Abstract
Silencing of the mating-type locus HMR in Saccharomyces cerevisiae requires DNA elements called silencers. To establish HMR silencing, the origin recognition complex binds the HMR-E silencer and recruits the silent information regulator (Sir)1 protein. Sir1 in turn helps establish silencing by stabilizing binding of the other Sir proteins, Sir2–4. However, silencing is semistable even in sir1Δ cells, indicating that SIR1-independent establishment mechanisms exist. Furthermore, the requirement for SIR1 in silencing a sensitized version of HMR can be bypassed by high-copy expression of FKH1 (FKH1hc), a conserved forkhead transcription factor, or by deletion of the S phase cyclin CLB5 (clb5Δ). FKH1hc caused only a modest increase in Fkh1 levels but effectively reestablished Sir2–4 chromatin at HMR as determined by Sir3-directed chromatin immunoprecipitation. In addition, FKH1hc prolonged the cell cycle in a manner distinct from deletion of its close paralogue FKH2, and it created a cell cycle phenotype more reminiscent to that caused by a clb5Δ. Unexpectedly, and in contrast to SIR1, both FKH1hc and clb5Δ established silencing at HMR using the replication origins, ARS1 or ARSH4, as complete substitutes for HMR-E (HMRΔE::ARS). HMRΔE::ARS1 was a robust origin in CLB5 cells. However, initiation by HMRΔE::ARS1 was reduced by clb5Δ or FKH1hc, whereas ARS1 at its native locus was unaffected. The CLB5-sensitivity of HMRΔE::ARS1 did not result from formation of Sir2–4 chromatin because sir2Δ did not rescue origin firing in clb5Δ cells. These and other data supported a model in which FKH1 and CLB5 modulated Sir2–4 chromatin and late-origin firing through opposing regulation of a common pathway.
INTRODUCTION
Chromatin structures vary with genome position, creating structural heterogeneity along chromosomes that modulates every aspect of DNA metabolism (Fischle et al., 2003). Specific DNA sequence elements form the foundation for this heterogeneity. For example, certain DNA sequences act to establish centromeric chromatin required for chromosome segregation (Cleveland et al., 2003; Henikoff and Dalal, 2005). In addition, some chromatin structures, such as heterochromatin, can dominate the functional capacity of DNA sequence elements across chromosomal domains or even over an entire chromosome. Heterochromatin, for example, can delay or repress initiation by DNA replication origins and initiation of transcription from promoters (Gomez and Brockdorff, 2004; Weinreich et al., 2004; Chang et al., 2006). Thus, a competing balance exists between DNA sequence elements that perform different functions or establish different types of chromatin structures (Kamakaka, 1997; Donze and Kamakaka, 2002; Valenzuela and Kamakaka, 2006).
Genetic analyses of transcriptional silencing of the HMRa locus in Saccharomyces cerevisiae reveal that perturbations in the cell cycle tip the balance between the types of chromatin that can form at a particular chromosomal domain (Laman et al., 1995; Fox and Rine, 1996; Ehrenhofer-Murray et al., 1999). HMRa silencing defines a form of transcription repression that requires the assembly of a specialized chromatin structure called silent chromatin (Fox and McConnell, 2005). Formation of silent chromatin requires small DNA elements called silencers that bind a collection of sequence-specific DNA binding proteins. HMRa contains two silencers, HMR-E and HMR-I that flank opposite ends of this ∼3000-base pair locus (Loo and Rine, 1995). HMR-E is necessary and sufficient for HMRa silencing; the importance of HMR-I can be observed only under conditions in which silencing has been compromised (Fox et al., 1995; Rivier et al., 1999). HMR-E, the better characterized of the two silencers, contains a binding site each for the origin recognition complex (ORC), and the Rap1 and Abf1 proteins. HMRa silencing is not essential for yeast cell viability, but ORC is because it functions as the eukaryotic initiator, the protein complex that marks chromosomal sites as DNA replication origins (reviewed in Bell, 2002). Rap1 and Abf1 are also essential multifunctional nuclear proteins (reviewed in Shore, 1994).
The role of the HMR-E silencer-binding proteins is to recruit, via multiple independent protein–protein interactions, four silent information regulator (Sir) proteins, Sir1, -2, -3, and -4, nonhistone chromatin-binding proteins dispensable for cell viability but necessary for silencing (reviewed in Gasser and Cockell, 2001; Rusche et al., 2003; Fox and McConnell, 2005). One important interaction occurs directly between Sir1 and ORC (Hou et al., 2005; Hsu et al., 2005). The Sir1-ORC complex helps recruit and/or stabilize the binding of the three other Sir proteins, Sir2, -3, and -4 to the silencer (Rusche et al., 2002). Another key set of interactions occurs between Rap1 and Sir3 and Sir4 proteins (Moretti and Shore, 2001). The multiple protein–protein interactions that recruit the four Sir proteins to HMR-E define the establishment/nucleation phase of silent chromatin assembly. In the second phase, Sir2, an enzyme, deacetylates nucleosomes neighboring HMR-E; deacetylated nucleosomes in turn promote binding of additional Sir2–4 complexes until a higher-order Sir2–4 silent chromatin structure forms at HMRa (reviewed in Rusche et al., 2003; Moazed et al., 2004; Fox and McConnell, 2005).
One key feature of this model is that Sir1 differs substantially from Sir2–4 because its role is confined to silencers where it enhances the assembly and/or reduces the disassembly of Sir2–4 chromatin, but is not intrinsically required either for its formation or function (Pillus and Rine, 1989, 2004; Xu et al., 2006). Thus, in contrast to Sir2–4, Sir1 is not a critical structural component of HMRa silent chromatin. Another notable feature that distinguishes SIR1 from the other SIRs is that its requirement in HMRa silencing can be partially bypassed by perturbations in the cell cycle (Laman et al., 1995). For example, a deletion of the major S phase cyclin CLB5 that is required for timely S phase progression (Donaldson et al., 1998; Gibson et al., 2004) can partially bypass the requirement for SIR1 (Laman et al., 1995). In addition, mutations in the HMR-E silencer are also rescued by cell cycle perturbations caused by drugs or certain mutations (Axelrod and Rine, 1991; Laman et al., 1995; Ehrenhofer-Murray et al., 1999). These data provide evidence that other molecular interactions can compensate for the role of Sir1 in transcriptional silencing and reveal that specific perturbations in the cell cycle can tip the balance between silent and permissive chromatin formation at HMRa. Thus, HMRa silencing serves as an experimentally tractable model for dissecting how the cell cycle can modulate the structure and function of a defined chromatin domain.
In a previous study, we identified forkhead homologue FKH1 as a high-copy suppressor of defects in HMRa silencing caused by a sir1Δ mutation (Hollenhorst et al., 2000). Fkh1 and its paralogue Fkh2 are evolutionarily conserved transcription factors that bind directly to promoters within a group of genes named the CLB2-cluster (reviewed in Breeden, 2000; Futcher, 2000). Transcription of these genes is repressed in late M, G1, and early S phases but is activated beginning in late S phase and through G2 and early M phases (Spellman et al., 1998). Genes within this cluster encode proteins including the major G2/M phase cyclin Clb2 that drive progress through M phase. Fkh2 is the primary regulator of CLB2-cluster genes, repressing transcription of these genes in G1 phase and stimulating their transcription in late S and G2/early-M phase. (Koranda et al., 2000; Kumar et al., 2000; Pic et al., 2000; Zhu et al., 2000; Reynolds et al., 2003). It is less clear how Fkh1 normally functions in CLB2-cluster transcription. FKH1 can partially compensate for loss of FKH2 because fkh1Δfkh2Δ cells exhibit a substantially greater reduction in CLB2-cluster transcription than FKH1fkh2Δ cells, indicating that Fkh1 can activate CLB2 transcription. However, paradoxically, the normal role for Fkh1 in wild-type cells seems to be as a negative regulator of CLB2-cluster transcription during G2/M, thus attenuating the level of transcription that can be achieved by Fkh2-mediated activation (Hollenhorst et al., 2000, 2001; Sherriff et al., 2007). Although it is unclear how the role(s) of FKH1 at the CLB2-cluster is related to its ability to affect silencing, it is known that the DNA binding domain of Fkh1 is critical for it ability to modulate SIR1-bypass HMR silencing (Hollenhorst et al., 2000).
In this report, we addressed the mechanism(s) by which FKH1hc bypasses the requirement for SIR1 in silencing. Unexpectedly (Hollenhorst et al., 2000), the genetic and cell cycle data reported here provide evidence against the role of FKH1 in CLB2 transcription serving as the primary factor in FKH1hc-dependent silencing. Specifically, although FKH1hc reduced CLB2 transcription, reductions in CLB2 were insufficient to mimic FKH1hc. Instead, the data generated through a combination of genetic analyses, Sir3-directed chromatin immunoprecipitations (ChIPs) and two-dimensional (2-D) origin mapping experiments were most consistent with a model in which FKH1 and the S phase cyclin CLB5 act as opposing regulators that converge on a common target(s) that inhibit late replication origin firing and promote Sir2–4 chromatin assembly. The data were also consistent with the idea that this pathway favors distinct SIR1-independent molecular interactions that contribute to Sir2–4 protein association with HMR.
MATERIALS AND METHODS
Strains and Plasmids
Yeast strains used in this study (Table 1) were constructed using standard yeast molecular genetics and recombinant DNA techniques (Sambrook et al., 1989; Guthrie and Fink, 1991). Five different plasmids were used in this study as indicated in the figure legends: pRS426 (Sikorski and Hieter, 1989), pCF99 (SIR1 in pRS316; Gardner et al., 1999; Hollenhorst et al., 2000), pCF345 (SIR1 in Yep24; Hollenhorst et al., 2000), pCF480 (FKH1 in pRS426; Hollenhorst et al., 2000), and pCF942 (FKH1 in pRS316). Cells were grown at 30°C in standard rich medium (YPD), in minimal medium supplemented with casamino acids (CAS) (to select for URA3-containing plasmids), or in synthetic media (SC) lacking defined supplements to select for diploids and/or plasmids as appropriate and described in the text (Guthrie and Fink, 1991). All strains were isogenic to W303-1A except the MATa cells used for mating lawns.
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| JRY19 | MATa his4 leu2 trp1 ura3 | Gift from Jasper Rine yeast collection |
| JRY2234 (W303-1A) | MATaade2-1, his3-11, 15 leu 2-3,112 trp1-1 ura3-1 can1-100 | Gift from Jasper Rine yeast collection |
| JRY3009 (W303-1B) | MATα ade2-1, his3-11, 15 leu 2-3,112 trp1-1 ura3-1 can1-100 | Gift from Jasper Rine yeast collection |
| CFY616 | MATahis4 leu2 trp1 ura3 | |
| CFY345 | JRY3009 HMR-SSa | Gift from Jasper Rine yeast collection |
| CFY762 | JRY3009 HMR-SSasir1Δ::LEU2 | |
| CFY1649 | JRY3009 HMR-SSasir1Δ::TRP1 | |
| CFY1819 | JRY3009 HMR-SSasir3Δ::LEU2 | |
| CFY1827 | JRY3009 HMR-SSasir1Δ::LEU2 sir2Δ::TRP1, LEU2::sir2N345A | Rusche et al. (2002) |
| CFY1265 | JRY2334 HMR-SSa ΔI bar1Δ::HIS3 | |
| CFY2016 | JRY2334 HMR-SSa ΔI bar1Δ::HIS3 fkh2Δ::HISG | |
| CFY527 | JRY3009 HMR-SSasir1Δ::LEU2 fkh1Δ::TRP1 | |
| CFY603 | JRY3009 HMR-SSasir1Δ::LEU2 fkh2Δ::HIS3 | |
| CFY1614 | JRY3009 HMR-SSasir1Δ::LEU2 clb1Δ::HIS3 | |
| CFY1124 | JRY3009 HMR-SSasir1Δ::LEU2 clb2Δ::TRP1 | |
| CFY2104 | JRY3009 HMR-SSasir1Δ::LEU2 clb5Δ::kanMx | |
| CFY2279 | HMR-SSasir1Δ::LEU2 clb6Δ::kanMx | Gibson et al. (2004) |
| CFY2268 | JRY3009 HMR-SSasir1Δ::LEU2 clb5Δ::HIS3 clb6Δ::kanMx | Gibson et al. (2004) |
| CFY2110 | JRY3009 HMR-SSasir1Δ::LEU2 clb5Δ::kanMx fkh1Δ::TRP1 | |
| CFY2446 | JRY3009 HMR-SSasir1Δ::LEU2 CLB5–3xHA | |
| CFY110 | JRY3009 HMR-SSa ΔI sir1Δ::TRP1 | |
| CFY2221 | JRY3009 HMR-SSarap1 sir1Δ::TRP1 | |
| CFY2133 | JRY3009 HMRΔE | |
| CFY2071 | JRY3009 HMRΔE::ARSH4 sir1Δ::LEU2 | |
| CFY2237 | JRY3009 HMRΔ::ARS1 sir1Δ::TRP1 | |
| CFY35 | JRY3009 HMR-SSa ΔI | |
| CFY2230 | JRY3009 HMR-SSa ΔI sir1Δ::LEU2 clb5Δ::kanMx | |
| CFY58 | JRY3009 HMR-SSarap1 | |
| CFY2218 | JRY3009 HMR-SSarap1 sir1Δ::TRP1 clb5Δ::kanMx | |
| CFY324 | JRY3009 HMRΔE::ARSH4 | |
| CFY2193 | JRY3009 HMRΔE::ARSH4 sir1Δ::LEU2 clb5Δ::kanMx | |
| CFY321 | JRY3009 HMRΔE::ARS1 | |
| CFY2236 | JRY3009 HMRΔE::ARS1 sir1Δ::LEU2 clb5Δ::kanMx | |
| CFY2481 | JRY2334 HMRΔE::ARS1 clb5Δ::kanMx sir1Δ::LEU2 sir2Δ::TRP1 | |
| CFY2613 | JRY2234 hmlΔ::URA3 sir2Δ::LEU2 | |
| CFY2615 | JRY2334 hmlΔ::URA3 sir2Δ::LEU2 clb5Δ::kanMx | |
| CFY2617 | JRY2334 hmlΔ::URA3 | |
| CFY2619 | JRY2334 hmlΔ::URA3 clb5Δ::kanMx HMRΔE::ARS1 |
Strains were from the laboratory collection or constructed during the course of this study. The strains noted were gifts from or derived from strains from the sources in the Reference column.
Semiquantitative Mating Assay
The MATα cells examined for silencing by mating efficiency were grown in log phase for 2 d, and then they were mixed with an excess of MATa cells (JRY19). The most concentrated samples of MATα cells analyzed were 5 × 106 cells/ml in a total volume of 50 μl. Tenfold serial dilutions of this concentration were generated in 50-μl final volumes. Six microliters of each dilution was analyzed per drop on either YPD or CAS solid agar medium, as appropriate, to determine cell counts. To the remainder of the cells, an excess of MATa cells (15 μl of log-phase cells concentrated to 5 OD) were added and mixed. Eight microliters of this mixture was plated to synthetic solid agar medium appropriately supplemented to select for diploids (and the retention of a URA3 plasmid, if appropriate). Plating efficiencies indicated that equivalent numbers of cells were being compared for every mating comparison shown.
RNA Blots
RNA was isolated from yeast as described previously (Fox et al., 1995), and 15 μg of total RNA was analyzed per lane. RNA blot hybridization was performed with multiprime-labeled DNA probes complementary to a1 mRNA, CLB2 mRNA, or SCR1 RNA. The probes for a1 mRNA and SCR1 RNA were described previously (Fox et al., 1995, 1997). Template for the CLB2 probe was generated by PCR by using the following primer pair: forward, GTCCAACCCAATAGAAAACAC and reverse, CATGCACCGTCTGTCTCTGATG. This probe was also used in a previous study (Hollenhorst et al., 2000).
Antibodies
Anti-Sir3 monoclonal antibodies were raised against full-length Sir3 purified from baculovirus-infected Sf9 cells as described previously (Georgel et al., 2001), and they are available from Neoclone at http://www.neoclone.com/ (the antibodies used to make the ChIP cocktail were 184A, 185A, and 184C). Anti-Fkh1 polyclonal antibodies were raised in rabbits (Harlan Labs, Madison, WI) to a 6xHIS–Fkh1 fusion protein encoding the N-terminal 302 amino acids of Fkh1 (pCF1564) expressed in Escherichia coli (BL21 cells) and purified using ion exchange and nickel-affinity chromatography. These antibodies were affinity purified and used at a 1:200 dilution for protein immunoblotting (Harlow and Lane, 1999). Anti-hemagglutinin (HA) monoclonal antibodies were from Covance Research Products (Princeton, NJ).
Chromatin Immunoprecipitations
ChIP was performed as described previously (Strahl-Bolsinger et al., 1997) except: Sir3 antibodies were cross-linked to protein A-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) by using standard methods before ChIP (Harlow and Lane, 1999). 1/50 of the immunoprecipitated DNA and 1/500 of the total DNA were subjected to 26 cycles of polymerase chain reaction (PCR) by using primers specific for HMRa or ADH4 or the appropriate ARS (Gardner and Fox, 2001). Primers to X/Ya, Ya/Z, and boundary elements were as described previously (Rusche et al., 2002). PCR products were separated on a 1% agarose gel, and band intensities were quantified using video densitometry analysis and Labworks analysis software (UVP, Upland, CA).
Protein Immunoblots
To compare levels of Fkh1 expressed from a chromosomal copy of FKH1 and from a 2μ plasmid (FKH1hc) quantitatively (Figure 2), crude yeast extracts were prepared as described previously (Gardner et al., 1999) from wild-type cells transformed with either empty vector (pRS426; pCF225) or with a 2μ plasmid expressing FKH1 (FKH1hc; pRS426 with FKH1; pCF480) and from a fkh1Δ cells transformed with vector. To avoid distortions in the gel that could affect quantification, lysates from fkh1Δ cells were used to dilute the lysates from FKH1 or FKH1hc cells, ensuring that the same amount of total protein was present in each lane. Four microliters of crude lysates was examined per lane by SDS-polyacrylamide gel electrophoresis (PAGE) by using a 10% acrylamide gel and standard protein blotting methods were used. To determine whether protein transfer to the blot was the same for all lanes, the blot was prestained with Poncea S before incubation with antibody.
Figure 2.
FKH1hc prolonged the cell cycle and attenuated the CLB2 mRNA expression peak. (A) MATa bar1Δ::HIS3 (CFY1265) cells harboring either a 2μ plasmid (vector; pRS426), a 2μ plasmid containing FKH1 (FKH1hc; pCF480), or a deletion of FKH2 (fkh2Δ; CFY2016) and a 2μ plasmid (vector) were harvested in log phase from medium lacking uracil and arrested in G1 with α-factor. The cells were then released from α-factor arrest into fresh medium, and every 15 min aliquots were harvested for analysis of bud morphology or CLB2 mRNA levels (see B). Unbudded cells were scored in G1 phase, small-budded cells were scored in S phase, and large-budded cells were scored in M phase. A representative graph for each experiment is shown. (B) The ratio of CLB2 mRNA to SCR1 RNA was determined for each of the strains and time points indicated in A. Total RNA (15 μg) as determined by absorbance at 260 λ was analyzed per lane, CLB2 mRNA and SCR1 RNA were probed independently, and the signals quantified by PhosphorImager analysis. Data from a representative experiment are shown. (C) Mating analysis of MATα HMR-SSa sir1Δ cells containing either wild-type FKH2 (CFY762) or a deletion of FKH2 (fkh2Δ; CFY603) and harboring 2μ plasmids with SIR1 (SIR1hc; pCF345), no insert (vector; pCF225) or FKH1 (FKH1hc; pCF480). The cells were mixed with an excess of MATa cells (CFY616) and 10-fold serial dilutions were plated on medium to select for the growth of a/α diploids as described in Materials and Methods. The most concentrated samples contained 5 × 103 cells/μl, and 8 μl was analyzed in each spot. Corresponding growth controls indicated that an equal number of cells were being compared between strains (LC; data not shown). (D) Levels of Fkh1 in MATα HMR-SSa sir1Δ (CFY1649) cells were determined by protein immunoblot with a polyclonal antibody raised against Fkh1 (α-Fkh1). Cells, harboring either an empty 2μ plasmid (pRS426; chromosomal; lanes 1–3) or a 2μ plasmid containing FKH1 (FKH1hc; pCF480; lanes 4–6), were harvested in log-phase growth in liquid CAS medium. Two-fold serial dilutions of each protein extract were made using protein extract from fkh1Δ cells as diluents (lane 7) to ensure that each lane contained similar total protein amounts that facilitated accurate quantification of Fkh1 levels. The fkh1Δ cells showed no protein band corresponding to Fkh1 in the immunoblot (lane 7). Equal amounts of protein were loaded per lane as determined by Ponceau S staining of the blot before immunoblotting.
Cell Cycle Arrest and Release Experiments
Cells were grown and harvested in log phase (23 or 30°C) from CAS liquid medium and arrested in G1 with α-factor for 150 min (5 μM for bar1Δ cells; 25 μM for BAR1 cells). The cells were then released from α-factor arrest by washing with and releasing into fresh medium. Every 15 min, aliquots were harvested for analysis of bud morphology and scored as indicated in Figure 2. Alternatively cells were harvested for analyses of DNA content by flow cytometry as described previously (Weinreich et al., 1999) except that Sytox Green (Invitrogen, Carlsbad, CA) at 1 μM was used to label DNA.
Two-Dimensional Origin Mapping
2-D origin mapping was performed as described previously (Fox et al., 1995). Cells were grown at 23°C in 2 liters of complete rich medium, or, if cells contained a URA3 plasmid, at 30°C in 2 liters of CAS medium. DNA from each sample was analyzed for replication intermediates (RIs) by digestion with HindIII for HMR-E::ARS1 and NcoI for ARS1. DNA was enriched for RIs with BND cellulose after restriction digest. DNA was separated in two dimensions and examined by DNA blot hybridization by using primers specific for each region (Fox et al., 1995). Probes were created using Megaprime DNA labeling system (GE Healthcare).
RESULTS
FKH1 is a high-copy suppressor of an HMRa silencing defect caused by a sir1Δ mutation (Hollenhorst et al., 2000). The role of FKH1, together with FKH2, in controlling CLB2-cluster transcription raised the possibility that FKH1-mediated silencing was related to perturbations of the cell cycle. To address this possibility and better define how Fkh1 modulated silencing, we compared high-copy FKH1-dependent (FKH1hc-dependent) and SIR1-dependent silencing at the molecular and genetic levels.
FKH1hc-dependent Silencing of HMRa Required Recruitment of Sir3 and the Catalytic Activity of Sir2
MATα HMR-SSa sir1Δ yeast cells are unable to mate because the sensitized synthetic silencer (HMR-SSa) requires SIR1 to silence the a-mating type genes at HMRa (McNally and Rine, 1991). Thus, these cells provide a genetically useful tool for examining SIR1 function and for identifying other mechanisms capable of transcriptional silencing. Expression of FKH1 on a 2μ vector (FKH1hc) partially restores silencing to these sir1Δ cells as seen by a reduction in a1 mRNA expressed from HMR-SSa (Hollenhorst et al., 2000; Figure 1A), thus creating SIR1-bypass silencing. Notably, even expression of FKH1 from a CEN plasmid contributed to a reproducible, albeit lower level of SIR1-bypass silencing (Figure 1A, lane 4) that was also evident in mating assays (Casey, unpublished data).
Figure 1.
FKH1hc-silencing required recruitment of Sir3 and the catalytic activity of Sir2. (A) Steady-state levels of a1 and SCR1 RNAs were measured in MATα HMR-SSa sir1Δ cells (CFY762) harboring a 2μ plasmid (lane 1, vector; pCF225), a CEN plasmid with SIR1 (lane 2, SIR1-CEN; pCF99), a 2μ plasmid with FKH1 (lane 3, FKH1-2μ; pCF480), or a CEN plasmid with FKH1 (lane 4, FKH1-CEN; pCF943). The average ratio of a1/SCR1 and SD for three independent experiments is indicated below each lane. FKH1-CEN also reproducibly enhanced silencing slightly as measured by mating assays (Casey, unpublished data). (B) Sir3 association with HMR-SSa and the control locus ADH4 was measured by ChIP with a monoclonal antibody cocktail against Sir3 (α-Sir3). Three isogenic MATα HMR-SSa strains were analyzed that differed only in terms of their SIR1 or SIR3 genotype: SIR1 sir3Δ (CFY1819), SIR1 SIR3 (CFY345), and sir1Δ SIR3 (CFY762). ChIPs were performed on each strain containing either a 2μ plasmid (vector, gray; pCF225) or a 2μ plasmid with FKH1 (FKH1hc, black; pCF480). The percentage of specific PCR fragment obtained in the immunoprecipitate was determined by videodensitometry analysis using Labworks analysis software (UVP). Data and standard deviations are reported for three independent ChIP experiments. (C) ChIPs were performed as described in B except that multiple regions within the HMR-SSa locus were examined using primer pairs at the positions indicated in the cartoon of HMRa above the figure as described previously (Rusche et al., 2002). Two isogenic MATα HMR-SSa sir1Δ strains were analyzed that differed only in terms of their SIR2 genotype; SIR2 (CFY762) or sir2N345A (CFY1827). Each strain harbored a plasmid containing SIR1 (pCF99) or a 2μ plasmid containing FKH1 (FKH1hc, pCF480).
To address the mechanism by which FKH1hc bypasses the requirement of SIR1 in silencing HMR-SSa, we first determined whether FKH1hc reestablished SIR-dependent chromatin (requiring SIR2–4) in sir1Δ cells (Figure 1B). These experiments were important because in some genetic backgrounds HMRa silencing can be established in the absence of Sir2–4 proteins (Rusche and Rine, 2001). To test whether Sir2–4 chromatin was reestablished by FKH1hc in sir1Δ cells, we performed ChIPs with a cocktail of monoclonal antibodies raised against Sir3 (α-Sir3). Sir3 binding requires the other Sir proteins, Sir2 and Sir4, and it is thus a good measure of association of the SIR-complex (Sir2–4) with a region of chromatin (Hoppe et al., 2002; Luo et al., 2002; Rusche et al., 2002).
These experiments provided evidence that FKH1hc-dependent silencing restored assembly of a Sir2–4 chromatin domain at HMRa in a substantial fraction of sir1Δ cells. Sir3-directed ChIPs specifically enriched HMR-SSa from SIR1 but not sir1Δ cells, indicating that Sir3-association with HMR-SSa required Sir1 as expected based on previous characterizations of HMR-SSa (Gardner and Fox, 2001; Bose et al., 2004) (Figure 1B). Significantly, FKH1hc-dependent silencing in sir1Δ cells, although less efficient than SIR1-dependent silencing (Figure 1A), reestablished measurable Sir3 binding to HMR-SSa (Figure 1B). The enrichment of HMR-SSa in these experiments required SIR3 as none was observed in sir3Δ cells, indicating that the Sir3 antibodies had the appropriate specificity (Figure 1B).
In SIR1-dependent silencing, Sir2–4 complexes bind regions of HMRa beyond the defined silencers (Rusche et al., 2002). This binding to distal regions, termed spreading, requires the deacetylase activity of Sir2, an NAD-dependent histone deacetylase (Denu, 2003). To further examine the molecular nature of FKH1hc-dependent silencing, we compared binding of Sir3 to multiple regions within and outside HMR-SSa by ChIPs (Figure 1C). Both SIR1 and FKH1hc produced similar patterns of Sir3 binding over the HMRa locus. Moreover, efficient Sir3 binding required the Sir2 deacetylase activity. Specifically, Sir3 binding was examined in cells harboring a mutant version of SIR2, sir2N345A, that produces a catalytically defective version of Sir2 (Imai et al., 2000). In these sir2N345A mutant cells, regardless of whether the silencing was mediated through SIR1 or FKH1hc, Sir3 failed to associate with regions distal to the silencer, although it retained some ability to bind to the silencer (Figure 1C). Together, these data provided evidence that FKH1hc- and SIR1-dependent silencing resulted in similar Sir2–4 silent chromatin domains at HMRa that initiated at the HMR-E silencer.
Fkh1hc Prolonged the Cell Cycle and Attenuated the CLB2 mRNA Expression Peak
Several observations provide evidence against the possibility that FKH1hc affects HMR silencing directly. First, Fkh1-3xHA does not bind HMRa as measured by ChIPs (Simon et al., 2001; Hollenhorst, unpublished data), and tethering a Fkh1–Gal4 fusion protein directly to HMRa via an engineered Gal4 binding site fails to restore silencing (Fox, unpublished data), although Gal4-fusions with other proteins known to function directly at the silencer (e.g., Sirs) or to recruit HMRa to the nuclear periphery do (Chien et al., 1993; Lustig et al., 1996; Andrulis et al., 1998, 2004). Second, certain cell cycle perturbations enhance silencing in strains containing mutations in SIR1 or within the HMR-E silencer (Axelrod and Rine, 1991; Laman et al., 1995; Ehrenhofer-Murray et al., 1999), and FKH1 and its closest paralogue FKH2 are direct regulators of transcription of the CLB2-cluster of genes (reviewed in Breeden, 2000) that includes CLB2, the major G2/M phase cyclin. Third a deletion of FKH1 (fkh1Δ) causes a small but measurable effect on cell cycle progression and CLB2 mRNA levels (Hollenhorst et al., 2000). Therefore, we hypothesized that FKH1hc would cause changes in cell cycle progression and CLB2 expression. To test this idea, cell bud index and CLB2 mRNA levels were monitored during cell cycle arrest-and-release experiments (Figure 2, A and B).
MATa bar1Δ cells harboring either an empty 2μ plasmid (vector) or a 2μ plasmid with FKH1 (FKH1hc) were arrested in G1 with α-factor, released from arrest into fresh medium, and at 15-min intervals the cell population was monitored by counting the number of cells in G1(no buds), S (small buds), and G2/M (large buds) (Figure 2A). CLB2 mRNA levels were also measured at each interval by RNA blot hybridization (Figure 2B). Compared with the vector control, FKH1hc entered S phase with similar kinetics but it exhibited a lengthened time in S phase, consistent with the slower growth rate of FKH1hc cells (Figure 2A). For example, although both the control cells (vector) and FKH1hc cells entered S phase 60 min after α-factor release, at 120 min after α-factor release >60% of the FKH1hc cells seemed to remain in S phase or early G2/M phase, whereas only ∼30% of the control cells did (Figure 2A). Differences between the vector control and the FKH1hc cells were also evident at the level of CLB2 mRNA expression (Figure 2B). In particular, although the temporal cycling pattern of CLB2 mRNA was similar in both types of cells, the FKH1hc cells produced lower levels of CLB2 mRNA relative to SCR1 loading control that were most evident during the peak of CLB2 mRNA expression. The overall trend was that FKH1hc dampened the amplitude but not the timing or duration of the CLB2 mRNA wave.
The aforementioned data were consistent with the idea that reduced CLB2 levels might explain the effect of FKH1hc on HMR silencing, but additional analyses of fkh2Δ cells provided evidence against this idea. Fkh2 is the primary transcriptional activator of CLB2-cluster genes (Koranda et al., 2000; Kumar et al., 2000; Pic et al., 2000; Zhu et al., 2000; Reynolds et al., 2003), and a deletion of FKH2 (fkh2Δ) also prolongs the cell cycle and reduces CLB2 mRNA expression (Hollenhorst et al., 2000). In addition, in fkh2Δ cells the level of Fkh1 bound to the promoters of CLB2-cluster genes increases as measured by ChIPs (Hollenhorst et al., 2001). Therefore, one reasonable explanation for the effects of FKH1hc on silencing was that the increased levels of Fkh1 outcompeted Fkh2 for binding to CLB2-cluster promoters, thus mimicking an fkh2Δ-like phenotype. This explanation made two predictions. First, FKH1hc should perturb the cell cycle and CLB2 expression similarly to fkh2Δ. Second, fkh2Δ should enhance silencing in MATα HMR-SSa sir1Δ cells similarly to FKH1hc.
Direct tests of these predictions provided evidence against this explanation. Deletion of FKH2 (fkh2Δ) caused a cell cycle defect distinct from that caused by FKH1hc; fkh2Δ cells showed delayed entry into S phase, but the duration of the remainder of the cell cycle was similar in fkh2Δ and wild-type cells (Hollenhorst et al., 2000; see Figure 2A, fkh2Δ). In terms of CLB2 mRNA expression, fkh2Δ reduced levels of CLB2 mRNA to an even greater extent than FKH1hc. In addition and in contrast to FKH1hc, fkh2Δ delayed the peak of CLB2 mRNA expression. Thus, compared with FKH1hc, fkh2Δ had qualitatively similar but quantitatively distinct effects on the timing and amplitude of CLB2 mRNA expression. Nevertheless, as was true for FKH1hc, the overall effect of fkh2Δ was to reduce CLB2 mRNA levels.
Therefore, we tested, in side-by-side comparisons whether fkh2Δ had any effect on HMRa silencing in MATα HMR-SSa sir1Δ cells or affected the efficiency of FKH1hc-dependent silencing (Figure 2C). As determined by semiquantitative mating assays fkh2Δ had no substantial effect on silencing in MATα HMR-SSa sir1Δ cells nor did fkh2Δ enhance FKH1hc-dependent silencing (Figure 2C). Therefore, FKH1hc was not restoring silencing in sir1Δ cells by creating an fkh2Δ-like phenotype. These data also provided evidence that reductions in CLB2 mRNA expression during the cell cycle were not sufficient to establish FKH1hc-silencing because fkh2Δ cells reduced CLB2 mRNA levels but failed to establish SIR1-bypass silencing. Thus, reduced levels of Clb2 were insufficient to explain FKH1hc-dependent silencing.
The modest levels of Fkh1 produced in FKH1hc cells were also consistent with the conclusion that FKH1hc cells were not achieving SIR1-bypass silencing by competing for Fkh2 target sites. Fkh1 levels were compared in wild-type and FKH1hc cells by using semiquantitative immunoblotting with a polyclonal antibody against Fkh1. These experiments revealed that FKH1hc led to an approximately fourfold increase in the steady-state levels of Fkh1 protein (Figure 2D), a relatively modest increase that was unlikely to be sufficient to compete with Fkh2 for bona fide Fkh2 target sites because Fkh2 binding, but not Fkh1 binding, is enhanced >10-fold through cooperative interactions with Mcm1 (Hollenhorst et al., 2001). Thus, a modest increase in Fkh1 was sufficient to cause both the cell cycle and silencing phenotypes. These data, combined with earlier analyses of fkh1Δ cells that produced cell cycle and CLB2 mRNA expression profiles that were the mirror opposite of those produced by FKH1hc (Hollenhorst et al., 2000) also provided evidence that the phenotypes detected in FKH1hc cells reflected a normal role for Fkh1 in vivo.
Genetic Analyses Revealed that FKH1 and CLB5 Were Functioning in a Common Pathway
Although the aforementioned data indicated that reduced CLB2 expression was insufficient to explain the FKH1hc phenotypes, they did not address whether CLB2 was necessary. In addition, the ability of FKH1hc to perturb the cell cycle (Figure 2) and the documented connection between cell cycle perturbations and enhanced silencing meant that the cell cycle perturbation caused by FKH1hc might be necessary for and/or closely associated with FKH1hc-dependent silencing. Therefore, we asked whether deletions of specific CLB genes affected FKH1hc-mediated silencing in MATα HMR-SSa sir1Δ cells by using RNA blot hybridization of a1 mRNA and mating assays (Figure 3). We focused on analyzing the effects of deletions in the G2/M phase cyclins, CLB1 and CLB2, because these genes are under direct FKH control. In addition, we analyzed the effects of deletions in the S phase cyclins CLB5 and CLB6 because clb5Δ enhances silencing in sir1Δ cells (Laman et al., 1995) and the FKH1hc cell cycle phenotype was consistent with the idea that FKH1hc elongated S phase (Figure 2).
Figure 3.
Genetic analyses revealed that FKH1 and CLB5 were functioning in a common pathway. (A) a1/SCR1 RNA ratios were determined by quantitative RNA blot hybridization for an isogenic series of MATα HMR-SSa sir1Δ strains that differed in terms of their CLB genotype (CLB (CFY762); clb1Δ (CFY1614); clb2Δ (CFY1124); clb5Δ (CFY2104); clb6Δ (CFY2279) and in terms of the plasmid they harbored (vector [pRS426], SIR1hc[pCF34], and FKH1hc[pCF48]). A normalized ratio of a1 mRNA/SCR1 RNA is shown on the y-axis with a value of 1.0 assigned to the a1/SCR1 ratio calculated for CLB cells transformed with a plasmid harboring SIR1 in one experiment. (B) Mating assays in an isogenic series of MATα HMR-SSa sir1Δ strains that differed in terms of their CLB genotype and in terms of the plasmid they harbored as described in A. In these experiments clb5Δclb6Δ (CFY2268) cells were also analyzed. (C) Because mating assays were more robust on rich media, the same cells analyzed in B were also analyzed in the absence of exogenous plasmids after growth in YPD. (D) An isogenic series of MATα HMR-SSa sir1Δ cells differing in their CLB5 and FKH1 genotypes (CLB5 FKH1 [CFY762]; clb5Δ FKH1 [CFY2104]; and clb5Δ FKH1Δ [CFY2120]) were assessed for silencing by mating assays as described above. Mating was assessed after growth of MATα cells on either synthetic CAS medium to retain a URA3 plasmid (vector) or rich medium in the absence of a plasmid (no plasmid) before selection for diploids. (E) The same cells were assessed for silencing by RNA blot hybridization of a1 mRNA. The a1 mRNA/SCR1 RNA ratio was normalized as in (A). The CLB5 FKH1hc cells were CFY762 containing a 2μ plasmid with FKH1 (pCF480). The other cells tested in this experiment contained an empty 2μ plasmid (pCF225).
In terms of the G2/M phase cyclins, CLB2 but not CLB1 was required for FKH1hc-dependent silencing (Figure 3, A and B). Specifically, FKH1hc failed to enhance silencing in MATα HMR-SSa sir1Δ clb2Δ cells as determined by a1/SCR1 RNA ratios (Figure 3A, compare vector and FKH1hc a1/SCR1 RNA ratios for clb2Δ and CLB). Mating assays provided independent evidence for this conclusion (Figure 3B). In contrast, FKH1hc enhanced silencing in MATα HMR-SSa sir1Δ clb1Δ cells based upon both RNA blot hybridization (Figure 3A) and mating assays (Figure 3B). Thus direct measurements of the a1/SCR1 RNA ratios and mating assays provided complementary evidence that CLB2 was necessary for establishing FKH1hc-dependent SIR1-bypass silencing at HMR. However, although CLB2 was necessary it was not sufficient; clb2Δ failed to enhance silencing in MATα HMR-SSa sir1Δ as measured by a1/SCR1 ratios (Figure 3A) or mating assays on cells grown either under selective conditions to retain plasmids (Figure 3B) or under rich conditions (Figure 3C). These data were consistent with the conclusion from the cell cycle experiments described in Figure 2 that reductions in CLB2 were insufficient to create the FKH1hc phenotype. Thus, even if the effect of FKH1hc on CLB2 expression contributes to FKH1hc-dependent silencing, or CLB2 functions in conjunction with FKH1hc, other CLB2-independent targets or mechanism(s) also must be relevant to FKH1hc-dependent silencing.
An earlier study demonstrated that clb5Δ could enhance silencing of sir1Δ cells (Laman et al.,1995). Although CLB2 is a direct target of Fkh2, our data provided evidence that reductions in CLB2 were insufficient to account for FKH1hc-dependent silencing. Therefore, we postulated that pathways or target genes affected by the S phase cyclin CLB5 might be more immediately relevant to FKH1hc-dependent SIR1-bypass silencing. In particular, if FKH1hc and clb5Δ achieved SIR1-bypass silencing through modulation of a common pathway, then FKH1hc and clb5Δ should cause quantitatively similar degrees of SIR1-bypass silencing, and, in addition, the two genes should fail to show additive interactions in terms of this phenotype. Therefore, we tested whether clb5Δ cells affected FKH1hc-dependent silencing and whether fkh1Δ cells affected clb5Δ-dependent silencing.
As expected, clb5Δ but not clb6Δ enhanced silencing in MATα HMR-SSa sir1Δ cells (Figure 3A; compare vector a1/SCR1 RNA ratios for clb5Δ and clb6Δ to CLB), consistent with the findings of a previous study (Laman et al., 1995). Based on RNA blot hybridization, clb5Δ and FKH1hc enhanced HMR-SSa silencing to similar degrees, and FKH1hc enhanced levels of silencing in clb5Δ only slightly but reproducibly (Figure 3A). The mating assays were consistent with the RNA blot hybridizations; FKH1hc provided for some level of silencing that was independent of CLB5 (Figure 3B, clb5Δ, compare vector and FKH1hc), but it was unable to enhance silencing of clb5Δ sir1Δ cells as well as it enhanced silencing of CLB5 sir1Δ cells. These data were consistent with the idea that FKH1hc and clb5Δ were impinging on a common pathway or target protein, to different extents or through different mechanisms, that was limiting for HMRa silencing in sir1Δ cells.
In contrast to CLB5 and CLB2, the CLB6 genotype had no substantial impact on FKH1hc-dependent silencing; clb6Δ did not enhance silencing in MATα HMR-SSa sir1Δ cells nor did it perturb the ability of FKH1hc to enhance silencing in these cells (Figure 3A, clb6Δ), consistent with an earlier analysis of the effect of clb6Δ on silencing (Laman et al., 1995). Interestingly, within the detection ranges of a1 mRNA, clb6Δ consistently reduced silencing by a small amount (Figure 3A, compare vector in clb6Δ to CLB cells), perhaps by enhancing the relative activity of CLB5 in vivo. Analyses of cells containing deletions of both CLB5 and CLB6 provided evidence that CLB5 was the S phase cyclin with the most significant impact on FKH1hc-silencing; clb5Δ clb6Δ produced silencing phenotypes similar to those produced by clb5Δ alone (Figure 3, B and C).
In these studies mating efficiencies were ∼10-fold lower and somewhat more variable when measured for cells containing plasmids and grown under selective growth conditions. Therefore, as an independent and additional assessment of the effect of CLB genotype on silencing, mating assays were performed on the same set of yeast cells described above that were instead grown on rich nonselective growth medium and in the absence of plasmids (Figure 3C). The data from these experiments were consistent with those obtained with cells harboring plasmids and grown on selective medium (compare to Figure 3B, vector).
These data provided evidence that FKH1hc affected silencing similarly to a clb5Δ and might function in a common pathway because FKH1hc only slightly enhanced the levels of silencing achieved in clb5Δ cells (Figure 3, A and B). A simple model was that FKH1hc somehow reduced the levels or activity CLB5. In this model, CLB5 function in silencing formally occurred downstream of FKH1 and if correct, a deletion of FKH1 (fkh1Δ) would have little or no effect on silencing achieved by a deletion of CLB5 (clb5Δ). To test this idea, we compared silencing of HMR-SSa in sir1Δ cells that were either clb5Δ FKH1 or clb5Δ FKH1Δ (Figure 3D). clb5Δ-dependent silencing was consistently ∼10-fold better in FKH1 cells compared with fkh1Δ cells as measured by mating assays (Figure 3D). As described above, mating assays were ∼10-fold more efficient when cells were grown on rich medium in the absence of plasmids before selective mating. Nevertheless, regardless of how cells were grown before the mating assay (harboring an empty plasmid and grown an selective media to retain the plasmid [vector] or grown on rich media in the absence of a plasmid [no plasmid]), the mating assays revealed that clb5Δ-dependent silencing was reduced ∼10-fold in fkh1Δ cells (Figure 3D). However, clb5Δ still clearly enhanced silencing in fkh1Δ cells. This effect was also evident in RNA blot hybridization experiments (Figure 3E); chromosomal FKH1 was required for the full level of clb5Δ-silencing. Thus these data provided evidence that chromosomal levels of FKH1 could affect HMRa silencing. Furthermore, the epistasis analyses provided evidence that CLB5 and FKH1 impinged on HMR silencing at least in part through a common pathway or target protein.
FKH1hc Did Not Affect Clb5 Levels nor Did CLB5 Genotype Affect Fkh1 Levels
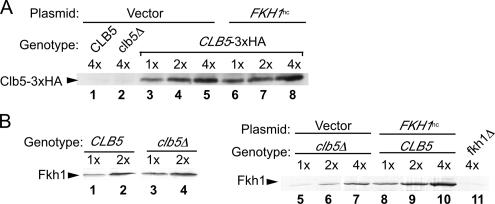
One possible explanation for some of the genetic data described above was that the FKH1hc reduced levels of Clb5 protein and thereby contributed to a clb5Δ-like phenotype. Conversely, perhaps clb5Δ-enhanced silencing was mediated by increases in the steady-state levels of Fkh1 protein. To test whether FKH1hc affected Clb5 protein levels, Clb5-3xHA was monitored by protein immunoblotting in MATα sir1Δ HMR-SSa cells harboring either an empty 2μ plasmid (vector) or a 2μ plasmid containing FKH1 (FKH1hc) (Figure 4A). These data provided evidence that FKH1hc had no effect on Clb5-3xHA levels. To test the converse possibility that clb5Δ-dependent silencing worked through increasing Fkh1 levels, Fkh1 was monitored in CLB5 or clb5Δ cells by protein immunoblotting (Figure 4B). Fkh1 levels were unaffected by a clb5Δ (Figure 4B, compare levels of Fkh1 in CLB5, lanes 1 and 2) to clb5Δ cells (lanes 3 and 4) and compare levels of Fkh1 in clb5Δ cells transformed with an empty plasmid (vector, lanes 5–7) to CLB5 cells transformed with FKH1hc (lanes 5–11). These data provided evidence that clb5Δ-dependent silencing did not result from increased levels of Fkh1 and that FKH1hc-dependent silencing did not result from reduced levels of Clb5.
Figure 4.
FKH1hc did not affect Clb5 levels nor did CLB5 genotype affect Fkh1 levels. (A) Levels of Clb5-3xHA protein were determined by protein immunoblot with anti-HA antibodies in CLB5 (CFY762), clb5Δ (CFY2104), and CLB5–3xHA (CFY2446) cells transformed with either an empty 2μ plasmid (vector) or a 2μ plasmid containing FKH1 (FKH1hc; pCF480). We analyzed 0.25 OD (optical density at 600 λ) cell equivalents per 4× lane, and 2× and 1× lanes contained twofold and fourfold reductions in that amount of extract, respectively. (B) Levels of Fkh1 protein were determined by protein immunoblot with anti-Fkh1 antibodies in CLB5 (CFY762) and clb5Δ (CFY2104) cells, as indicated (lanes 1–4) and in clb5Δ cells transformed with an empty 2μ plasmid (vector [pCF225]; lanes 5–7) or CLB5 cells transformed with a 2μ plasmid containing FKH1 (FKH1hc [pCF480]; lanes 8–10). fkh1Δ cells (CFY527) containing vector (pCF225) were analyzed as a negative control (lane 11). We analyzed 0.25 OD cell equivalents per 4× lane and 2× and 1× were twofold and fourfold reductions, respectively, in that amount of extract.
FKH1hc Could Use Nonsilencer Replication Origins in Place of the HMR-E Silencer
As shown above, SIR1- and FKH1hc-dependent silencing differed in terms of their relationship to the major G2 and S phase cyclin genes, indicating that at some level these two forms of silencing relied on different sets of molecular interactions to ultimately establish the same SIR2–4 chromatin structures. It is well established that Sir1 performs its silencing function primarily through its interactions with the HMR-E silencer, an element with a distinct arrangement of protein binding sites, most notably a binding site for ORC juxtaposed next to a binding site for Rap1 (Fox and McConnell, 2005) (Figure 5A, HMR-SS). Both ORC and Rap1 play important roles at the HMR-E silencer, with ORC recruiting Sir1 and Rap1 recruiting Sir3 and Sir4 through direct protein–protein interactions. To gain further insight into how SIR1- and FKH1hc-dependent silencing differed in terms of their ability to establish Sir2–4 chromatin, their silencer requirements were compared.
Figure 5.
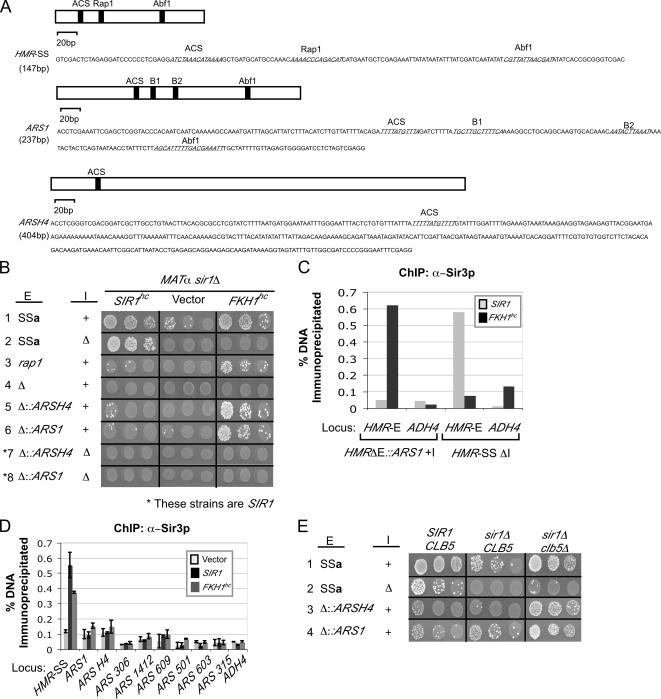
FKH1hc or clb5Δ could use nonsilencer replication origins in place of the HMR-E silencer. (A) Structures and sequences of the E-silencer variations used in these experiments. HMRΔE:ARS contains either ARS1 or ARSH4 in the orientation shown as a complete substitute for HMR-SS. (B) Mating assays were performed with an isogenic series of MATα sir1Δ strains that differed in terms of their HMR-E (E) and/or HMR-I (I) silencers at HMRa, as indicated to the left of the figure and described in the text. The cells contained the following silencers: HMR-SSa (CFY1649) in place of an 800-base pair deletion that includes HMR-E (McNally and Rine, 1991; Palacios DeBeer and Fox, 1999); HMR-SSa as described above, and a 335-base pair deletion of that includes HMR-I (CFY110) (Fox et al., 1995); HMR-SSa with a mutation in the Rap1 binding site (CFY2221) (McNally and Rine, 1991); a deletion of an 800-base pairs region, including HMR-E (CFY2133); deletion of an 800-base pairs region including HMR-E replaced with ARSH4 (CFY2071) or ARS1 (CFY2237) as shown in A. Each strain was transformed with a 2μ plasmid containing SIR1 (SIR1hc [pCF345]), an empty 2μ plasmid (vector [pCF225]), or a 2μ plasmid containing FKH1 (FKH1hc [pCF480]). Mating was assessed by drop tests in which 10-fold dilutions of MATα cells being tested were mixed with an excess of MATa cells (CFY616) and grown on medium that selected for diploids retaining the plasmids. (C) Sir3-directed ChIPs were performed on MATα sir1Δ HMRΔE::ARS1 + HMR-I cells (CFY2071) or on MATα sir1Δ HMR-SSa + HMRΔI (CFY35) cells transformed with either SIR1 or FKH1hc as indicated. (D) Sir3-directed ChIPs were used to determine Sir3 association with a number of ARSs compared with HMR-SSa in sir1Δ cells transformed with vector, SIR1, or FKH1hc. (E) Mating assays were performed with an isogenic series of MATα cells that differed in terms of their silencers at HMRa (as indicated on left of figure and described in A) and their SIR1 or CLB5 genotypes (as indicated at the top of the figure). The various strains used in these experiments were as follows: SIR1 CLB5 with HMR-SSa (CFY345); HMR-SSaΔI (CFY35); HMR containing an 800-base pair deletion that includes HMR-E replaced with either ARSH4 or ARS1 (CFY325 and CFY321, respectively). sir1Δ CLB5 versions with the silencers as listed for SIR1 CLB5 (CFY1649, CFY110, CFY2071 and CFY2237); and sir1Δ clb5Δ with versions with the same silencers as listed (CFY2104, CFY2230, CFY2193, CFY2236).
Appropriate MATα sir1Δ yeast cells that varied only in terms of their silencers at HMRa were transformed with an empty 2μ vector (vector) or 2μ plasmids harboring SIR1 (SIR1hc) or FKH1 (FKH1hc). The transformed cells were compared for their ability to silence HMRa in mating assays (Figure 5B). As expected, SIR1hc-dependent silencing did not require the presence of the HMR-I silencer (Figure 5B, line 2, SIR1hc). In contrast, FKH1hc-dependent silencing was abolished by deletion of HMR-I (Figure 5B, line 2, FKH1hc). Thus, the two forms of silencing showed different dependencies on HMR-I. However, it is important to note that SIR1hc- (and SIR1 at chromosomal levels) is more effective than FKH1hc at silencing HMRa (Figure 1A), and that deletion of HMR-I weakens silencing under a variety of conditions (Fox et al., 1995; Rivier et al., 1999). Therefore, the different requirement for HMR-I might simply reflect the ability of SIR1 to silence more robustly than FKH1hc. To test this possibility, we examined mutant silencers that we knew were defective in SIR1-dependent silencing for their ability to support FKH1hc-dependent silencing (Figure 5B, lines 3–5). Importantly, these experiments revealed that several silencers existed that were better at supporting FKH1hc- than SIR1-dependent silencing.
First, a mutation in the Rap1 binding site (Figure 5B, rap1) in HMR-SSa abolishes silencing even when SIR1 is overexpressed (McNally and Rine, 1991); Figure 5B), line 3, SIR1hc), presumably because Rap1 is so critical for stabilizing the binding of Sir3 and Sir4 proteins (Moretti and Shore, 2001). It was surprising, given that FKH1hc silencing clearly required Sir3 and Sir4, that FKH1hc could restore some silencing to cells that contained a mutation in the Rap1 binding site within HMR-SSa, whereas SIR1 could not (Figure 5B, line 3). It was clear that both FKH1hc- and SIR1hc-dependent silencing required some feature of an E-silencer because an entire deletion of E (HMRΔE) failed to support either forms of silencing (Figure 5B, line 4). This experiment established that FKH1hc differed from SIR1 in terms of a requirement for a Rap1 site in the HMR-E silencer.
As mentioned above, the role for ORC in SIR1-dependent silencing is well established (Fox and McConnell, 2005). Sir1 shows a high specificity for silencer-bound ORCs in vivo; Sir1 cannot be detected at several nonsilencer replication origins such as ARS1, even though such elements bind ORC (Gardner and Fox, 2001). Furthermore, substitution of the entire E-silencer with the nonsilencer replication origins ARS1 or ARSH4 (Figure 5A, sequences of “HMR-E” silencers used in Figure 5B) fails to support SIR1-dependent silencing (Fox et al., 1995). Even when SIR1 was overexpressed ARS1 or ARSH4 failed to function as effective “E-silencers” (Figure 5B, lines 5 and 6, SIR1hc and vector). Thus, it was unexpected that efficient FKH1hc-dependent silencing could be achieved with either ARS1 or ARSH4 as substitutes for the HMR-E-silencer (Figure 5B, lines 5 and 6, FKH1hc). What was particularly striking was that the level of FKH1hc-dependent silencing achieved in cells in which either ARS1 or ARSH4 served as the HMR-E silencer was equivalent to that achieved in the HMR-SSa silencer background that was used in the original isolation of FKH1 as a high-copy suppressor of a sir1Δ mutation (Hollenhorst et al., 2000) (Figure 5B, line 1). That is, for FKH1hc-dependent silencing and in contrast to SIR1-dependent silencing, either ARS1 or ARSH4 was as good as HMR-E in terms of functioning as an E-silencer.
As an independent assessment of the unexpected silencer requirements described above, a Sir3-directed ChIP experiment was performed (Figure 5C). These data were consistent with the silencing data; Sir3 could bind to HMRΔE::ARS1 in FKH1hc cells but not SIR1 cells. In contrast, Sir3 could bind to HMR-E within an HMR locus that lacked HMR-I in SIR1 cells but not in FKH1hc cells. These ChIP data added supporting evidence to the conclusion that FKH1hc cells could use an ordinary ARS in place of HMR-E for Sir2–4–dependent silencing.
The data described above indicated that in FKH1hc cells, the Sir3 protein was exhibiting some affinity for ARS1 or ARSH4 when present at HMR in place of HMR-E. To test whether Sir3 was exhibiting some affinity for these and other ARSs in their native locations, we used ChIP to examine Sir3 binding to a number of ARSs in sir1Δ (vector), SIR1, and sir1Δ FKH1hc cells (Figure 5D). These data revealed that the level of Sir3 binding detected by ChIP to ARSs at their native location was only slightly above background and far below the level of Sir3 that could be detected by ChIP at HMR. Because HMR-I was critical for FKH1hc-dependent silencing (Figure 5B, lines 7 and 8), these data were not surprising because HMR-I is an element unique to the HMR locus. Thus, although FKH1hc may enhance the affinity for Sir3 at some nonsilencer ARSs, stable binding of Sir3 to these elements that is detectable by ChIP requires features unique to HMR, including but not necessarily only HMR-I.
In summary, FKH1hc- and SIR1hc-dependent silencing differed measurably in terms of silencer requirements optimal for producing Sir2–4–dependent chromatin at HMRa. Most remarkably, FKH1hc could establish Sir2–4 chromatin by using ARS1 or ARSH4 in place of the HMR-E silencer, whereas SIR1hc could not.
clb5Δ Also Could Use ARS1 or ARSH4 as a Substitute for HMR-E
The data concerning the relationship between clb5Δ and FKH1hc in terms of SIR1-bypass silencing (Figure 3) raised the possibility that clb5Δ and FKH1hc exerted their effects on silencing through a common pathway. If this were true, then clb5Δ-dependent silencing should share the same unusual silencer requirements as FKH1hc. To test this idea, we determined whether clb5Δ-dependent silencing could be achieved with the same mutant silencers that effectively established FKH1hc-dependent silencing (Figure 5E). As was true for FKH1hc-dependent silencing, clb5Δ-dependent silencing required HMR-I (Figure 5E, lines 1 and 2). More strikingly, however, clb5Δ cells were able to use ARSH4 or ARS1 as effective substitutes for the HMR-E silencer (Figure 5E, compare lines 3 and 4 with line 1 for sir1Δclb5Δ column), whereas SIR1 could not (Figure 5E, compare lines 3 and 4 with line 1 for SIR1CLB5 column). In summary, clb5Δ and FKH1hc could each establish SIR1-independent silencing at HMRa by using replication origins as substitutes for HMR-E.
clb5Δ or FKH1hc Could Suppress HMRΔE::ARS1 Origin Activity
The relationship between origin firing and SIR2–4–dependent chromatin is, in general, antagonistic; wherever SIR2–4 silent chromatin forms, origin firing is suppressed (Stevenson and Gottschling, 1999; Zappulla et al., 2002). Conversely, origin firing at the HM loci is inefficient regardless of SIR genotype, in part because the silencers are intrinsically ineffective replication origins (Dubey et al., 1991; Palacios DeBeer and Fox, 1999; Vujcic et al., 1999; Sharma et al., 2001; Palacios DeBeer et al., 2003).
CLB5 is the S phase cyclin most critical for activating replication origins that fire late in S phase (Donaldson et al., 1998; Gibson et al., 2004). Thus, in clb5Δ cells early S phase origins fire efficiently, whereas late S phase origins do not, and overall S phase progression is slowed because replication forks emanating from early origins must now replicate a greater fraction of the genome. In the ARS1 substitution experiment described above (Figure 5), the entire HMR-E silencer was deleted and replaced with ARS1. In its native location, ARS1 fires efficiently during early- to mid-S phase and is CLB5 independent (Donaldson et al., 1998; Gibson et al., 2004). Given this collection of observations, we asked how the ARS1 origin that replaced HMR-E was affected in clb5Δ cells by performing 2-D origin mapping experiments on ARS1 at its native location and at HMR where it substituted for the HMR-E silencer (HMRΔE::ARS1) (Figure 6).
Figure 6.
clb5Δ or FKH1hc suppressed HMRΔE::ARS1 origin activity. (A) 2-D origin mapping experiments were performed in MATα HMRΔE::ARS1 sir1Δ cells that contained either CLB5 (CFY2237) or clb5Δ (CFY2236). Origin activity at HMRa was assessed with an HMR-specific probe, whereas origin activity at the native ARS1 locus was assessed with an ARS1-specific probe. Two different exposures of HMRΔE::ARS1 blots are shown (dark and light) to facilitate comparison of this origin activity in CLB5 versus clb5Δ cells. (B) 2-D origin mapping experiments with the cells used in A were repeated side by side with clb5Δ sir2Δ cells (CFY2481). (C) The DNA content in actively dividing populations of isogenic yeast cells that varied in terms of their SIR2 or CLB5 genotypes as indicated was determined by flow cytometry. (D) 2-D origin mapping experiments were performed in MATα HMRΔE::ARS1 sir1Δ CLB5 cells containing either an empty 2μ plasmid (vector; pCF225) or a 2μ plasmid with FKH1 (FKH1hc; pCF480).
These 2-D origin-mapping experiments revealed that in wild-type (CLB5) cells ARS1 functioned as a robust, efficient origin whether it was present at its native location (ARS1) or at HMR, as a substitute for the deleted HMR-E silencer (HMRΔE::ARS1) (Figure 6A, CLB5). However, only ARS1 at HMRa (HMRΔE::ARS1) was sensitive to CLB5 genotype; clb5Δ reduced the ability of ARS1 to function as an origin when it substituted for HMR-E (clb5Δ, HMRΔE::ARS1) whereas it had no effect on ARS1 firing at its native locus (clb5Δ, ARS1) (Figure 6A, clb5Δ).
Two different mechanisms could contribute to clb5Δ-mediated suppression of origin firing by HMRΔE::ARS1. First, clb5Δ reestablished silencing at HMRΔE::ARS1 and thus silencing, or Sir2–4 chromatin formation, might be inhibiting firing by HMRΔE::ARS1. If this scenario were true, then a deletion of SIR2 (sir2Δ), a gene essential for silencing, should restore efficient firing by HMRΔE::ARS1 in clb5Δ cells as is observed for other cases of Sir2–4–mediated origin suppression (Zappulla et al., 2002). Second, some other SIR-independent feature of this chromosomal region might suppress origin firing by HMRΔE::ARS1 in clb5Δ cells. As discussed above, the HM loci fail to allow for more robust origin firing by their native silencers even in sir mutant cells that cannot silence. To distinguish between these mechanisms, we performed 2-D origin-mapping experiments on HMRΔE::ARS1 in clb5Δ sir2Δ cells, thereby precluding the ability of clb5Δ to reestablish Sir2–4 chromatin at HMRa. The sir2Δ mutation failed to restore robust firing to the HMRΔE::ARS1 origin in clb5Δ cells (Figure 6B), suggesting that SIR2–4 chromatin was not the cause of CLB5-sensitive firing by HMRΔE::ARS1. However, a complicating factor in interpreting these experiments was that large replication forks were also consistently underrepresented in sir2Δ clb5Δ cells compared with CLB5 SIR2 or clb5Δ SIR2 cells raising the possibility that sir2Δ was having additional effects on genome replication. Nevertheless, the replication pattern generated for HMRΔE::ARS1 in sir2Δ clb5Δ cells indicated that sir2Δ did not simply restore robust replication to HMRΔE::ARS1. Thus, the dependence on CLB5 that ARS1 acquired when it replaced HMR-E was independent of Sir2–4 chromatin formation.
A recent study raises the possibility that SIR genes, in particular SIR2, can negatively regulate many replication origins in the yeast genome, most probably at the level of pre-RC assembly (Pappas et al., 2004). Therefore, in addition to examining the effect of a sir2Δ on CLB5 sensitivity of a particular origin (Figure 6B), the effect of a sir2Δ on DNA replication was examined in clb5Δ cells by flow cytometry (Figure 6C). In the simplest case, if the majority of CLB5-sensitive origins were negatively regulated by SIR2, then a sir2Δ mutation would suppress the defects of a clb5Δ. However, examination of the DNA content in asynchronous populations of cells indicated that a sir2Δ did not reestablish a wild-type CLB5 DNA-content profile. In addition, cell-cycle arrest and release experiments indicated that sir2Δ clb5Δ cells showed a delayed entry into M phase compared with sir2Δ CLB5 cells, suggesting that many of the global replication defects caused by a clb5Δ were not rescued by a sir2Δ (Fox, unpublished data). Together, these data provided evidence that SIR2 likely did not regulate many CLB5-sensitive origins, because sir2Δ did not suppress a clb5Δ, consistent with our data from directed analyses of HMRΔE::ARS1 in sir2Δ clb5Δ cells (Figure 6B).
The genetic and cell cycle data concerning FKH1hc were consistent with the notion that clb5Δ and FKH1hc functioned in part through a shared pathway or target to modulate HMRa silencing. As another test of this shared relationship, we determined the effect that FKH1hc had on origin firing by HMRΔE::ARS1 (Figure 6D). Significantly, FKH1hc also reduced the efficiency of origin firing by HMRΔE::ARS1, producing a molecular phenotype similar to that caused by a clb5Δ (Figure 6A, compare vector with FKH1hc), albeit the effect of FKH1hc on origin firing by HMRΔE::ARS1 was less drastic than the effect of clb5Δ on this origin. This observation was consistent with clb5Δ having a more significant impact on cell growth compared with FKH1hc (Casey, unpublished data). Nevertheless these data supported the idea that FKH1hc and clb5Δ bypassed the requirement for SIR1 in establishing Sir2–4 chromatin through effects on a similar target or pathway that can impinge on or is associated with S phase progression.
DISCUSSION
In this report, we addressed the hypothesis that Fkh1, an evolutionarily conserved transcription factor implicated in transcriptional control of CLB2-cluster genes (reviewed in Breeden, 2000; Futcher, 2000), modulated Sir2–4 chromatin formation at HMRa through an effect on cell cycle progression. Several lines of evidence presented here linked FKH1 and the major S phase cyclin CLB5 to a common pathway(s) or target(s). First, FKH1hc and clb5Δ each bypassed the requirement for SIR1 in silencing HMRa to a similar extent. Second, FKH1hc and clb5Δ each had the unexpected ability to use a replication origin, ARS1 or ARSH4, as an effective substitute for the HMR-E silencer in silencing HMRa. And third, FKH1hc and clb5Δ each reduced origin firing by the normally robust ARS1 origin when it substituted for the HMR-E silencer. Together, this set of shared phenotypes supports a model in which FKH1 and CLB5 impinge on a common pathway relevant to both the control of late-firing replication origins and the cell-cycle sensitive formation of Sir2–4–repressive chromatin at HMRa.
It was conceivable that FKH1 and CLB5 were part of a linear pathway in which FKH1 affected expression of CLB5 or vice versa as such a relationship could explain the similar phenotypes caused by FKH1hc and clb5Δ. Fkh1 is, after all, a transcription factor that could directly or indirectly affect levels of Clb5. However, direct analyses of Fkh1 and Clb5 protein levels provided evidence against the simplest version of this possibility. In particular, FKH1hc had no effect on the steady-state levels of Clb5, and, conversely, clb5Δ had no effect on either chromosomally produced levels of Fkh1 or the ability to overexpress Fkh1 to levels sufficient to establish HMRa silencing. Moreover, double mutant analyses also provided evidence against a simple linear relationship between FKH1 and CLB5. If FKH1hc enhanced HMRa silencing entirely through CLB5, we would have predicted that FKH1hc would fail to enhance silencing at all in clb5Δ cells. However, FKH1hc did enhance silencing of clb5Δ cells to a small but reproducible extent as determined by both mating assays and RNA blot hybridizations. Similarly, although clb5Δ cells required a chromosomal copy of FKH1 for maximal HMRa silencing, these cells still silenced HMRa more effectively than corresponding CLB5 cells. A simple model consistent with these data describes a forked pathway in which FKH1 and CLB5 have opposing regulatory roles on common target(s) (T) in this pathway or at least targets with shared functions (Figure 7). That is, the final activity of T depends on contributions from both FKH1 and CLB5. Given what is known about the proteins encoded by FKH1 and CLB5, a reasonable idea is that FKH1 controls the levels of T, whereas CLB5 negatively regulates the activity of T through phosphorylation (e.g., by the Clb5/Cdc28 kinase). Target T in turn impinges on the activity of late-firing replication origins (or at least a subset of these origins) and the assembly of Sir2–4 chromatin.
Figure 7.
Two models for how FKH1 and CLB5 modulated origin activity and silencing at HMRa (A) FKH1 positively regulates (arrow) and CLB5 negatively regulates (flat head) a common pathway or target referred to as T. T in turn negatively regulates origin firing by normally late-firing replication origins (or origins placed within chromosomal domains that cause late firing). Robust origin firing is incompatible with the assembly of Sir2–4 chromatin. (B) FKH1 and CLB5 regulate target T as described in A, but origin firing is not causally linked to Sir2–4 chromatin assembly. Rather firing by late replication origins is negatively regulated, whereas Sir2–4 is positively regulated by target T.
Role of CLB2
Our genetic and molecular studies provided evidence for how FKH1 and CLB5 were related, but based on our current data it is difficult to determine how the M phase cyclin CLB2 fits into the model (Figure 7). Because clb2Δ can rescue some HMR silencing defects (Laman et al., 1995) and Fkh1 is a direct transcriptional regulator of CLB2, it was unexpected that a clb2Δ failed to mimic the silencing phenotype caused by FKH1hc. Although FKH1hc clearly reduced levels of CLB2 mRNA, the inability of fkh2Δ cells, which also reduced the levels of CLB2 mRNA, as well as clb2Δ cells to establish SIR1-bypass silencing provided strong evidence that reductions in CLB2 were not sufficient to explain FKH1hc-dependent silencing. The observation that CLB2 was required for FKH1hc-silencing could be because CLB2 acts positively on target T through an FKH1-independent mechanism, and that the effects of FKH1hc on CLB2 mRNA levels were unrelated to how FKH1 established SIR1-bypass silencing at HMR. Alternatively reduced levels of CLB2 may be an essential component of FKH1hc silencing, but the precise pattern and amplitude of CLB2 mRNA expression caused by FKH1hc is itself critical, such that a clb2Δ or the distinct pattern of CLB2 reduction caused by fkh2Δ does not suffice to create SIR1-bypass silencing. A better understanding of precisely how Fkh1 controls CLB2 expression and/or a FKH-independent mode for regulating CLB2 mRNA levels would help address this issue.
Sir2–4 Chromatin Formation and the Cell Cycle
It is well established that perturbations in the cell cycle can enhance or reduce the efficiency of silencing assessed in certain mutant backgrounds, but the mechanisms behind these effects are unclear (Axelrod and Rine, 1991; Laman et al., 1995; Ehrenhofer-Murray et al., 1999). In addition, studies in which expression of individual SIR genes is controlled with inducible promoters or temperature-sensitive mutations indicate that multiple phases of the cell cycle may modulate different distinct steps required for the ultimate assembly of functionally silenced chromatin (Lau et al., 2002; Kirchmaier and Rine, 2006; Matecic et al., 2006). For example, studies using a temperature-sensitive sir3 allele or an inducible version of SIR1 provide evidence that Sir2–4 can rebind HMRa in G1 phase yet fail to establish silencing (Lau et al., 2002; Kirchmaier and Rine, 2006). Rather silencing, as defined by repression of transcription of the a1 gene, is not established until passage through the following S and M phases. Thus some of the cell cycle-regulated steps that ultimately lead to a silenced transcriptional state at HMRa must occur downstream of the binding of Sir proteins to HMRa. It was conceivable, therefore, that the effects of FKH1hc and by inference those of several cell cycle mutants on HMRa silencing were mediated through similar steps and that they did not affect the efficiency of Sir binding to chromatin. Our use of the synthetic silencer (HMR-SSa) (McNally and Rine, 1991) allowed us to address this question. Unlike natural HMRa where substantial levels of Sir3 binding can be detected by ChIP even in sir1Δ cells (Rusche et al., 2002), Sir3 binding to HMR-SSa was completely dependent on Sir1. This feature allowed us to demonstrate unequivocally that FKH1hc reestablished Sir2–4 nucleation at HMR-E even in the absence of Sir1. Thus FKH1hc, and by inference clb5Δ, bypass the requirement for SIR1 in HMRa silencing at least in part by promoting efficient Sir2–4 nucleation at a silencer.
Linking Late Origin Firing and HMRa Silencing
The close association between silencers and DNA elements with the capacity to function as origins at the HM loci has spurred studies over the past two decades aimed at trying to explain the mechanistic nature of this link. Recent data focused on HMR-E has raised the possibility that there exists a competing relationship between the roles of ORC at origins and silencers and that the close association between these elements is a by-product of the dual functions of ORC (Palacios DeBeer and Fox, 1999; Palacios DeBeer et al., 2003; McConnell et al., 2006). In this model for the silencer–origin relationship, HML and HMR silencers are weak or inactive origins at least in part because robust and/or early origin firing inhibits silencing. Therefore, it was unexpected that ARS1 and ARSH4, two independent and robust replication origins with no documented silencer activity, could act as effective substitutes for the HMR-E silencer. Indeed in a silencer-trap screen in which the 2μ ARS was shown to possess weak silencer activity, direct tests of ARS1 demonstrated it could not substitute for HMR-E in silencing HMRa (Grunweller and Ehrenhofer-Murray, 2002). This observation was confirmed in this report because neither SIR1 cells nor sir1Δ cells could use ARS1 as effectively as they used HMR-E for silencing HMRa. However, what was new and surprising was that ARS1 could function as effectively as HMR-E in sir1Δ cells that also harbored FKH1hc or clb5Δ. Thus, ARS1 became an effective SIR1-independent silencer under conditions that also, notably, reduced the ability of ARS1 to act as a replication origin. Therefore, this observation was consistent with the idea that robust origin and silencer activity were incompatible and that target T enhanced Sir2–4 chromatin formation because it inhibited origin firing by HMRΔE::ARS1 (Figure 7A). Sir2–4 chromatin did not create a late-firing CLB5-sensitive origin at HMRa because even in the absence of silent chromatin formation (sir2Δ) the HMRΔE::ARS1 origin remained suppressed in clb5Δ cells. However, another explanation consistent with the observations reported here was that late origin firing and silencer function were not linked causally but instead could be regulated in opposing directions by the same target T (Figure 7B). This second version of the model is actually more consistent with the observation that clb5Δ clb6Δ cells could silence as effectively as clb5Δ clb6 cells, because clb5Δ clb6Δ cells restore a normal S phase (Schwob and Nasmyth, 1993), suggesting that suppression of late origin firing is not essential for Sir2–4 chromatin formation in clb5Δ cells.
Because silencing of HMRa weakened by silencer mutations can be enhanced by artificially tethering the mutant HMRa locus to the nuclear membrane (Andrulis et al., 1998), we propose that a reasonable candidate for target T is a cell cycle-regulated protein(s) or protein complex that modulates the association of HMRa with the nuclear periphery, where Sir2,-3, and -4 proteins are concentrated (Palladino et al., 1993; Maillet et al., 1996; Taddei et al., 2005). In addition, late-firing replication origins show an enhanced association with the inner nuclear membrane in both yeast and mammals (Dimitrova and Gilbert, 1999; Heun et al., 2001), which also supports the hypothesis that target T represents a protein or complex that affects the association of HMRa with the nuclear membrane and that this association in turn can affect both origin firing and silencing.
In this interpretation, ARS1 and ARSH4 were equivalent to an HMR-E silencer weakened by mutation. That is, these origins acted as “proto-silencers” (Boscheron et al., 1996), too weak to function on their own but efficient enough when combined with other Sir-stabilizing influences, such as another weak proto-silencer, HMR-I. This must mean that ARS1 and ARSH4 possess an intrinsic affinity, albeit substantially reduced compared with the native silencer, for the Sir2–4 protein complex. Thus even in the absence of Sir1, Sir2–4 proteins must have some inherent affinity for origins, or at least a component of origins, such as ORC. Consistent with this possibility, a recent study provides evidence that SIR2, and less effectively SIR3 and SIR4, can affect the activity of many genomic origins, suggesting that these proteins may interact with many nonsilencer origins in the genome (Pappas et al., 2004). This interaction may be fleeting and thus not detectable by ChIP. However, when this weak interaction is combined with additional weak Sir interaction sites provided by the HMR-I silencer and a higher concentration of Sir proteins that might be provided by association with the nuclear membrane, the result is sufficient stabilization energy that allows Sir2–4 proteins to establish silencing. Thus HMRa silencing in clb5Δ (or FKH1hc) cells measured here might be effectively trapping transient interactions that occur throughout the genome between Sir2–4 complexes and origins.
In summary, the cell cycle perturbation caused by a clb5Δ (or FKH1hc) associated with enhanced silencing could involve an increase in the levels and/or activity of a target T. Although it is possible that target T acts to suppress late-firing replication origins that in turn directly interfere with silencers (Figure 7A), it is also possible that both silencers and late-firing replication origins are connected to the common target T that in turn regulates the function of each of these DNA elements in opposing directions and to different degrees (Figure 7B).
The FKH1hc Effect Involved Gene Targets and/or Target Sites Distinct from Fkh2 Targets
In terms of gene expression FKH1hc had a modest but reproducible dampening effect on the wave of CLB2-mRNA expression during the cell cycle. These data were consistent with the idea, supported by earlier analysis of fkh1Δ cells (Hollenhorst et al., 2000) that Fkh1 acts as a negative regulator of CLB2 transcription at least during the stages of the cell cycle when CLB2 mRNA expression peaks. Because Fkh2 is a positive regulator of CLB2 transcription, it was somewhat surprising that an fkh2Δ affected neither silencing on its own nor the ability of FKH1hc to enhance silencing. Therefore, FKH1 must act through gene targets and/or binding sites distinct from those controlled by FKH2 to cause the phenotypes documented here. Whether FKH1hc enhances Fkh1 association with its normal targets or also causes significant binding to new targets, and whether one or more of these targets is also critical for the clb5Δ-like phenotype remains to be determined.
Forkhead transcription factors (forkhead box, FOX in metazoans) have evolutionary conserved roles in cell cycle regulation, proliferation and differentiation, and modest changes in forkhead transcription factor levels have clear and pivotal effects on complex multicellular phenomena such as organogenesis and disease states (Carlsson and Mahlapuu, 2002; Gaudet and Mango, 2002). Studies of the effects of Fkh1 dosage in the single-celled yeast should contribute to unraveling how forkhead transcription factors contribute to remodeling a eukaryotic transcriptional program. Transcriptional silencing serves as sensitive barometer for examining this issue and others associated with cell cycle regulation of chromosome structure and function.
ACKNOWLEDGMENTS
We thank Zhonggang Hou for help in producing Fkh1 antibodies, Kristopher H. McConnell and Bonita Brewer (University of Washington, Seattle) for advice on 2-D origin mapping, Philipp Müller for help with figures, and Oscar Aparicio (University of Southern California) for generously sharing yeast strains. We also thank Erika Shor for comments on the manuscript. LC was funded in part by a fellowship from the American Heart Association (AHA) (0135265Z). EEP was funded in part by the NIH Predoctoral Training Program in Genetics to the Laboratory of Genetics (5 T32 GM07133) and an AHA fellowship (0615552Z). This work was supported primarily by grants from the American Cancer Society (ACS-RSG-02-164-02-GMC) and the National Institutes of Health (RO1 GM56890) to CAF.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-04-0323) on November 28, 2007.
REFERENCES
- Andrulis E. D., Neiman A. M., Zappulla D. C., Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Andrulis E. D., Zappulla D. C., Alexieva-Botcheva K., Evangelista C., Sternglanz R. One-hybrid screens at the Saccharomyces cerevisiae HMR locus identify novel transcriptional silencing factors. Genetics. 2004;166:631–635. doi: 10.1534/genetics.166.1.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod A., Rine J. A role for CDC7 in repression of transcription at the silent mating-type locus HMR in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:1080–1091. doi: 10.1128/mcb.11.2.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- Boscheron C., Maillet L., Marcand S., Tsai-Pflugfelder M., Gasser S. M., Gilson E. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 1996;15:2184–2195. [PMC free article] [PubMed] [Google Scholar]
- Bose M. E., McConnell K. H., Gardner-Aukema K. A., Muller U., Weinreich M., Keck J. L., Fox C. A. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol. Cell. Biol. 2004;24:774–786. doi: 10.1128/MCB.24.2.774-786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L. L. Cyclin transcription: timing is everything. Curr. Biol. 2000;10:586–588. doi: 10.1016/s0960-9822(00)00634-5. [DOI] [PubMed] [Google Scholar]
- Carlsson P., Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- Chang S. C., Tucker T., Thorogood N. P., Brown C. J. Mechanisms of X-chromosome inactivation. Front. Biosci. 2006;11:852–866. doi: 10.2741/1842. [DOI] [PubMed] [Google Scholar]
- Chien C. T., Buck S., Sternglanz R., Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Mao Y., Sullivan K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Denu J. M. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem. Sci. 2003;28:41–48. doi: 10.1016/s0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- Dimitrova D. S., Gilbert D. M. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Donaldson A. D., Raghuraman M. K., Friedman K. L., Cross F. R., Brewer B. J., Fangman W. L. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Donze D., Kamakaka R. T. Braking the silence: how heterochromatic gene repression is stopped in its tracks. Bioessays. 2002;24:344–349. doi: 10.1002/bies.10072. [DOI] [PubMed] [Google Scholar]
- Dubey D. D., Davis L. R., Greenfeder S. A., Ong L. Y., Zhu J. G., Broach J. R., Newlon C. S., Huberman J. A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol. Cell. Biol. 1991;11:5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., Kamakaka R. T., Rine J. A role for the replication proteins PCNA, RF-C, polymerase epsilon and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics. 1999;153:1171–1182. doi: 10.1093/genetics/153.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Wang Y., Allis C. D. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Fox C. A., Ehrenhofer-Murray A. E., Loo S., Rine J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science. 1997;276:1547–1551. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- Fox C. A., Loo S., Dillin A., Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- Fox C. A., McConnell K. H. Toward biochemical understanding of a transcriptionally silenced chromosomal domain in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:8629–8632. doi: 10.1074/jbc.R400033200. [DOI] [PubMed] [Google Scholar]
- Fox C. A., Rine J. Influences of the cell cycle on silencing. Curr. Opin. Cell Biol. 1996;8:354–357. doi: 10.1016/s0955-0674(96)80009-3. [DOI] [PubMed] [Google Scholar]
- Futcher B. Microarrays and cell cycle transcription in yeast. Curr. Opin. Cell Biol. 2000;12:710–715. doi: 10.1016/s0955-0674(00)00156-3. [DOI] [PubMed] [Google Scholar]
- Gardner K. A., Fox C. A. The Sir1 protein's association with a silenced chromosome domain. Genes Dev. 2001;15:147–157. doi: 10.1101/gad.852801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K. A., Rine J., Fox C. A. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics. 1999;151:31–44. doi: 10.1093/genetics/151.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S. M., Cockell M. M. The molecular biology of the SIR proteins. Gene. 2001;279:1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- Gaudet J., Mango S. E. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Georgel P. T., Palacios DeBeer M. A., Pietz G., Fox C. A., Hansen J. C. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. USA. 2001;98:8584–8589. doi: 10.1073/pnas.151258798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Aparicio J. G., Hu F., Aparicio O. M. Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:10208–10222. doi: 10.1128/MCB.24.23.10208-10222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M., Brockdorff N. Heterochromatin on the inactive X chromosome delays replication timing without affecting origin usage. Proc. Natl. Acad. Sci. USA. 2004;101:6923–6928. doi: 10.1073/pnas.0401854101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunweller A., Ehrenhofer-Murray A. E. A novel yeast silencer. the 2mu origin of Saccharomyces cerevisiae has HST3-, MIG1- and SIR-dependent silencing activity. Genetics. 2002;162:59–71. doi: 10.1093/genetics/162.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Harlow E., Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- Henikoff S., Dalal Y. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Heun P., Laroche T., Raghuraman M. K., Gasser S. M. The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 2001;152:385–400. doi: 10.1083/jcb.152.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst P. C., Bose M. E., Mielke M. R., Muller U., Fox C. A. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle: overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics. 2000;154:1533–1548. doi: 10.1093/genetics/154.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst P. C., Pietz G., Fox C. A. Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: implications for regulating the cell cycle and differentiation. Genes Dev. 2001;15:2445–2456. doi: 10.1101/gad.906201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe G. J., Tanny J. C., Rudner A. D., Gerber S. A., Danaie S., Gygi S. P., Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Bernstein D. A., Fox C. A., Keck J. L. Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc. Natl. Acad. Sci. USA. 2005;102:8489–8494. doi: 10.1073/pnas.0503525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H. C., Stillman B., Xu R. M. Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc. Natl. Acad. Sci. USA. 2005;102:8519–8524. doi: 10.1073/pnas.0502946102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C. M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Kamakaka R. T. Silencers and locus control regions: opposite sides of the same coin. Trends Biochem. Sci. 1997;22:124–128. doi: 10.1016/s0968-0004(96)10074-8. [DOI] [PubMed] [Google Scholar]
- Kirchmaier A. L., Rine J. Cell cycle requirements in assembling silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:852–862. doi: 10.1128/MCB.26.3.852-862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koranda A., Schleiffer A., Endler L., Ammerer G. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature. 2000;406:94–98. doi: 10.1038/35017589. [DOI] [PubMed] [Google Scholar]
- Kumar R., Reynolds D. M., Shevchenko A., Shevchenko A., Goldstone S. D., Dalton S. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 2000;10:896–906. doi: 10.1016/s0960-9822(00)00618-7. [DOI] [PubMed] [Google Scholar]
- Laman H., Balderes D., Shore D. Disturbance of normal cell cycle progression enhances the establishment of transcriptional silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:3608–3617. doi: 10.1128/mcb.15.7.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A., Blitzblau H., Bell S. P. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 2002;16:2935–2945. doi: 10.1101/gad.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S., Rine J. Silencing and domains of heritable gene expression. Annu. Rev. Cell Dev. Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- Luo K., Vega-Palas M. A., Grunstein M. Rap1-Sir4 binding independent of other Sir, y Nospacekuqq, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig A. J., Liu C., Zhang C., Hanish J. P. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2483–2495. doi: 10.1128/mcb.16.5.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., Gasser S. M. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Matecic M., Martins-Taylor K., Hickman M., Tanny J., Moazed D., Holmes S. G. New alleles of SIR2 define cell cycle specific silencing functions. Genetics. 2006;173:1939–1950. doi: 10.1534/genetics.106.055491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell K. H., Muller P., Fox C. A. Tolerance of Sir1p/Origin recognition complex-dependent silencing for enhanced origin firing at HMRa. Mol. Cell. Biol. 2006;26:1955–1966. doi: 10.1128/MCB.26.5.1955-1966.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F. J., Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:5648–5659. doi: 10.1128/mcb.11.11.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Rudner A. D., Huang J., Hoppe G. J., Tanny J. C. A model for step-wise assembly of heterochromatin in yeast. Novartis Found. Symp. 2004;259:48–56. discussion 56–62, 163–169. [PubMed] [Google Scholar]
- Moretti P., Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios DeBeer M. A., Fox C. A. A role for a replicator dominance mechanism in silencing. EMBO J. 1999;18:3808–3819. doi: 10.1093/emboj/18.13.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios DeBeer M. A., Muller U., Fox C. A. Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev. 2003;17:1817–1822. doi: 10.1101/gad.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., Gasser S. M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- Pappas D. L., Jr, Frisch R., Weinreich M. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 2004;18:769–781. doi: 10.1101/gad.1173204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pic A., Lim F. L., Ross S. J., Veal E. A., Johnson A. L., Sultan M. R., West A. G., Johnston L. H., Sharrocks A. D., Morgan B. A. The forkhead protein Fkh2 is a component of the yeast cell-cycle transcription factor SFF. EMBO J. 2000;19:3750–3761. doi: 10.1093/emboj/19.14.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L., Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Pillus L., Rine J. SIR1 and the origin of epigenetic states in Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. 2004;69:259–265. doi: 10.1101/sqb.2004.69.259. [DOI] [PubMed] [Google Scholar]
- Reynolds D., Shi B. J., McLean C., Katsis F., Kemp B., Dalton S. Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev. 2003;17:1789–1802. doi: 10.1101/gad.1074103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier D. H., Ekena J. L., Rine J. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics. 1999;151:521–529. doi: 10.1093/genetics/151.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Rusche L. N., Rine J. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 2001;15:955–967. doi: 10.1101/gad.873601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schwob E., Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Sharma K., Weinberger M., Huberman J. A. Roles for internal and flanking sequences in regulating the activity of mating-type-silencer-associated replication origins in Saccharomyces cerevisiae. Genetics. 2001;159:35–45. doi: 10.1093/genetics/159.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherriff J. A., Kent N. A., Mellor J. The Isw2 chromatin-remodeling ATPase cooperates with the Fkh2 transcription factor to repress transcription of the B-type cyclin gene CLB2. Mol. Cell. Biol. 2007;27:2848–2860. doi: 10.1128/MCB.01798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D. RAP 1, a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I., et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K. Comprehensive identification of cell-cycle regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J. B., Gottschling D. E. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. Sir2 and Sir4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Taddei A., Gartenberg M. R., Neumann F. R., Hediger F., Gasser S. M. Multiple pathways tether telomeres and silent chromatin at the nuclear periphery: functional implications for sir-mediated repression. Novartis Found. Symp. 2005;264:140–156. discussion 156–165, 227–230. [PubMed] [Google Scholar]
- Valenzuela L., Kamakaka R. T. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- Vujcic M., Miller C. A., Kowalski D. Activation of silent replication origins at autonomously replicating sequence elements near the HML locus in budding yeast. Mol. Cell. Biol. 1999;19:6098–6109. doi: 10.1128/mcb.19.9.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M., Liang C., Stillman B. The Cdc6p nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc. Natl. Acad. Sci. USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M., Palacios DeBeer M. A., Fox C. A. The activities of eukaryotic replication origins in chromatin. Biochim. Biophys. Acta. 2004;1677:142–157. doi: 10.1016/j.bbaexp.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Xu E. Y., Zawadzki K. A., Broach J. R. Single-cell observations reveal intermediate transcriptional silencing states. Mol. Cell. 2006;23:219–229. doi: 10.1016/j.molcel.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Zappulla D. C., Sternglanz R., Leatherwood J. Control of replication timing by a transcriptional silencer. Curr. Biol. 2002;12:869–875. doi: 10.1016/s0960-9822(02)00871-0. [DOI] [PubMed] [Google Scholar]
- Zhu G., Spellman P. T., Volpe T., Brown P. O., Botstein D., Davis T. N., Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]