Abstract
In eukaryotic cells, mRNAs encoding signal sequence-bearing proteins undergo translation-dependent trafficking to the endoplasmic reticulum (ER), thereby restricting secretory and integral membrane protein synthesis to the ER compartment. However, recent studies demonstrating that mRNAs encoding cytosolic/nucleoplasmic proteins are represented on ER-bound polyribosomes suggest a global role for the ER in cellular protein synthesis. Here, we examined the steady-state protein synthesis rates and compartmental distribution of newly synthesized proteins in the cytosol and ER compartments. We report that ER protein synthesis rates exceed cytosolic protein synthesis rates by 2.5- to 4-fold; yet, completed proteins accumulate to similar levels in the two compartments. These data suggest that a significant fraction of cytosolic proteins undergo synthesis on ER-bound ribosomes. The compartmental differences in steady-state protein synthesis rates correlated with a divergent regulation of the tRNA aminoacylation/deacylation cycle. In the cytosol, two pathways were observed to compete for aminoacyl-tRNAs—protein synthesis and aminoacyl-tRNA hydrolysis—whereas on the ER tRNA deacylation is tightly coupled to protein synthesis. These findings identify a role for the ER in global protein synthesis, and they suggest models where compartmentalization of the tRNA acylation/deacylation cycle contributes to the regulation of global protein synthesis rates.
INTRODUCTION
Early studies into the mechanism of protein secretion established the endoplasmic reticulum (ER) as the site of secretory and integral membrane protein synthesis (Palade and Siekevitz, 1956a,b; Siekevitz and Palade, 1958). The subsequent discovery of the signal recognition particle pathway provided important insight into the mRNA partitioning events that enable the compartmentalization of secretory/integral membrane protein biogenesis to the ER (Blobel and Dobberstein, 1975a,b; Walter and Blobel, 1981a,b). This process is initiated when cytosolic mRNA/ribosome/nascent chain complexes (RNCs) engaged in the synthesis of secretory or integral membrane proteins undergo high-affinity interactions with the signal recognition particle (SRP). SRP binding elicits a suppression of translation and enables the trafficking of mRNA/ribosome/nascent chain complexes to the ER (Meyer and Dobberstein, 1980; Walter and Blobel, 1981a,b; Walter et al., 1981; Gilmore et al., 1982a,b). At the ER, sequential RNC binding interactions with the SRP receptor and Sec61p yield SRP release, ribosome binding to Sec61p, and an ER-restricted process of coupled protein synthesis/protein translocation (Blobel and Dobberstein, 1975b; Blobel et al., 1979; Walter and Lingappa, 1986; Gorlich et al., 1992). Consistent with this “positive selection” model, genome-wide studies of mRNA partitioning between cytosolic and membrane-bound ribosomes, conducted in yeast, fly, rodent, and human model systems demonstrate the expected enrichment of mRNAs encoding secretory/integral membrane proteins on the ER membrane (Mechler and Rabbitts, 1981; Mueckler and Pitot, 1981; Kopczynski et al., 1998; Diehn et al., 2000; Lerner et al., 2003). These studies also demonstrated that mRNAs encoding cytosolic/nucleoplasmic proteins are broadly represented on ER-bound polyribosomes with a small cohort of mRNAs being highly ER enriched (Mechler and Rabbitts, 1981; Mueckler and Pitot, 1981; Kopczynski et al., 1998; Diehn et al., 2000, 2006). Because these mRNAs do not encode signal sequences or transmembrane domains, these observations indicate that additional mechanisms function in the partitioning of mRNAs in eukaryotic cells, and they suggest possible roles for the ER in global protein synthesis.
In addition to studies of mRNA partitioning, biochemical and molecular genetic analyses of cellular protein synthesis also suggest a more global role for the ER in protein synthesis regulation. For example, studies conducted using an in vitro rough microsome-directed translation system have demonstrated that membrane-bound ribosomes can initiate de novo mRNA translation; moreover, they are capable of synthesizing cytosolic and secretory/integral membrane proteins (Potter and Nicchitta, 2000). Furthermore, molecular genetic analyses of the SRP pathway in Saccharomyces cerevisiae suggest that the loss of SRP/SRP receptor function can be compensated for by enhanced protein synthesis initiation on ER-bound ribosomes (Mutka and Walter, 2001). A physiological role for the ER in the synthesis of cytosolic/nucleoplasmic proteins has been demonstrated to occur in response to induction of the unfolded protein response (UPR) or picornavirus infection, stress conditions that elicit the suppression of protein synthesis via different mechanisms (Stephens et al., 2005; Lerner and Nicchitta, 2006). In these studies, cell stress-dependent inactivation of eukaryotic initiation factor (eIF)2α activity or proteolytic inactivation of eIF4G, resulted in a marked inhibition of cytosolic protein synthesis. In contrast, protein synthesis on the ER was sustained, with cytosolic/nucleoplasmic- and secretory/integral membrane-encoding mRNAs undergoing ER-restricted translation (Stephens et al., 2005; Lerner and Nicchitta, 2006).
In this report, we examined the steady-state protein synthesis activities of the cytosol and ER compartments. Using in situ radioisotope pulse-chase labeling studies of ribosome-bound nascent chains, we report that the protein synthesis activity of ER-bound ribosomes is 2.5- to 4-fold higher than cytosolic ribosomes. Consistent with this observation, using a globin reporter system, we observed a 3.5-fold increase in the level of globin protein synthesized on the ER membrane relative to synthesis directed by cytosolic ribosomes. Complementing these findings, the cytosol and ER compartments displayed distinct pathways of tRNA deacylation; in the cytosol, aminoacyl-tRNA deacylation could occur independent of protein synthesis; in contrast, on the ER, the deacylation of aminoacyl-tRNAs was tightly coupled to protein synthesis. This study demonstrates that the cytosol and ER compartments differ substantially in their protein synthesis capacities, and they suggest that the compartmental regulation of tRNA deacylation pathways may contribute to these differences.
MATERIALS AND METHODS
Reagents
[35S]Methionine/cysteine (1175 Ci/mmol) was obtained from MP Biomedicals (Irvine, CA). [(3,4,5)-3H]Leucine (100 Ci/mmol) was from MP Biomedicals and PerkinElmer Life and Analytical Sciences (Boston, MA). Digitonin was from Calbiochem (San Diego, CA). Cycloheximide was from Sigma-Aldrich (St. Louis, MO). Hybond membrane was from GE Healthcare (Chalfont St. Giles, United Kingdom). Cell culture reagents were from Invitrogen (Carlsbad, CA) with the exception of leucine-deficient media, which was provided by Millipore Specialty Media (Billerica, MA). RNase OUT, pcDNA6.1B-myc, Lipofectamine 2000, and Alexa Fluor-conjugated antibodies were from Invitrogen. β-Tubulin antibody (E7) was from the Iowa Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). Anti-myc epitope antibody was from Abcam (Cambridge, MA).
Cell Culture
HeLa, human embryonic kidney (HEK)293, and Cos7 cells were cultured in DMEM supplemented with 10% fetal calf serum at 37°C with 5% CO2. For most experiments, cells were used at ∼80% confluence. In transfection experiments, conducted in Cos7 cells, cell density was at 85–95% confluence, plasmid concentrations were 250 ng/well (six-well plate), and transfections were conducted according to the manufacturer's instructions. At 4–5 h after transfection, cells were trypsinized and split into paired wells of a six-well dish for overnight culture. Experiments were performed at 24 h after transfection.
Sequential Detergent Extraction
The cytosol and ER compartments were obtained by a sequential detergent extraction protocol (Lerner et al., 2003; Stephens et al., 2005, 2007; Lerner and Nicchitta, 2006). Briefly, cell monolayers were washed with phosphate-buffered saline, and they were incubated with 0.3–0.5 ml (12-well) or 0.6–1.0 ml (six-well) of permeabilization buffer [110 mM KOAc, 25 mM K-HEPES, pH 7.2, 2.5 mM Mg(OAc)2, 1 mM EGTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 10 U/ml RNase Out] containing 0.015% digitonin for 5 min on ice. The supernatant (cytosol) was recovered, and cells were washed once with permeabilization buffer containing 0.004% digitonin. Permeabilized cell monolayers were then solubilized with an equivalent volume of NP-40/sodium deoxycholate (DOC) lysis buffer [400 mM KOAc, 25 mM K-HEPES, pH 7.2, 15 mM Mg(OAc)2, 1% NP-40, 0.5% DOC, 1 mM DTT, 1 mM PMSF, and 10 U/ml RNase Out] for 5 min on ice. The supernatant (membrane-bound fraction) was recovered, and both cytosol and membrane-bound fractions were clarified by centrifugation at 7500 × g for 10 min at 4°C. Buffers were supplemented with 0.2 mM cycloheximide where applicable. For ribosome isolation, equal volumes of cytosol and membrane-bound fractions were aliquoted, and cytosol fractions were adjusted to 1% NP-40 and 12. 5 mM Mg(OAc)2. Lysates were layered over a 1.0 M sucrose cushion, and ribosomes were collected by centrifugation at 90,000 rpm in a TLA100.2 (Beckman Coulter, Fullerton, CA) rotor for 40 min at 4°C.
Metabolic Radiolabeling of Cell Cultures
Cell media were replaced with prewarmed methionine-deficient DMEM (supplemented with 1 mM sodium pyruvate, 1 mM glutamine, and 25 mM K-HEPES, pH 7.5) containing 0.2 mCi/ml [35S]methionine/cysteine (Met/Cys) (or leucine deficient DMEM containing 0.3–0.5 mCi/ml [3H]Leu) and cultured for the indicated times (without prior amino acid starvation). Where indicated, protein synthesis was inhibited by addition of 0.2 mM cycloheximide, and chase protocols were initiated by rapid replacement of the cell media with normal growth media supplemented with 1 mM unlabeled methionine (or leucine). For tRNA-based experiments, the time points represent the absolute time of labeling (and cycloheximide treatment). For pulse-chase tRNA labeling experiments, the chase was initiated by addition of 1 mM unlabeled methionine (or leucine) to the labeling media. Cycloheximide (0.2 mM) was added as indicated.
Velocity Sedimentation Analysis
HeLa cells cultured in a 175 cm2 flask at ∼80% confluence were starved for 10 min in methionine-deficient media and pulse labeled for 2 min with 1 mCi/ml [35S]Met/Cys and subsequently treated with 0.2 mM cycloheximide. Clarified cytosol and membrane-bound fractions were resolved by centrifugation through 10 ml 15–50% linear sucrose gradients at 151,000 × g for 3 h in a SW41 Beckman rotor. Fractions were manually collected, and the UV (260 nm) absorbance was determined. Then, 15-μl aliquots of fractions were analyzed by liquid scintillation spectrometry. For SDS-PAGE/phosphorimager analysis, total protein was isolated by TCA precipitation, resuspended in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (0.5 M Tris base, pH ∼11, 5% SDS, 4% glycerol, 0.02% bromphenol blue, and 100 mM 2-mercaptoethanol), heated at 65°C for 15 min, and resolved by SDS-PAGE on 12.5% gels. Radioisotope incorporation was determined by phosphorimager analyses of the dried gels.
Radiolabel Incorporation into Total Protein and Ribosome-bound Nascent Polypeptide Chains
Ribosome pellets were resuspended in 50 μl of 0.5 M Tris base, pH ∼11, 5% SDS, and heated at 65°C for 10 min to hydrolyze aminoacyl-tRNA ester linkages. Ribosome samples and 10- to 20-μl aliquots of total protein (removed before ultracentrifugation) were spotted onto 1-inch squares of 3MM filter paper and immersed in 10% ice-cold trichloroacetic acid (TCA) for 15 min. Filters were then placed in boiling 5% TCA for 15 min, rinsed with 5% TCA, immersed in ethanol:ether (1:1) for 5 min, and transferred to ether for 5 min. Filters were dried and quantified by liquid scintillation spectrometry. For kinetic experiments, data were graphed using one-phase exponential decay nonlinear regression curve fitting (GraphPad Software, San Diego, CA).
For the in vitro aminoacylation reaction, 0.1 ml of Met-tRNA reaction buffer [100 mM KOAc, 50 mM K-HEPES, pH 7.2, 10 mM Mg(OAc)2, 5 mM ATP, 5 mM DTT, 0.1 mg/ml calf liver tRNA, 10 μM methionine, and 1 μCi of 1 μM [35S]methionine/cysteine] was added to 10 μl of cell lysate (0.5–1.0 × 104 cell equivalents) and incubated at 37°C for 10 min. Reactions were stopped by the addition of 0.4 ml of cold 10% TCA and iced for 10 min. Samples were collected and washed by vacuum filtration and quantified by liquid scintillation spectrometry.
Plasmid Construction
The vector pTetBBB containing a genomic sequence of rabbit β-globin was a kind gift from Dr. A.-B. Shyu (University of Texas Houston Medical School, Houston, TX). The myc epitope was inserted at the BamHI site by using phosphorylated primers (sense, 5′-GATCCTGAACAAAAACTCATCTCAGGAGAGGATCTCGG-3′; antisense, 5′-GATCCGAGATCCTCTTCTGAGATGAGTTTTTGTTCAG-3′). The full-length globin sequence was subcloned into a HindIII/XhoI site in pcDNA6.1B after generation of restriction site bearing coding region via polymerase chain reaction (PCR), by using the primers (sense, 5′-GTGACTAAGCTTACCATGGTGCATCTGTCCAG-3′; antisense, 5′-GAGACACTCGAGTGCAATGAAAATAAATTTCC-3′). The immunoglobulin κ signal sequence was inserted into the HindIII site at the amino terminus of the globin-coding region by using overlapping phosphorylated primers (sense, 5′-AGCTTACCATGGAGACAGACACACTCCTGCTATGGGTACTGC-3′ and 5′-TGCTCTGGGTTCCAGGTTCCACTGGTGACA-3′; antisense, 5′-AGCAGGAGTGTGTCTGTCTCCATGGTA-3′ and 5′-AGCTTGTCACCAGTGGAACCTGGAACCCAGAGCAGCAGTACCCAT-3′). All sequences were verified (Duke University Comprehensive Cancer DNA Sequencing Facility, Durham, NC).
RNA Analysis
RNA was isolated from cell lysates or ribosome fractions by using a phenol-chloroform based extraction protocol (Stephens et al., 2007). RNA was resolved on 1% agarose/3% formaldehyde denaturing gels and stained with ethidium bromide. Ethidium bromide fluorescence was determined using a Typhoon Trio 9400 (GE Healthcare). Agarose gels were transferred to Hybond membranes in 5× SSC + 10 mM NaOH by downward capillary flow for 2 h and UV cross-linked at 1200 mJ. For tRNA-radiolabeling studies, membranes were directly exposed to phosphorimager plates ([35S]Met) or quantified by liquid scintillation ([3H]Leu). For Northern blots, membranes were probed with an 32P-end-labeled antisense oligonucleotide (directed against the myc epitope encoding domain).
Immunoprecipitations
Total cell lysates were generated by solubilization of cell monolayers in 25 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.05% SDS, 1 mM DTT, and 0.5 mM PMSF. Lysates were clarified by centrifugation at 7500 × g for 10 min at 4°C. Globin proteins was immunoprecipitated using a 1:200 antibody dilution and 20 μl of protein A-Sepharose slurry (50%) rotating end over end for 1 h at room temperature. Beads were washed in lysis buffer and heated at 95°C for 10 min in SDS-PAGE sample buffer. Proteins were resolved by 12.5% SDS-PAGE or 18% SDS-PAGE (globin), and dried gels were exposed to phosphorimager plates.
Phosphorimager plates were scanned using a Typhoon 9400 (GE Healthcare) and quantified using ImageQuantTL version 2003 software (GE Healthcare). Images were assembled in Adobe Photoshop 7.0 (Adobe Systems, Mountain View, CA). Graphs were generated using GraphPad Prism 4.0 software (GraphPad Software).
RESULTS
It has recently been reported that cytosolic and ER membrane-directed protein synthesis can undergo differential regulation in mammalian cells (Stephens et al., 2005; Lerner and Nicchitta, 2006). For example, cell stress arising from either activation of the UPR or picornaviral infection results in suppression of cytosolic mRNA translation, whereas translation on ER membrane-bound ribosomes is sustained (Stephens et al., 2005; Lerner and Nicchitta, 2006). In these scenarios, the ER functions as the predominant site of global protein synthesis, and it engages in the synthesis of cytosolic and nucleoplasmic proteins, such as ATF4 and XBP-1(s), and secretory and integral membrane proteins. Extending from these studies, we postulated that under normal homeostatic growth conditions, the protein synthesis capacities of the two cellular compartments may differ. To explore this possibility, we used a radioisotope pulse-labeling/cell fractionation assay to assess the steady-state in situ protein synthesis activities of the cytosol and ER compartments. In this assay, cells are pulse labeled for 10 min with [35S]methionine/cysteine, and radiolabeling is blocked by addition of excess unlabeled methionine. To document isotope incorporation at time points during the labeling and chase periods, cell aliquots were treated with the elongation inhibitor cycloheximide (0.2 mM) at varying times. In Figure 1, cell samples (n = 3) were treated with cycloheximide at three times: 1) at the beginning of the pulse (t = 0 min), to assess cycloheximide-insensitive background isotope recovery; 2) at the end of the 10-min pulse (t = pulse), to assess the maximal level of isotope incorporation; and 3) at the end of the 15-min chase (t = chase), to assess the completion/release of newly synthesized proteins. After the pulse-chase procedure, cells were harvested and processed to yield distinct cytosolic and membrane-bound subcellular fractions by using a sequential detergent extraction method (Seiser and Nicchitta, 2000; Potter and Nicchitta, 2002; Lerner et al., 2003; Stephens et al., 2005, 2007). To validate that the described fractionation methods were appropriate to these investigations, pulse-labeled cells were fractionated at increasing time periods after isotope addition, and the subcellular fractions were analyzed for the accumulation of a newly synthesized cytosolic (tubulin) and ER-resident (GRP94) protein. These data are depicted in Supplemental Figure S1, and they demonstrate that the two fractions are highly enriched in their respective newly synthesized resident proteins, with little to no cross-contamination. To directly assay protein synthesis activity, ribosome-bound nascent chains (RNCs) were collected from the two subcellular fractions by ultracentrifugation of the subcellular fractions, and the isotopic protein content was analyzed by liquid scintillation spectrometry (Figure 1, A, C, and E). The labeling profiles of RNCs were compared with that of newly synthesized (i.e., completed) proteins accumulating in the respective compartment (Figure 1, B, D, and F).
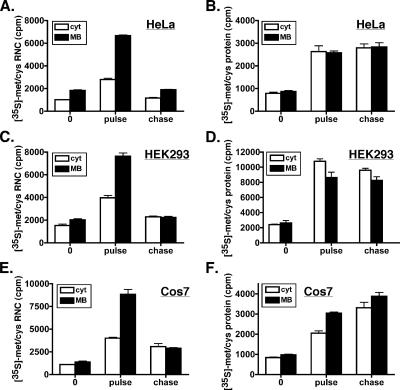
Figure 1.
Protein synthesis activity is enhanced in the ER compartment. HeLa (A and B), HEK293 (C and D), and Cos7 (E and F) cells were pulse labeled in methionine-deficient media with [35S]Met/Cys for 10 min, and then they were chased for 15 min in normal growth media supplemented with 1 mM methionine. No prior amino acid starvation was performed. Cycloheximide (0.2 mM) was added to individual cell aliquots at the beginning of the pulse (0), the end of the pulse (pulse), or at the end of the chase (chase). Cytosol (cyt) and MB fractions were generated by sequential detergent extraction. Aliquots were removed for total protein (B, D, and F) and RNC fractions subsequently isolated by ultracentrifugation (A, C, and E). (A–F) Isotope incorporation was determined by liquid scintillation of the TCA-insoluble, alkaline-insensitive material. Mean values of replicate (n = 3) labeling experiments ± SE are displayed.
Using the methodology described above, the rates of protein synthesis and the compartmental fates of newly synthesized proteins were determined for the cytosol and ER fractions of HeLa, HEK293, and Cos7 cells (Figure 1). As shown in Figure 1, 35S-amino acid incorporation into RNCs was substantially higher in the membrane-bound ribosome fractions of HeLa (Figure 1A), HEK293 (Figure 1C), and Cos7 (Figure 1E) cells. After the chase period, the quantities of 35S-labeled RNCs returned to basal levels, demonstrating the efficient release of the nascent polypeptide chain fraction. Quantification of these data indicated that the magnitude of amino acid incorporation in the membrane-bound RNC fractions was 2.5- to 4-fold higher than cytosolic RNC fractions (t = pulse), suggesting that steady-state protein synthesis rates on the ER are substantially higher than cytosolic rates. To determine whether differences in the ribosome content of the two compartments were responsible for this phenomenon, we determined the relative ribosome distribution between the cytosol and ER by quantifying 18S and 28S rRNA levels. Here, we observed that ∼45% of cellular ribosomes were recovered in the cytosol fraction in HeLa (Figure 2, A and C) and Cos7 (Figure 2, B and D) cells. Thus, the observed differences in the amino acid incorporation into the RNC fractions could not be explained by differences in ribosome distribution. Furthermore, given the very minor amount of mitochondrial ribosomes present in the membrane-bound (MB) fraction (compare relative intensities of ethidium bromide fluorescence of 28S/18S rRNA with 23S/16S mitochondrial rRNA in Figure 2, A and B, top, MB), it is unlikely that mitochondrial protein synthesis contributed significantly to these findings. To account for potential differences in ribosome recovery, we also assessed relative protein synthesis activities by using ribosome equivalents (Martin et al., 1969). Although this latter method of analysis does not account for the presence of inactive ribosomes/ribosomal subunits, it does provide for a more direct comparison of protein synthesis activity. When normalized to a fixed quantity (500 fmol) of ribosomes, ER protein synthesis activity was again 2.5- to 4-fold greater than cytosol activity in HeLa, HEK293, and Cos7 cells (Supplemental Figure S2A).
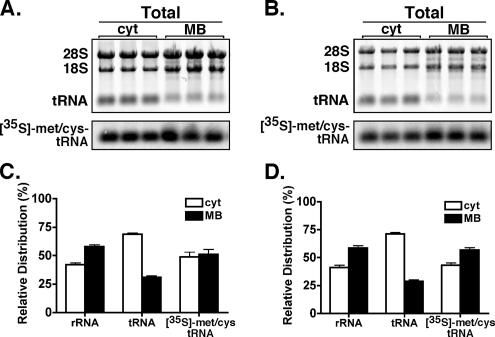
Figure 2.
Subcellular distribution of ribosomes, total tRNA, and newly charged tRNA. HeLa (A and C) and Cos7 (B and D) cells were pulse labeled with [35S]Met/Cys for 5 min in methionine-deficient media. RNA from cyt and MB fractions was resolved on denaturing agarose (1%) gels and transferred to Hybond membrane. Relative rRNA and tRNA levels were determined by quantitative fluorescent imaging of the ethidium bromide-stained gels. Isotope incorporation was determined by phosphorimager analysis. Mean values of replicate labeling experiments (n = 3) ± SE are displayed.
In contrast to the isotope incorporation rates of RNCs, the levels of accumulating (completed) newly synthesized proteins were comparable between the cytosol and ER compartments (Figure 1, B, D, and F). These data suggest that a fraction of the proteins synthesized on the ER were released to the cytosol. This interpretation is consistent with previous reports demonstrating that a broad array of mRNAs encoding cytosolic and nucleoplasmic proteins can be partitioned to and translated on membrane-bound ribosomes (Diehn et al., 2000, 2006; Lerner et al., 2003; Stephens et al., 2005). In addition, several reports have also demonstrated that specific secretory proteins such as calreticulin and the plasminogen-activated inhibitor-2 protein (PAI-2) can be inefficiently translocated, and as a result, accumulate in the cytosol compartment (Belin et al., 1989; Levine et al., 2005). As an alternative possibility, nascent polypeptides in the cytosol could be preferentially degraded as a consequence of the fractionation procedure. To test the latter hypothesis, unlabeled and isotopically labeled cytosolic and ER fractions were reciprocally mixed (i.e., labeled cytosol + unlabeled MB and vice versa), and the radioactivity of RNCs and total protein fractions was subsequently assayed as described above. As shown in Figure 3, reciprocal mixing of subcellular fractions did not yield significant changes in recovered radiolabeled RNCs or proteins. Furthermore, to assess whether our observations were influenced by rapid protein (or nascent chain) turnover in situ, we determined the relative levels of recovered radioactivity in the RNC fractions obtained directly after the 10-min pulse or in cells treated with cycloheximide after the 10-min pulse and harvested after the 15-min chase. Under both conditions, we consistently observed 2.5- to 4-fold higher isotope incorporation into the membrane-bound RNC fraction (data not shown). Similarly, we compared the relative levels of total newly synthesized protein, and again we did not observe any significant differences in the level of recovered radioactivity. These data indicate that the degradation of nascent chains and/or proteins either in situ or during the processing procedure did not bias our observations.
Figure 3.
Newly synthesized proteins display identical stabilities in endoplasmic reticulum and cytosol fractions. HeLa cells were pulse labeled with [35S]Met/Cys for 10 min in methionine-deficient media. Isotopically labeled cyt and MB fractions were mixed with cell equivalent quantities (volume) of either the reciprocal cellular fraction isolated from unlabeled cells or buffer alone and incubated for 20 min on ice. Aliquots were removed for total isotopic protein determinations, and the RNC fraction was isolated by ultracentrifugation. Isotope incorporation was determined by liquid scintillation of the TCA-insoluble, alkaline-insensitive material. Mean values of replicate (n = 3) labeling experiments ± SE are displayed. Recovered radioactivity was normalized to buffer alone.
To further characterize the differences in relative protein synthesis activities of the cytosol and ER membrane, detailed time course studies of (isotopic) amino acid incorporation into completed protein and RNCs were performed. Cycloheximide treatment was used to halt protein synthesis at varying times of the pulse-chase procedure. In the experiments presented in Figure 4, A and D and C and F, HeLa cells were pulse labeled with [35S]Met/Cys or [3H]Leu, respectively, for 12 min. The time points reflect addition of cycloheximide (0.2 mM) to individual cell aliquots (n = 3) at the indicated times of the pulse labeling. In Figure 4, A ([35S]Met/Cys) and C ([3H]Leu), comparisons of in situ pulse-labeling kinetics of subcellular RNC fractions identified significantly higher rates and magnitudes of protein synthesis activity on the ER membrane, consistent with data presented in Figure 1. In addition, the appearance of newly synthesized proteins was comparable in rate and magnitude between the two compartments (Figure 4, D and F); no statistical difference in the level of isotope incorporation into proteins was observed between the two compartments by using either amino acid tracer. Notably, the differences in RNC incorporation were independent of labeling conditions used (i.e., amino acid-deficient vs. enriched culture media; Figure 4, A and D, and Supplemental Figure S2, B and C) or radiolabeled amino acid (methionine vs. leucine; Figure 4, A and D vs. C and F). Potentially, our observations could result from delayed released kinetics of nascent chains from membrane-bound ribosomes. To evaluate this possibility, we examined the turnover kinetics (i.e., release) of nascent chains by using a pulse-chase procedure (Figure 4, B and E). In these experiments, cells were pulse labeled for 10 min and chased with excess unlabeled amino acid for 12 min. Here, cycloheximide (0.2 mM) was added to cell aliquots (n = 3) at varying times of the chase, analogous to the procedure described above used for pulse labeling. As shown in Figure 4B, both compartments exhibited rapid decay (release) kinetics of ribosome-bound nascent chains, demonstrating that delayed termination (i.e., nascent chain release) did not make a substantial contribution to the greater magnitude of membrane-bound RNC labeling.
Figure 4.
Protein synthesis is kinetically favored on the endoplasmic reticulum. HeLa cells were pulse labeled with [35S]Met/Cys (A and D) for 12 min or [3H]Leu (C and F) for 15 min. (B and E) HeLa cells were pulse labeled with [35S]Met/Cys for 10 min and chased in normal growth media supplemented with 1 mM methionine for 12 min. (A–F) Cycloheximide was added at the indicated times (closed squares and triangles) during the pulse (A, C, D, and F) or chase procedures (B and E). Aliquots were removed for total protein (D–F), and RNC fractions were subsequently isolated by ultracentrifugation (A–C). (A–F) Isotope incorporation was determined by liquid scintillation of the TCA-insoluble, alkaline-insensitive material. Mean values of replicate (n = 3) labeling experiments ± SE are displayed. The cyt is depicted as the solid line; MB fractions are the dotted lines. Background labeling, which was assessed by cycloheximide addition at time 0 of the pulse, was subtracted from all time points.
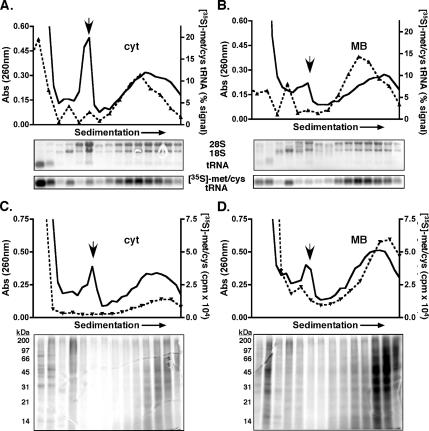
To provide additional experimental evidence that the analyses described above indeed corresponded to polyribosomes, the cytosol and membrane-bound fractions of pulse-radiolabeled HeLa cells were resolved by sucrose gradient velocity sedimentation, and the radiolabeled polypeptide composition of gradient fractions was analyzed. As shown in Figure 5, the gradient sedimentation procedure resolved the ribosomal subunits (18S and 28S rRNA), the 80S monosome (downward arrow) and large polyribosome-containing fractions. As assessed by UV absorbance values (260 nm), approximately equal quantities of polyribosomes were recovered from both subcellular fractions. To assess amino acid incorporation into ribosome-bound nascent chains, gradient fractions were acid precipitated, resolved by SDS-PAGE, and the incorporated radioactivity was determined by phosphorimager analyses (Figure 5, C and D). Consistent with the data depicted in Figures 1 and 2 and Supplemental Figure S2, when polyribosome-bound radiolabeled nascent polypeptide levels were evaluated, the membrane-derived fraction displayed considerably higher radioisotope incorporation, and the enhanced incorporation was present throughout the polyribosome profile. Summation of the total radioactivity present in the peak polyribosome-containing fractions indicated that the membrane-bound nascent chains had approximately fivefold greater isotope levels than the cytosol fraction. As an additional component of this experiment, we also evaluated the levels of isotopically charged (aminoacylated)-tRNA between the two compartments. For this analysis, total RNA was isolated from individual gradient fractions, resolved by denaturing agarose gel, transferred, and radioactivity corresponding to the tRNA determined by phosphorimager analysis. Here, we observed approximately equivalent levels of [35S]Met/Cys-tRNA in the polyribosome fractions from the two subcellular (cytosol and ER) compartments.
Figure 5.
Membrane-bound ribosomes display elevated rates of protein synthesis: velocity sedimentation of polyribosome-associated nascent polypeptides. HeLa cells were pulse labeled with [35S]Met/Cys for 2 min. Cytosol (A and C) and membrane-bound (B and D) fractions were prepared and sedimented through 15–50% linear sucrose gradients for 3 h at 151,000 × g. Ribosomes were monitored by UV absorbance (solid black line), and the downward facing arrow denotes the 80S monosome peak. (A and B) RNA extracted from individual gradient fractions was resolved on denaturing agarose gels and transferred to Hybond membrane. Total rRNA (28S and 18S) and tRNA levels were determined by ethidium bromide fluorescence. Isotope incorporation was analyzed by phosphorimager analysis. The dotted line represents the percent radioactivity of the total [35S]Met/Cys-tRNA signal per gradient. (C and D) Total radioactivity (dotted line) of individual gradient fractions was determined by liquid scintillation. Proteins were resolved by SDS-PAGE, and radioisotope incorporation was determined by phosphorimager analysis.
To determine whether the differences in the steady-state rates of protein synthesis in the cytosol and ER compartments reflected global synthesis rates, rather than variations in the translation rates of discrete subsets of mRNAs, we evaluated the synthesis rate of a reporter protein whose mRNA was targeted to either the ER or the cytosol. For these experiments, myc-tagged globin protein, expressed from the genomic rabbit β-globin sequence, was used as a common reporter. To direct globin (Gbn) mRNA to the ER, an encoded N-terminal signal sequence derived from the immunoglobulin (Ig) κ chain was appended 5′ to the globin open reading frame (=ssGbn). To validate the activity of the signal peptide in directing the partitioning of the globin mRNA to the ER, Northern blot analyses of the cytosol and membrane fractions of reporter-transfected cells were performed. As shown in Figure 6, A and B, globin mRNA (Gbn) strongly copurified (≥80%) with the cytosol fraction, whereas addition of a signal sequence localized globin mRNA to the ER.
Figure 6.
Targeting globin mRNA to the ER membrane yields enhanced globin protein expression. Cos7 cells were either mock transfected (M) or transfected with plasmid encoding myc-tagged wild-type globin (Gbn) or myc-tagged globin containing an N-terminal signal sequence (ssGbn). (A and B) Northern blot analysis of RNA isolated from cytosol and membrane-bound fractions. (C and D) Cells were transfected in triplicate and analyzed for RNA and protein content. (C) RNA was isolated from total cell lysates and analyzed by Northern blot (Gbn) with ethidium bromide fluorescence of the rRNA as a loading control. (D) Cells were metabolically labeled with [35S]Met/Cys and myc-tagged globin and GRP94 (loading control) isolated by indirect immunoprecipitation from total cell lysates. Samples were resolved by SDS-PAGE, and dried gels were analyzed by phosphorimager analysis. (E) Quantification of immunoprecipitations was performed using ImageQuantTL version 5.2 software. It should be noted that of the two ER forms of globin, the higher molecular weight species (ssGbn-myc), which consists of approximately half of the globin product, contains an additional methionine residue, totaling three methionine and one cysteine residue (Gbn-myc has 2 met and 1 cys); thus, there could be a maximal increase in specific activity of 33% of this band, or 15% of total recovered globin from pssGbn-transfected cells.
Using the two globin reporter constructs, the translation rates of the two cellular compartments were compared. Cos7 cells transfected with equivalent levels of plasmid DNA encoding either wild-type (Gbn) or signal sequence globin (ssGbn) displayed similar mRNA expression levels (Figure 6C; three independent transfection experiments are depicted). To assay protein synthesis levels, transfected cells (n = 3) were pulse labeled, and newly synthesized globin and GRP94 (loading control) proteins were isolated by immunoprecipitation. As shown in Figure 6D with quantification in Figure 6E, phosphorimager analysis of newly synthesized globin protein demonstrated that 3.5-fold more globin protein was produced when globin mRNA was translated on ER-bound ribosomes (ssGbn) compared with globin protein produced on cytosolic ribosomes (Gbn). These data recapitulate the observations obtained in biochemically fractionated cells (Figures 1–3 and Supplemental Figure S2), and they are consistent with a model of enhanced protein synthesis on the ER membrane.
As noted above, the observation that the protein synthesis rates on the ER compartment were substantially higher than the cytosol, yet approximately similar quantities of completed proteins were recovered from the two fractions, suggests a net flux of protein from the site of synthesis on the ER to the site of residence, the cytosol. To directly test for the existence of such a pathway, the signal sequence of an abundant, resident ER chaperone protein, GRP94, was deleted and the mRNA/protein expression patterns of myc epitope-tagged versions of control (wild type; WT), and signal sequence-deleted (ΔSS) GRP94 was determined (Supplemental Figure S3, B–F). A schematic illustration of these constructs is shown in Supplemental Figure S3A. As depicted in Supplemental Figure S3, B and C, the mRNAs encoding the WT and ΔSS GRP94 and the protein products were expressed at near identical levels. Immunoblot analysis of the compartmental distribution of the two proteins demonstrated efficient partitioning of the WT form to the ER and the ΔSS form to the cytosol (Supplemental Figure S3D). As additional controls for the subcellular fractionation, paired immunoblots for the cytosolic protein 90-kDa heat-shock protein and the ER-resident protein TRAPα confirmed that the two fractions were highly enriched in their respective proteins. Unexpectedly, deletion of the signal sequence did not substantially alter GRP94 mRNA localization to ER, with nearly 100% of the WT mRNA and 90% of the ΔSS mRNA copurifying with the ER fraction of HeLa (Supplemental Figure S3E) and Cos7 cells (Supplemental Figure S3F). For comparison, the mRNAs encoding the resident ER lumenal proteins BiP and calreticulin and the cytosolic/nucleoplasmic protein histone H3.3 displayed the expected enrichment in the ER and cytosol fractions, respectively (Supplemental Figure S3, E and F). The observation that deletion of the signal sequence of a resident ER lumenal protein does not substantially alter its mRNA partitioning pattern is further developed in a recent communication from this laboratory (Pyhtila et al., 2008). The data presented in Supplemental Figure S3 demonstrate that in the absence of a signal sequence, ER-directed translation of ΔSS GRP94-encoding mRNAs results in complete recovery of GRP94 protein in the cytosol, thus directly demonstrating an ER-to-cytosol pathway of protein synthesis and residence. It should also be mentioned that previous investigations have observed a related phenomenon for the signal sequence-bearing proteins calreticulin and PAI-2, where both ER lumenal and cytosolic forms of the proteins are observed as the result of inefficient translocation (Belin et al., 1989; Levine et al., 2005).
The precise mechanism(s) contributing to the observed kinetic advantage for protein synthesis on the ER is under investigation. From simple topological considerations, cytosolic and ER-bound ribosomes would be expected to use common pools of protein synthesis initiation, elongation, and termination factors, aminoacyl-tRNAs, and nucleotides, and so it is not clear how divergent regulation of protein synthesis in the cytosol and ER could be established. Potentially, localizing the complex reactions of protein synthesis to the two-dimensional plane of the ER membrane would alone be expected to provide a substantial kinetic advantage (Berry, 2002). Of related interest, it has been reported that in detergent-permeabilized tissue culture cells lacking cytosol, aminoacyl-tRNAs can be used by the protein synthesis machinery and subsequently recharged without undergoing free exchange with a soluble pool (Negrutskii and Deutscher, 1991, 1992; Stapulionis and Deutscher, 1995). This phenomenon, termed tRNA channeling, would be expected to significantly enhance protein synthesis rates (Negrutskii and Deutscher, 1991; Stapulionis and Deutscher, 1995). Extending from these findings, we sought to determine whether the cycle of tRNA aminoacylation and deacylation in the cytosol and on the ER are under common or distinct regulation in intact cells. To examine this, we first determined the relative distribution of total and newly charged (aminoacylated) tRNA from cytosol and membrane-bound fractions of HeLa and Cos7 cells. Total tRNA levels were quantified from ethidium bromide fluorescence; newly charged [35S]Met/Cys-tRNA levels were determined by phosphorimager analysis. As depicted in Figure 2, the distribution of newly charged [35S]Met/Cys-tRNA between the cytosol and ER compartments of HeLa and Cos7 cells was approximately equal, and it mirrored the ribosome (rRNA) distribution (Figure 2). Total tRNA, in contrast, was highly enriched in the cytosol fraction, comprising 70–90% of the cellular pool (Figure 2). Expressed relative to the total tRNA, these data identify a substantial enrichment of newly charged tRNA on the ER. Assays of the subcellular partitioning of methionyl-/cysteinyl-tRNA synthetase activities indicated that the enrichment of newly charged tRNAs on the ER could not be attributed to the subcellular distribution of tRNA synthetase activity, which favored the cytosol (Supplemental Figure S4).
The subcellular regulation of the tRNA acylation/deacylation cycle was further examined in studies of the discharge kinetics (deacylation) of aminoacylated-tRNA in the context of the protein synthesis reaction. In these experiments, cells were either pulse labeled for 10 min or pulse labeled for 5 min and chased for 10 min, and the radioactivity present in tRNA was determined by phosphorimager analysis as stated above for Figure 5. In this system, the deacylation reaction can be monitored as either the discharge and subsequent recharging (aminoacylation) of tRNA (pulse labeling) or directly as the immediate discharge of isotopically labeled tRNA, via a pulse-chase experiment. To determine the contribution of protein synthesis to aminoacyl-tRNA turnover, paired experiments were performed in the presence of cycloheximide (0.2 mM). Importantly, cycloheximide was added to individual cell aliquots in concert with addition of isotope (i.e., beginning of the pulse) for pulse labeling (Figure 7, C and D) or addition of excess unlabeled amino acid (i.e., beginning of chase) for pulse-chase experiments (Figure 7, A, B, E, and F). Thus, in this assay, the time points reflect the total time of labeling (Figure 7, C and D) or chase (Figure 7, A, B, C, and F).
Figure 7.
tRNA aminoacylation/deacylation is subject to divergent regulation in the cytosol and ER compartments. Untreated (UT) and cycloheximide (CHX)-treated Cos7 (A and B) and HeLa (E and F) cultures were pulse labeled for 5 min with [35S]Met/Cys or [3H]Leu, respectively, and chased with the appropriate unlabeled amino acid (1 mM) for periods up to 10 min. (C and D) UT and CHX-treated HeLa cultures were pulse labeled with [35S]Met/Cys for periods up to 10 min. (A–F) Cycloheximide was administered in concert with either addition of isotope during the pulse (C and D) or addition of unlabeled amino acid during the chase (A, B, E, and F). The time points reflect the total time of pulse (C and D) or chase (A, B, E, and F) (open and closed triangles and squares). Total RNA from cyt and MB cell lysates was resolved on denaturing agarose gels and transferred to Hybond membrane. Total tRNA was analyzed by ethidium bromide fluorescence. Isotope incorporation was analyzed by phosphorimager analysis (A–D) or liquid scintillation spectrometry (E and F). (A–F) Control (untreated) data points are represented by the solid line, and cycloheximide-treated data points by the dotted line.
In the absence of cycloheximide (untreated [UT]), the kinetics of tRNA deacylation (chase) was similar in the cytosol and membrane-bound compartments. Using either [35S]Met/Cys (Figure 7, A and B) or [3H]Leu (Figure 7, E and F), charged tRNA was almost completely deacylated in 2 min (chase). Correspondingly, tRNA was (re)-aminoacylated (pulse) in ∼2 and 4 min in HeLa and Cos7 cells, respectively (Figure 7, C and D; data not shown). Pulse and pulse-chase experiments performed in concert with the inhibition of translation elongation (cycloheximide; identical results obtained with emetine) revealed marked differences in aminoacyl-tRNA turnover in the cytosol and ER compartments. In the cytosol, the inhibition of protein synthesis only modestly affected tRNA discharge rates (chase) (Figure 7, A and E); similarly the cytosolic aminoacylation reaction (pulse) was only slightly delayed by cycloheximide, suggesting that tRNA discharge can occur independent of the protein synthesis reaction (Figure 7C). In contrast, aminoacyl-tRNA deacylation on the ER membrane was tightly coupled to protein synthesis; in the presence of cycloheximide, the rate of aminoacyl-tRNA discharge and subsequent (re)-acylation were dramatically suppressed, as demonstrated for both [35S]Met/Cys- and [3H]leucyl-tRNA (Figure 7, B, D, and F). These results indicate that in the cytosol, aminoacyl-tRNAs are accessible to an as yet unidentified tRNA hydrolase activity and thus may participate in futile aminoacylation/deacylation cycles. On the ER membrane, in contrast, tRNA discharge is efficiently coupled to protein synthesis. These data identify a complex subcellular organization of the metabolic processes associated with tRNA aminoacylation and deacylation. The findings that aminoacyl tRNA use is tightly coupled to protein synthesis on the ER, relative to the cytosol, suggests that compartmental regulation of the tRNA aminoacylation cycle contributes to enhanced rates of protein synthesis on the ER.
DISCUSSION
In this study, we report three primary findings: 1) steady-state protein synthesis rates on the endoplasmic reticulum are 2.5- to 4-fold higher than in the cytosol. This was evident at both a global level, by evaluating amino acid incorporation kinetics into ribosome/nascent chain complexes, and at a message-specific level, by using globin reporter mRNAs displaying opposing subcellular localizations; 2) a substantial fraction of proteins undergo synthesis on ER-bound ribosomes, yet reside in the cytosol compartment. Although this conclusion was deduced by comparing the relative levels of amino acid incorporation into nascent polypeptides versus completed newly synthesized proteins in the cytosol and ER, and so is inferred, a direct demonstration of this pathway by using a signal sequence deletion mutant of the ER chaperone GRP94 confirmed that such a pathway can operate in mammalian cells (Pyhtila et al., 2008); 3) the tRNA aminoacylation/deacylation cycle is subject to divergent regulation between the cytosol and ER compartments of higher eukaryotic cells. In the cytosol, aminoacyl-tRNAs are discharged via two pathways: protein synthesis and an as yet poorly understood aminoacyl-tRNA hydrolase function. Conversely, tRNA deacylation on the ER is tightly coupled to protein synthesis. Together, these studies implicate the ER membrane as the primary site of cellular protein synthesis, and in this context it may be a critical site for regulating posttranscriptional gene expression.
Whereas it is generally accepted that the ER compartment functions uniquely in the biogenesis of secretory and integral membrane proteins, the data presented here suggest that the ER membrane also contributes a substantial portion of newly synthesized proteins to the cytosol. This proposal is consistent with the established patterns of mRNA partitioning between the cytosol and ER compartments, where subsets of mRNAs encoding cytosolic/nucleoplasmic proteins are partitioned to and translated on ER-bound ribosomes (Mechler and Rabbitts, 1981; Mueckler and Pitot, 1982; Kopczynski et al., 1998; Diehn et al., 2000, 2006; Lerner et al., 2003; Stephens et al., 2005). For example, the mRNAs encoding the key stress response transcription factors ATF4 and XBP-1, which are translationally silent under normal growth conditions, are highly enriched in ER-bound small polyribosomes after activation of the UPR (Stephens et al., 2005). Extending from the data in this report, ER-directed protein synthesis may afford the cell the most efficient platform for the biogenesis of key stress response proteins during a cellular crisis. In addition, it may be anticipated that nucleoplasmic regulatory proteins and transcription factors, may be synthesized on the ER/outer nuclear envelope. Indeed, messages encoding the mitogenic proteins cyclins-B3 and -E2 have also been reported to be enriched on membrane-bound fractions; however, the biological function of such events has yet to be determined (Diehn et al., 2006).
In studying the mechanism(s) responsible for the observed differences in ER and cytosolic steady-state protein synthesis, similar global termination rates were observed in the two compartments, indicating that the kinetic advantage displayed by the ER must reflect enhanced rates of initiation and/or elongation. Potentially, restriction of the complex, multistage reactions of protein synthesis to the two-dimensional plane of the ER could yield significant kinetic enhancements (Berry, 2002). However, the magnitude of this effect is difficult to predict. In part, direct coupling of the tRNA deacylation reaction and protein synthesis may contribute to this relative increase in translation rate. These observations are strongly reminiscent of previous reports demonstrating tRNA channeling in detergent-permeabilized cells (Negrutskii and Deutscher, 1991; Stapulionis and Deutscher, 1995). In these prior studies, it was observed that aminoacyl tRNA deacylation/reacylation was coupled to protein synthesis and that it occurred without requisite transfer of the deacylated tRNA to the soluble phase (Negrutskii and Deutscher, 1991; Stapulionis and Deutscher, 1995). Under the experimental conditions described in these prior studies, the cytosol fraction was not assayed, and thus the cytosolic aminoacyl tRNA hydrolysis pathway reported here would not have been observed. With regard to the phenomenon of tRNA channeling, several reports have demonstrated that the release of aminoacyl-tRNA from its cognate tRNA synthetase and the subsequent association with elongation factor complexes are concerted events (Dang et al., 1983; Reed et al., 1994; Reed and Yang, 1994; Yang et al., 2006). Extending from data presented here, this process may be strongly favored on the ER membrane allowing for ER ribosomes to maintain a kinetic advantage in the protein synthesis reaction.
In summary, protein synthesis is a highly organized process that likely occurs in discrete subcellular locales in which the assembly of translation complexes can be efficiently regulated. In this report, we have identified the ER membrane as such a locale, and we suggest that the previously observed and highly complex patterns of mRNA partitioning between the cytosol and ER are a reflection of this organization. In this view, mRNA partitioning between the cytosol and ER serves as a novel form of posttranscriptional gene regulation, of particular relevance to cell activation, and cell stress and recovery. Given the magnitude of the differences in the steady-state mRNA translation rates of the cytosol and ER compartments reported herein, relatively modest alterations in cytosol/ER mRNA partitioning ratios would be predicted to yield substantial differences in steady state protein levels. The identification of the mechanisms governing the compartmental regulation of mRNA partitioning and the tRNA aminoacylation/deacylation cycle will provide needed insight into the supramolecular organization of cellular protein synthesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Rebecca D. Dodd, Brook Pyhtila, Brigid Hogan, and Jon Yewdell for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM-077382 (to C.V.N.) and American Heart Association predoctoral fellowship 0515333U (to S.B.S.).
Abbreviations used:
- ER
endoplasmic reticulum.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0677) on December 12, 2007.
REFERENCES
- Belin D., Wohlwend A., Schleuning W. D., Kruithof E. K., Vassalli J. D. Facultative polypeptide translocation allows a single mRNA to encode the secreted and cytosolic forms of plasminogen activators inhibitor 2. EMBO J. 1989;8:3287–3294. doi: 10.1002/j.1460-2075.1989.tb08489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry H. Monte Carlo simulations of enzyme reactions in two dimensions: fractal kinetics and spatial segregation. Biophys. J. 2002;83:1891–1901. doi: 10.1016/S0006-3495(02)73953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 1975a;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 1975b;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Walter P., Chang C. N., Goldman B. M., Erickson A. H., Lingappa V. R. Translocation of proteins across membranes: the signal hypothesis and beyond. Symp. Soc. Exp. Biol. 1979;33:9–36. [PubMed] [Google Scholar]
- Dang C. V., Yang D. C., Pollard T. D. Association of methionyl-tRNA synthetase with detergent-insoluble components of the rough endoplasmic reticulum. J. Cell Biol. 1983;96:1138–1147. doi: 10.1083/jcb.96.4.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M., Bhattacharya R., Botstein D., Brown P. O. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M., Eisen M. B., Botstein D., Brown P. O. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Blobel G., Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982a;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R., Walter P., Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol. 1982b;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Prehn S., Hartmann E., Kalies K. U., Rapoport T. A. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Kopczynski C. C., Noordermeer J. N., Serano T. L., Chen W. Y., Pendleton J. D., Lewis S., Goodman C. S., Rubin G. M. A high throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc. Natl. Acad. Sci. USA. 1998;95:9973–9978. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner R. S., Nicchitta C. V. mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. RNA. 2006;12:775–789. doi: 10.1261/rna.2318906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner R. S., Seiser R. M., Zheng T., Lager P. J., Reedy M. C., Keene J. D., Nicchitta C. V. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine C. G., Mitra D., Sharma A., Smith C. L., Hegde R. S. The efficiency of protein compartmentalization into the secretory pathway. Mol. Biol. Cell. 2005;16:279–291. doi: 10.1091/mbc.E04-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T. E., Rolleston F. S., Low R. B., Wool I. G. Dissociation and reassociation of skeletal muscle ribosomes. J. Mol. Biol. 1969;43:135–149. doi: 10.1016/0022-2836(69)90084-9. [DOI] [PubMed] [Google Scholar]
- Mechler B., Rabbitts T. H. Membrane-bound ribosomes of myeloma cells. IV. mRNA complexity of free and membrane-bound polysomes. J. Cell Biol. 1981;88:29–36. doi: 10.1083/jcb.88.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. I., Dobberstein B. Identification and characterization of a membrane component essential for the translocation of nascent proteins across the membrane of the endoplasmic reticulum. J. Cell Biol. 1980;87:503–508. doi: 10.1083/jcb.87.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M. M., Pitot H. C. Structure and function of rat liver polysome populations. I. Complexity, frequency distribution, and degree of uniqueness of free and membrane-bound polysomal polyadenylate-containing RNA populations. J. Cell Biol. 1981;90:495–506. doi: 10.1083/jcb.90.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M. M., Pitot H. C. Structure and function of rat liver polysome populations. II. Characterization of polyadenylate-containing mRNA associated with subpopulations of membrane-bound particles. J. Cell Biol. 1982;94:297–307. doi: 10.1083/jcb.94.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutka S. C., Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol. Biol. Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii B. S., Deutscher M. P. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. USA. 1991;88:4991–4995. doi: 10.1073/pnas.88.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii B. S., Deutscher M. P. A sequestered pool of aminoacyl-tRNA in mammalian cells. Proc. Natl. Acad. Sci. USA. 1992;89:3601–3604. doi: 10.1073/pnas.89.8.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. E., Siekevitz P. Liver microsomes; an integrated morphological and biochemical study. J. Biophys. Biochem. Cytol. 1956a;2:171–200. doi: 10.1083/jcb.2.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. E., Siekevitz P. Pancreatic microsomes; an integrated morphological and biochemical study. J. Biophys. Biochem. Cytol. 1956b;2:671–690. doi: 10.1083/jcb.2.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter M. D., Nicchitta C. V. Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J. Biol. Chem. 2000;275:33828–33835. doi: 10.1074/jbc.M005294200. [DOI] [PubMed] [Google Scholar]
- Potter M. D., Nicchitta C. V. Endoplasmic reticulum-bound ribosomes reside in stable association with the translocon following termination of protein synthesis. J. Biol. Chem. 2002;277:23314–23320. doi: 10.1074/jbc.M202559200. [DOI] [PubMed] [Google Scholar]
- Pyhtila B. M., Zheng T., Lager P. J., Keene J. D., Reedy M. C., Nicchitta C. V. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008 doi: 10.1261/rna.721108. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed V. S., Wastney M. E., Yang D. C. Mechanisms of the transfer of aminoacyl-tRNA from aminoacyl-tRNA synthetase to the elongation factor 1 alpha. J. Biol. Chem. 1994;269:32932–32936. [PubMed] [Google Scholar]
- Reed V. S., Yang D. C. Characterization of a novel N-terminal peptide in human aspartyl-tRNA synthetase. Roles in the transfer of aminoacyl-tRNA from aminoacyl-tRNA synthetase to the elongation factor 1 alpha. J. Biol. Chem. 1994;269:32937–32941. [PubMed] [Google Scholar]
- Seiser R. M., Nicchitta C. V. The fate of membrane-bound ribosomes following the termination of protein synthesis. J. Biol. Chem. 2000;275:33820–33827. doi: 10.1074/jbc.M004462200. [DOI] [PubMed] [Google Scholar]
- Siekevitz P., Palade G. E. A cyto-chemical study on the pancreas of the guinea pig. III. In vivo incorporation of leucine-[14C] into the proteins of cell fractions. J. Biophys. Biochem. Cytol. 1958;4:557–566. doi: 10.1083/jcb.4.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapulionis R., Deutscher M. P. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. USA. 1995;92:7158–7161. doi: 10.1073/pnas.92.16.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens S. B., Dodd R. D., Brewer J. W., Lager P. J., Keene J. D., Nicchitta C. V. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol. Biol. Cell. 2005;16:5819–5831. doi: 10.1091/mbc.E05-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens S. B., Dodd R. D., Lerner R. S., Pyhtila B. M., Nicchitta C. V. Analysis of the partitioning of mRNAs between cytosolic and ER membrane-bound compartments in mammalian cells. In: Wilusz J., editor. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2007. (in press) [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981a;91:551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 1981b;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Ibrahimi I., Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Lingappa V. R. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- Yang X. L., Otero F. J., Ewalt K. L., Liu J., Swairjo M. A., Kohrer C., RajBhandary U. L., Skene R. J., McRee D. E., Schimmel P. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.