Virol. 2003b;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Alam S. L., Eckert D. M., Gygi S. P., Sundquist W. I. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2007;2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich A. J., Davies B. A., Payne J. A., Katzmann D. J. Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol. Biol. Cell. 2007;18:646–657. doi: 10.1091/mbc.E06-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Mammucari C., Nenci A., Cabodi S., Cohen S. N., Dotto G. P. Negative regulation of cell growth and differentiation by TSG101 through association with p21(Cip1/WAF1) Proc. Natl. Acad. Sci. USA. 2002;99:5430–5435. doi: 10.1073/pnas.082123999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh K. B., Stanton M. J., West W. W., Todd G. L., Wagner K. U. Tsg101 is upregulated in a subset of invasive human breast cancers and its targeted overexpression in transgenic mice reveals weak oncogenic properties for mammary cancer initiation. Oncogene. 2007;26:5,950–5,959. doi: 10.1038/sj.onc.1210401. [DOI] [PubMed] [Google Scholar]
- Pineda-Molina E., Belrhali H., Piefer A. J., Akula I., Bates P., Weissenhorn W. The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic. 2006;7:1007–1016. doi: 10.1111/j.1600-0854.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- Pornillos O., Alam S. L., Davis D. R., Sundquist W. I. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 2002a;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- Pornillos O., Alam S. L., Rich R. L., Myszka D. G., Davis D. R., Sundquist W. I. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002b;21:2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O., Higginson D. S., Stray K. M., Fisher R. D., Garrus J. E., Payne M., He G. P., Wang H. E., Morham S. G., Sundquist W. I. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 2003;162:425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruland J., Sirard C., Elia A., MacPherson D., Wakeham A., Li L., de la Pompa J. L., Cohen S. N., Mak T. W. p53 accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101. Proc. Natl. Acad. Sci. USA. 2001;98:1859–1864. doi: 10.1073/pnas.98.4.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B., Calistri A., Craig S., Popova E., Gottlinger H. G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Stuchell M. D., Garrus J. E., Muller B., Stray K. M., Ghaffarian S., McKinnon R., Krausslich H. G., Morham S. G., Sundquist W. I. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 2004;279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- Teo H., Gill D. J., Sun J., Perisic O., Veprintsev D. B., Vallis Y., Emr S. D., Williams R. L. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- VerPlank L., Bouamr F., LaGrassa T. J., Agresta B., Kikonyogo A., Leis J., Carter C. A. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc. Natl. Acad. Sci. USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U. K., et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Wagner K. U., Krempler A., Qi Y., Park K., Henry M. D., Triplett A. A., Riedlinger G., Rucker I. E., Hennighausen L. Tsg101 is essential for cell growth, proliferation, and cell survival of embryonic and adult tissues. Mol. Cell. Biol. 2003;23:150–162. doi: 10.1128/MCB.23.1.150-162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. L., Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Young T. W., Mei F. C., Rosen D. G., Yang G., Li N., Liu J., Cheng X. Up-regulation of tumor susceptibility gene 101 protein in ovarian carcinomas revealed by proteomics analyses. Mol. Cell Proteom. 2007;6:294–304. doi: 10.1074/mcp.M600305-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhu G., et al. Combination of microdissection and microarray analysis to identify gene expression changes between differentially located tumour cells in breast cancer. Oncogene. 2003;22:3742–3748. doi: 10.1038/sj.onc.1206428. [DOI] [PubMed] [Google Scholar]