Abstract
Type III phosphatidylinositol (PtdIns) 4-kinases (PI4Ks) have been previously shown to support plasma membrane phosphoinositide synthesis during phospholipase C activation and Ca2+ signaling. Here, we use biochemical and imaging tools to monitor phosphoinositide changes in the plasma membrane in combination with pharmacological and genetic approaches to determine which of the type III PI4Ks (α or β) is responsible for supplying phosphoinositides during agonist-induced Ca2+ signaling. Using inhibitors that discriminate between the α- and β-isoforms of type III PI4Ks, PI4KIIIα was found indispensable for the production of phosphatidylinositol 4-phosphate (PtdIns4P), phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2], and Ca2+ signaling in angiotensin II (AngII)-stimulated cells. Down-regulation of either the type II or type III PI4K enzymes by small interfering RNA (siRNA) had small but significant effects on basal PtdIns4P and PtdIns(4,5)P2 levels in 32P-labeled cells, but only PI4KIIIα down-regulation caused a slight impairment of PtdIns4P and PtdIns(4,5)P2 resynthesis in AngII-stimulated cells. None of the PI4K siRNA treatments had a measurable effect on AngII-induced Ca2+ signaling. These results indicate that a small fraction of the cellular PI4K activity is sufficient to maintain plasma membrane phosphoinositide pools, and they demonstrate the value of the pharmacological approach in revealing the pivotal role of PI4KIIIα enzyme in maintaining plasma membrane phosphoinositides.
INTRODUCTION
Activation of cell surface receptors by a variety of stimuli initiates a cascade of molecular events ultimately eliciting a response characteristic of the target cell. One of the most studied and best-characterized signal transduction pathways is initiated by the phospholipase C-mediated breakdown of phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] to generate the Ca2+-mobilizing messenger inositol trisphosphate (InsP3) and the protein kinase C activator diacylglycerol (Berridge and Irvine, 1984). It has long been recognized that the sustained production of these messengers requires continuous phosphorylation of phosphatidylinositol (PtdIns) to phosphatidylinositol 4-phosphate (PtdIns4P) and PtdIns(4,5)P2 by phosphoinositide (PI) 4-kinase (PI4K) and PIP 5-kinase enzymes, due to the limited amount of PtdIns(4,5)P2 present in the plasma membrane (Creba et al., 1983). Several isoforms within the PI 4-kinase and PIP 5-kinase families have been described previously (Doughman et al., 2003; Balla and Balla, 2006), and a recent study identified a splice variant of PIP 5-kinase γ as the enzyme responsible for PtdIns4P to PtdIns(4,5)P2 conversion in agonist-induced Ca2+ signaling (Wang et al., 2004). In contrast, the PI 4-kinase that generates PtdIns4P for this process so far has eluded identification.
Four PI 4-kinase enzymes have been identified in mammalian cells that belong to either the type II or type III families (Balla and Balla, 2006). Type II PI 4-kinase enzymes (α- and β-forms) are small, 56-kDa proteins that are abundant in almost all cellular membranes, and they are enriched in plasma membrane preparations. They are kept in the membrane by palmitoylation, and recent studies suggest that these enzymes have a role in post-trans-Golgi network trafficking (Wang et al., 2003, 2007) and the endocytic processing of the epidermal growth factor receptors (Minogue et al., 2006). Type III PI 4-kinases (α- and β-forms), in contrast, are soluble enzymes structurally related to PI 3-kinases, and they can be inhibited by higher concentrations of PI 3-kinase inhibitors, such as wortmannin (Wm). We have reported over 10 years ago that production of the agonist-sensitive phosphoinositide pools required the activity of Wm-sensitive type III PI 4-kinase enzymes (Nakanishi et al., 1995). However, it is still not known which of the type III enzymes participates in the formation of the agonist-regulated PtdIns4P pools.
In the present study, we used both pharmacological and genetic approaches to interfere with the activity of all of the PI 4-kinase enzymes, and we monitored phosphoinositide changes both by conventional metabolic labeling of inositol lipids and -phosphates and by using green fluorescent protein (GFP)-tagged pleckstrin homology (PH) domains recognizing PtdIns4P and PtdIns(4,5)P2. These results show that the PH domain of the yeast OSH2 protein is a valuable tool to monitor plasma membrane PtdIns4P pools and that PI 4-kinase IIIα is required for sustained InsP3 production and Ca2+ signaling. Although this conclusion was supported by small interfering RNA (siRNA)-mediated gene silencing of the individual PI4K enzymes, the present studies also highlight the difficulties in obtaining conclusive data with siRNA on enzymes whose functions are near essential and emphasize the value of inhibitors to dissect the roles of these proteins in cellular signaling.
MATERIALS AND METHODS
Materials
Wortmannin and ionomycin were purchased from Calbiochem (San Diego, CA). Phenylarsine oxide (PAO), β-mercaptoethanol, dithiothreitol, and poly-lysine were obtained from Sigma-Aldrich (St. Louis, MO). Fura-2/acetoxymethyl ester (AM) and Pluronic acid were from Invitrogen (Carlsbad, CA). PIK93 was synthesized as described previously (Knight et al., 2006). Myo-[3H]inositol (60 Ci/mmol) was purchased from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom) and ortho-[32P]phosphate (9000 Ci/mmol) was from General Electric (PerkinElmer Life and Analytical Sciences, Boston, MA). The polyclonal antibody against PI4KIIIβ was purchased from UBI (Lake Placid, NY), the polyclonal antibody against PI4KIIα was a kind gift from Dr. Pietro De Camilli (Yale School of Medicine, New Haven, CT), and the anti PI4KIIIα antibody was raised in New Zealand rabbits as described previously (Balla et al., 2005).
DNA Constructs and Transfections
The OSH2–2x-GFP construct was generated by amplifying the PH domain sequence of OSH2 (residues 256–424) from Saccharomyces cerevisiae cDNA (American Type Culture Collection, Manassas, VA) by using two primer pairs to obtain fragments flanked by XhoI/EcoRI and EcoRI/KpnI sites. These fragments were then cloned in tandem between the XhoI/KpnI sites of the pEGFP-C1 plasmid (Clontech, Mountain View, CA), with a linker (VNSKL) in between them following the design of Roy and Levine (2004). The single PH domain version of the PH domain also has been created as well as the cyan and yellow fluorescent versions of the tandem construct. The PLCδ1PH-GFP construct (Várnai and Balla, 1998) and its color variants have been described previously (Varnai et al., 2002). The OSH1-PH-GFP was kindly provided by Dr. Mark Lemmon (University of Pennsylvania, Philadelphia, PA) (Yu et al., 2004), and the type IV phosphoinositide 5-phosphatase was a kind gift of Dr. Philip Majerus (Washington University, St. Louis, MO) (Kisseleva et al., 2000). The constructs used for the rapamycin-inducible translocation of the type-IV phosphoinositide 5-phosphatase have been described recently (Varnai et al., 2006).
The human embryonic kidney (HEK)-293AT1 cells used for these studies have been stably transfected with the hemagglutinin (HA)-AT1a and FLAG-AT1a angiotensin receptors in two rounds of selection by using G-418 and Zeocine (Zeo) (Invitrogen, Carlsbad, CA) for the two constructs, respectively. These cells were kindly provided by Drs. Alberto Jesus Olivares-Reyes and Kevin J. Catt (NICHD, NIH, Bethesda, MD). Cells were cultured in DMEM with Pen/Strep (Invitrogen) and 10% fetal bovine serum (FBS), and they have been subjected to a G-418/Zeo selection from time to time but not during cultures for the experiments.
For RNA interference (RNAi)-mediated knockdown, the cells were cultured either in 10-cm dishes (for suspension Ca2+ measurements), 12-well plates (for metabolic labeling studies), or 25-mm glass coverslips treated with poly-lysine (for single-cell Ca2+ or confocal studies). The duplexes used for treatment have been described previously (Balla et al., 2005). Cells were treated with 100 nM siRNA twice in consecutive days, and they were analyzed on the fourth day after the first treatment.
Analysis of Single Cells for Fluorescence Resonance Energy Transfer (FRET), Cytoplasmic Ca2+ Measurements, and Confocal Microscopy
HEK-293AT1 cells were cultured on glass coverslips (3 × 105 cells/35-mm dish) pretreated with poly-lysine and transfected with the various constructs (0.5–2 μg DNA/dish) by using Lipofectamine 2000 (Invitrogen) for 24 h as described previously (Varnai et al., 2005). For calcium measurements, cells were loaded with 3 μM Fura-2/AM in medium 199/Earle's balanced salt solution containing 1.2 mM CaCl2, 3.6 mM KCl, 25 mM HEPES containing 1 mg/ml bovine serum albumin (BSA), 0.06% Pluronic acid, and 200 μM sulfinpyrazone, for 45 min at room temperature. Calcium measurements were performed at room temperature in a modified Krebs-Ringer buffer containing 120 mM NaCl, 4.7 mM KCl, 1.2 mM CaCl2, 0.7 mM MgSO4, 10 mM glucose, and Na-HEPES 10 mM, pH 7.4. An Olympus IX70 inverted microscope equipped with a Lamda-DG4 illuminator and a MicroMAX-1024BFT digital camera and the appropriate filter sets was used for Ca2+ analysis. The same microscope equipped with a beam-splitter (Optical Insights, Photometrics, Tucson, AZ) with a 505-nm dichroic mirror was used for FRET analysis with 435/25-nm excitation and 475/30- and 535/30-nm emission filters, respectively. Images acquired simultaneously in the two emission wavelengths in the two halves of the camera sensor chip were used to form the ratios. These ratio values were then normalized, taking their control initial values as 100 and the minimum values obtained after ionomycin (or agonist treatment if the latter was smaller) as zero.
Cells expressing the OSH2–2x-PH-GFP and PLCδ1PH-monomeric red fluorescent protein (mRFP) constructs were studied in the same modified Krebs-Ringer solution at 35°C by using a Zeiss 510Meta laser-scanning confocal microscope (Carl Zeiss, Thornwood, NY) in multitrack mode, and pictures were taken in time-series mode. Membrane localization was assessed by forming membrane/cytosol intensity ratio values from line-intensity histograms of cells in the whole series of images after acquisition. Data acquisitions for Ca2+ and FRET measurements and processing were performed by the MetaFluor software (Molecular Devices, Sunnyvale, CA). Postacquisition data analysis of the confocal images was performed with either the MetaView (Carl Zeiss) or the MetaMorph (Molecular Devices) software.
Cytoplasmic Ca2+ Measurements in Cell Suspensions
Cells grown on 10-cm culture plates were removed by mild trypsinization and loaded with 0.5 μM Fura-2/AM in the same solution described above for single Ca2+ measurements. Cells were then washed with the same medium without Fura-2/AM and stored at room temperature in the dark. Aliquots of cells (∼5 × 105 cells) were centrifuged rapidly before the measurements and dispersed in 2.5 ml of the modified Krebs-Ringer buffer used for all other analysis. Ca2+ measurements were performed at 35°C in a PTI Deltascan spectrofluorometer (Photon Technology International, Princeton, NJ).
Analysis of Myo-[3H]Inositol- or [32P]Phosphate-labeled Lipids and 3H-labeled Inositol Phosphates
Cultured cells were labeled with myo-[3H]inositol (20 μCi/ml) on 12-well culture plates in inositol-free DMEM supplemented with 2% dialyzed FBS and 1 mg/ml BSA for 24 h. For 32P-phopshate labeling, the cells were labeled with 2 μCi/ml o-[32P]phosphate for 3 h in phosphate-free DMEM supplemented with 1 mg/ml BSA. After myo-inositol labeling, the cells were washed twice with a medium without inositol, and after a 10-min preincubation, inhibitors were added for a further 10 min before stimulation with 100 nM angiotensin II (AngII) for the indicated times. Reactions were terminated by the addition of ice-cold perchloric acid (5% final concentration), and cells were kept on ice for 30 min. After scraping, and freezing/thawing, the cells were centrifuged, and the supernatant was processed for perchloric acid (PCA) extraction and analysis by high-performance liquid chromatography as described previously (Nakanishi et al., 1995). The cell pellet was also processed to extract the phosphoinositides by an acidic chloroform/methanol extraction followed by thin layer chromatography (TLC) analysis essentially as described previously (Nakanishi et al., 1995). 32P-labeled cells were not washed after the labeling period, but they were treated in the same medium with inhibitors for 10 min followed by AngII (10−7 M) stimulation. Reactions were terminated by PCA, and lipids were extracted and separated as with the inositol-labeled cells.
RESULTS
Pharmacological Manipulations of Type III PI 4-Kinase Activities Affect Agonist-regulated PtdIns4P Pools
High (micromolar) concentrations of Wm have been valuable tools to demonstrate a role of type III PI4Ks in InsP3/Ca2+ signaling. However, it is not possible to discriminate between the α- and β-forms of these PI4Ks based on their Wm sensitivities (Balla et al., 1997; Knight et al., 2006). Therefore, we took advantage of the recent characterization of isoform-specific PI 3-kinase inhibitors that revealed one of the inhibitors, PIK93, as being significantly more potent against PI4KIIIβ than PI4KIIIα (Knight et al., 2006). This inhibitor inhibits PI 3-kinases and clearly discriminates between the two PI 4-kinase enzymes as assessed by an in vitro PI kinase assay on the purified mammalian proteins (Knight et al., 2006). Unfortunately, none of the tested inhibitors showed the opposite selectivity, i.e., inhibition of PI4KIIIα more potently than the PI4KIIIβ enzyme. Phenylarsine oxide (PAO), a sulfhydryl (SH)-reactive agent has been shown earlier to inhibit PI4Ks (Wiedemann et al., 1996). PAO was found to inhibit type III PI 4-kinases, whereas it is relatively ineffective against the type II enzymes based on in vitro kinase assays of the purified proteins (Balla et al., 2002; Barylko et al., 2002). Among the type III enzymes, PI4KIIIα is significantly more sensitive to PAO than the type IIIβ form (Balla et al., 2002). This difference was also exploited to discriminate between the two enzymes in their involvement to generate the hormone-sensitive PtdIns4P pools. It is important to emphasize that PAO is a SH-reactive agent that probably has many targets in a cell. However, in the present study, the effects of PAO have only been tested on parameters that immediately reflect PI 4-kinase functions (see below); therefore, in this narrow context the effects of PAO on the purified PI 4-kinases and PtdIns4P formation in the intact cell can be correlated with somewhat higher confidence.
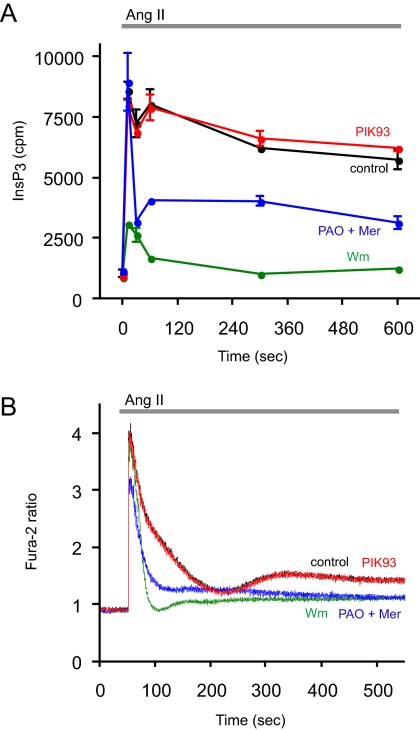
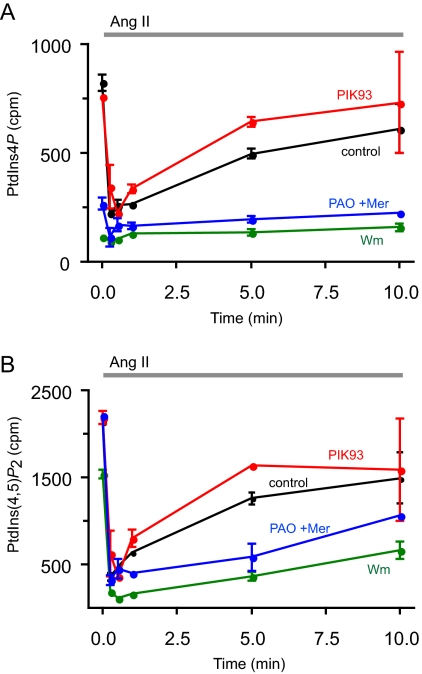
Several parameters were tested with these inhibitors in HEK-293 cells stably expressing AT1a angiotensin receptors (HEK-293-AT1). Cellular phosphoinositides were monitored after [32P]phosphate or myo-[3H]inositol labeling, and InsP3 formation was followed from cells prelabeled with myo-[3H]inositol. Finally, the effects of the inhibitors on AngII-induced changes in cytoplasmic Ca2+ concentration [Ca2+]i were measured in cell suspensions with Fura-2. Figure 1 shows the effects of inhibitors on InsP3 levels analyzed from myo-[3H]inositol-labeled cells and on Ca2+ signaling. As shown earlier in AngII-stimulated adrenal glomerulosa cells (Nakanishi et al., 1995), 10 μM Wm pretreatment greatly reduced the size of AngII-induced InsP3 elevation in HEK-293-AT1 cells, and it eliminated its sustained increase. This was also reflected in the cytoplasmic Ca2+ changes that became transient lacking the plateau phase of Ca2+ rise in Wm treated cells (Figure 1B). These effects of Wm were not observed at lower concentrations (<300 nM) that already inhibit PI 3-kinases (data not shown), and they were clearly caused by the limited supply of PtdIns4P and PtdIns(4,5)P2, consistent with the inhibition of a PI4K that helps replenishing these pools during a sustained phospholipase C (PLC) activation. This is demonstrated in Figure 2, where the [32P]phosphate-labeled PtdIns4P and PtdIns(4,5)P2 were analyzed in AngII-stimulated cells. Without the inhibitors, AngII stimulation evokes a robust PLC activation, resulting in the rapid depletion of PtdIns(4,5)P2 that slowly returns toward prestimulatory levels. A similar change is observed in PtdIns4P levels due to the conversion of this lipid to PtdIns(4,5)P2 by the PIP 5-kinases and its relatively slower synthesis by the PI4Ks. Pretreatment of the cells with 10 μM Wm greatly reduced the 32P-labeled PtdIns4P and, hence, the AngII-induced changes observed in HEK-293-AT1 cells (Figure 1A), but it only slightly reduced basal PtdIns(4,5)P2 levels. However, this treatment significantly enhanced the AngII-induced decrease in PtdIns(4,5)P2 and strongly inhibited its resynthesis (Figure 2B).
Figure 1.
Effects of PI4K inhibitors on the kinetics of InsP3 and [Ca2+]i changes in HEK-293-AT1 cells stimulated with AngII. (A) HEK-293-AT1 cells were labeled with myo-[3H]inositol for 24 h in inositol-free medium as described under Materials and Methods. After washing, AngII (10−7 M) was added to the cells for the indicated times, and reactions were terminated by PCA. Labeled inositol phosphates were extracted from the soluble fraction and separated by high-performance liquid chromatography connected to a scintillation flow detector as described previously (Nakanishi et al., 1995). Data are shown are means ± range of duplicate determinations. The concentrations of the inhibitors added 10 min before AngII were 250 nM PIK93, 10 μM PAO with 1 mM β-mercaptoethanol (Mer), and 10 μM Wm. Two additional experiments were performed with the 10-min time points with similar results. (B) HEK-293-AT1 cells were loaded with Fura-2/AM, and their fluorescence monitored in a fluorescent spectrophotometer as described under Materials and Methods. AngII (10−7 M) was added at the indicated time. Pretreatment with the inhibitors for 10 min was as described in A. This result is a representative of three similar observations.
Figure 2.
Effects of PI4K inhibitors on the kinetics of phosphoinositide changes in HEK-293-AT1 cells stimulated with AngII. HEK-293-AT1 cells were labeled with [32P]phosphate for 3 h in a phosphate-free medium as described under Materials and Methods. AngII (10−7 M) was added to the cells at the indicated times, and reactions terminated by the addition of PCA. Lipids were extracted from the cell pellets, separated by TLC, and analyzed both by a PhosphorImager and scintillation counting of the spots cut out from the plates. Data shown are means ± range of duplicate determinations. The concentrations of the inhibitors added 10 min before AngII were 250 nM PIK93, 10 μM PAO with 1 mM Mer, and 10 μM Wm. This full time course experiment was repeated from myo-[3H]inositol-labeled cells with similar results, supporting the same conclusion.
These effects of Wm were not reproduced by 250 nM PIK93, which inhibits PI4KIIIβ (and the PI 3-kinases) but not PI4KIIIα (Knight et al., 2006), and which was found as effective as 10 μM Wm in inhibiting the endoplasmic reticulum (ER)-to-Golgi transport of ceramide, a process linked to PI4KIIIβ function (Toth et al., 2006). PIK93 failed to affect either the InsP3 and Ca2+ changes or those of the 32P-labeled phosphoinositides during AngII stimulation (Figures 1 and 2). The effect of PAO was tested at a concentration of 10 μM (applied in the presence of 1 mM β-mercaptoethanol to reduce its side effects). Although at this concentration PAO only partially inhibits PI4KIIIα (∼80%), it was chosen because it does not yet inhibit PI4KIIIβ (Balla et al., 2002). As shown in Figure 1, PAO partially mimicked the effects of Wm on both the InsP3 and Ca2+ responses, substantially reducing the sustained phases of both signals. Also, after 10 μM PAO treatment the 32P-labeled phosphoinositides showed changes similar to those observed with Wm treatment (Figure 2). All of these data were consistent with the limited supply of PtdIns4P in the presence of Wm or PAO but not PIK93, implicating the PI4KIIIα enzyme in the process, and the incomplete effects of PAO relative to Wm were consistent with the incomplete inhibition of PI4KIIIα at this concentration of the inhibitor.
Analysis of PtdIns(4,5)P2 Changes in the Plasma Membrane with the PLCδ1PH Domain
These metabolic data provided information on the overall inositide pools of the cells but only limited information on the question of which membranes contributed to these changes. In fact, when cells were labeled with myo-[3H]inositol, a smaller fraction of the inositides showed changes with AngII and the inhibitors (data not shown), suggesting the presence of additional metabolic pools that become labeled with myo-[3H]inositol in 24 h but not with [32P]phosphate in 3 h. Therefore, further experiments were designed to monitor the phosphoinositide changes in single cells by using PH domains that recognize PtdIns4P and PtdIns(4,5)P2. PtdIns(4,5)P2 changes were followed in single cells by either confocal microscopy or by FRET analysis using yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP)-tagged versions of the PLCδ PH domain (van Der Wal et al., 2001).
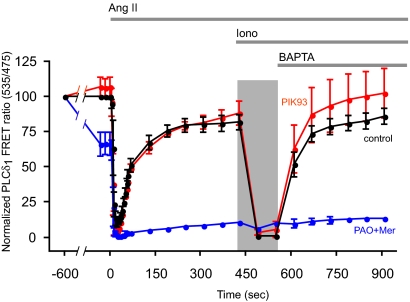
Cells were stimulated with AngII for the indicated times followed by the addition of high concentration (10 μM) of ionomycin to activate PLC and to eliminate PLCδ1PH-GFP localization and FRET (Várnai and Balla, 1998; Balla et al., 2005). Subsequent addition of 1,2-bis(2-aminophenoxy)ethane- N,N,N′,N′-tetraacetic acid (BAPTA) (or EGTA) was used to reverse the Ca2+ change and allow the PtdIns(4,5)P2 pools to recover. As shown in Figure 3, the PLCδ1PH translocation was rapid and complete after AngII treatment, and the FRET signal rapidly returned close to the baseline in control cells (black trace). Ionomycin treatment then caused the complete translocation of the probe to the cytosol from which the cells recovered slowly after Ca2+ chelation. These changes were not affected by preincubation of the cells with 250 nM PIK93 (Figure 3, red trace), consistent with the previous data with metabolic labeling and suggesting that PI4KIIIβ plays no significant role in this process. In contrast, 10 μM PAO (with 1 mM β-mercaptoethanol) decreased the basal FRET by ∼40% and prevented the relocalization of the probe after the AngII-induced complete translocation (Figure 3, blue trace).
Figure 3.
Effects of PI4K inhibitors on the kinetics of redistribution of PLCδ1PH domain in HEK-293-AT1 cells stimulated with AngII. HEK-293 cells stably transfected with the AT1a angiotensin receptors were cotransfected with the PLCd1PH-CFP and -YFP constructs for 24 h. The FRET signal from individual cells was then monitored in a wide-field fluorescent microscope equipped with an emission beam splitter using 430-nm excitation and 535- and 475-nm emissions as described under Materials and Methods. To minimize light exposure, after the first minute of stimulation, data were collected in longer intervals. AngII (10−7 M) was added at the indicated time followed by 10 μM inonomycin (Iono) and 5 mM BAPTA (or EGTA). The dark bar represents the exposure of the cells to the high (extracellular) Ca2+ concentration after ionomycin treatment. The concentrations of the inhibitors added 10 min before AngII were 250 nM PIK93 and 10 μM PAO with 1 mM Mer. FRET was expressed as fluorescent emission ratio values (535/475 nm), and it was normalized so that the ratio value recorded before the addition of inhibitors was taken as 100% and the lowest values (after AngII or ionomycin) were taken as 0%. Means ± SEM (or range) from five, two, and three cells are shown for control, PAO + Mer, and PIK93, respectively. Similar changes were observed in two additional experiments.
When the effects of Wm were tested in similar experiments, we encountered several technical difficulties. First, the inhibitory effects of Wm showed a strong light-induced reversal in single-cell experiments. At shorter excitation wavelengths, these effects were stronger, making it essentially impossible to monitor the effect of Wm on single cell Ca2+ responses with Fura-2. (The photon flux density is significantly lower during Ca2+ measurements in cell suspension; therefore, those measurements are less sensitive to this effect). In addition, a significant drift was observed in the FRET baseline during the 10 min Wm preincubation period (without illumination). Although this was partially eliminated rapidly after two to three light scans, it was difficult to assess the basal FRET values after Wm treatment. Therefore the data on the effects of Wm (still partially inhibitory) were not included in this analysis.
Evaluation of the OSH2 PH Domain as a Probe of Plasma Membrane PtdIns4P Levels
None of the type III PI4K enzymes can be detected by immunofluorescence at the plasma membrane; yet, PtdIns4P has to be present in the plasma membrane to be converted to PtdIns(4,5)P2. To obtain information on the activity rather than the steady-state cellular distribution of the PI4K enzymes, it was highly desirable to use reporter probes that monitor PtdIns4P production in single cells. PH domain-GFP chimeras with high in vitro PtdIns4P binding specificities, such as the four phosphate adaptor protein (FAPP)1 or oxysterol binding protein (OSBP) PH domains have been used for this purpose, but these probes mostly report on PtdIns4P formed in the Golgi as they also require Arf1-GTP for their Golgi recruitment (Levine and Munro, 2002; Godi et al., 2004; Balla et al., 2005). Although both of these PH domains can detect PtdIns4P formation in the plasma membrane under special circumstances (Balla et al., 2005), they are not optimal for monitoring PtdIns4P in the plasma membrane. It has recently been shown by two independent studies that the PH domain of the yeast oxysterol binding protein homologue, OSH2 recognizes PtdIns4P in the plasma membrane, in spite of its limited specificity to bind PtdIns4P in vitro (Roy and Levine, 2004; Yu et al., 2004). Therefore, first, we wanted to determine whether the OSH2 PH domain could, in fact, be used to follow PtdIns4P changes in the plasma membrane of mammalian cells.
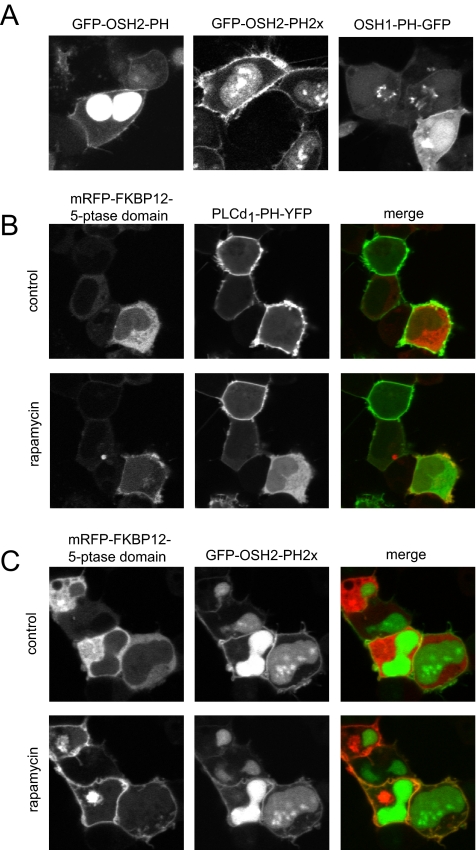
Expression of the single PH domain of OSH2 fused to GFP showed clear plasma membrane localization and a strong nuclear staining as described previously (Roy and Levine, 2004; Yu et al., 2004). Using two PH domains in tandem fused to GFP (GFP-OSH2-PH2x) greatly reduced the nuclear localization of the construct and clearly labeled the plasma membrane (Figure 4A). Importantly, unlike in yeast cells, neither the tandem nor the single OSH2 PH domain decorated the Golgi membrane in COS-7 or HEK-293 cells (Figure 4A). In contrast, the yeast OSH1 PH domain labeled both the plasma membrane (at high expression levels) and the Golgi (Figure 4A). Because the OSH2 PH domain has limited in vitro specificity to PtdIns4P (Yu et al., 2004), we were concerned that it may be recruited to the plasma membrane by PtdIns(4,5)P2. The specificity question was examined with our recently developed method of acute depletion of PtdIns(4,5)P2 in the plasma membrane. This approach is based on a drug induced plasma membrane recruitment of a truncated type-IV phosphoinositide 5-phosphatase (5-ptase) that was rendered cytoplasmic by mutations in its localization sequences (Varnai et al., 2006). Recruitment of this cytoplasmic 5-ptase to the plasma membrane rapidly eliminated PtdIns(4,5)P2 as monitored by PLCδ1PH-GFP localization (Figure 4B), without elimination of the GFP-OSH2-PH2x localization (Figure 4C). This question was further analyzed in total internal reflection fluorescence experiments where the release of the PH domain from the membrane can be better quantified. These experiments showed no effect of the phosphatase recruitment on GFP-OSH2-PH2x localization while eliminating PLCδ1PH-GFP localization in COS-7 cells (Korzeniowski and Balla, unpublished observations).
Figure 4.
Localization of OSH1- and OSH2-PH domain-GFP fusion proteins in HEK-293-AT1 cells. (A) The PH domains of the yeast oxysterol-binding protein homologues OSH2 and OSH1 were fused to GFP as described under Materials and Methods. These constructs were transfected into HEK-293-AT1 cells and examined 24 h after transfection with live cell confocal microscopy. Note that the OSH2-PH domain binds to the plasma membrane and strongly accumulates in the nucleus but does not bind to the Golgi. A tandem PH domain of the OSH2 protein shows smaller signal in the nucleus and a strong plasma membrane binding. The OSH1-PH domain binds both the plasma membrane and the Golgi. (B) Elimination of the plasma membrane localization of PLCδ1PH-GFP [monitoring PtdIns(4,5)P2] by plasma membrane recruitment of a phosphoinositide 5-phosphatase. HEK-293-AT1 cells were transfected with a truncated type-IV phosphoinositide 5-phosphatase fused to mRFP and FKBP12, a plasma membrane-targeted FRB-CFP construct and the PLCδ1PH-YFP reporter described in Varnai et al. (2006). Addition of rapamycin for 3 min recruits the otherwise cytoplasmic 5-ptase construct to the plasma membrane (left) with a concomitant elimination of PtdIns(4,5)P2 and loss of PLCδ1PH-YFP localization (middle). (C) The same manipulations do not eliminate the plasma membrane localization of the OSH2-PH2x-GFP, suggesting that this construct is not kept at the membrane by PtdIns(4,5)P2.
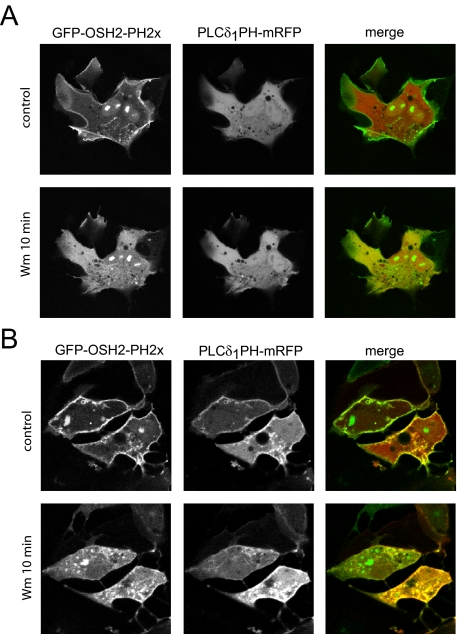
In a separate set of studies performed in COS-7 cells, the wild-type 5-ptase enzyme was expressed together with the mRFP-fused PLCδ1PH domain and the GFP-OSH2-PH2x construct. This triple transfection yielded many cells in which the plasma membrane localization of the PLCδ1PH-mRFP construct was eliminated indicating the depletion of PtdIns(4,5)P2.; yet, the localization of the OSH2-PH2x was still preserved (Figure 5A). These studies also confirmed that the OSH2-PH2x was not recruited to the membrane by PtdIns(4,5)P2. When such cells were treated with 10 μM Wm, the localization of OSH2-PH2x was rapidly eliminated (Figure 5A). Lower concentrations of Wm specific for PI 3-kinases had no such effect (data not shown), indicating that the plasma membrane pool of PtdIns4P monitored by OSH2-PH2x requires the activity of type III PI 4-kinases. Notably, Wm exerted a much slower effect on OSH2-PH2x localization in cells not expressing the 5-phosphatase (Figure 5B; see below) indicating that the dephosphorylation of PtdIns(4,5)P2 probably contributes to maintaining PtdIns4P levels in the membrane for a period of time when PI4K is inhibited.
Figure 5.
Localization of OSH2-PH2x-GFP to the plasma membrane is wortmannin sensitive. (A) COS-7 cells were transfected with OSH2-PH2x-GFP together with PLCδ1PH-mRFP and the wild-type type IV phosphoinositide 5-phosphatase for 24 h. Cells were selected so that the PLCδ1PH-mRFP showed no localization, indicating the lack of PtdIns(4,5)P2 as a result of phosphatase expression. These cells still showed plasma membrane localization of OSH2-PH2x-GFP, indicating that the construct is kept in the membrane not by PtdIns(4,5)P2. Addition of 10 μM Wm to such cells caused a rapid translocation of the OSH2-PH2x-GFP domain construct from the membrane to the cytosol. (B) Release of the OSH2-PH2x-GFP construct from the membrane after Wm treatment is significantly slower in control cells where PtdIns(4,5)P2 is present in the membrane.
The PH Domain of OSH2 Follows Agonist-induced Changes of PtdIns4P Levels
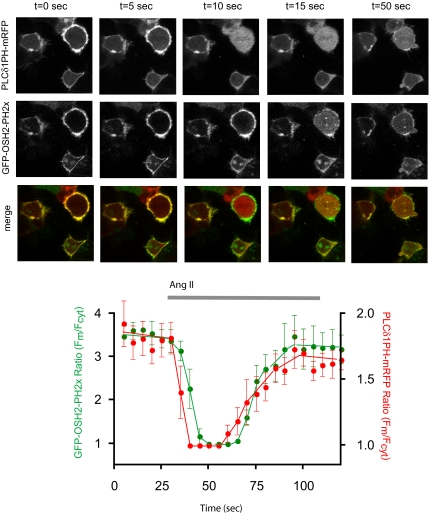
Next, we determined whether GFP-OSH2-PH2x localization is affected by agonist-induced PLC activation. HEK-293-AT1 cells were cotransfected with the PLCδ1PH-mRFP and GFP-OSH2-PH2x for simultaneous monitoring of PtdIns(4,5)P2 and PtdIns4P. As shown in Figure 6, both constructs showed a rapid and transient translocation from the plasma membrane to the cytosol after AngII addition, but with notable kinetic differences. The translocation of the GFP-OSH2-PH2x occurred with a clearly measurable (∼5-s) delay compared with that of PLCδ1PH-mRFP. The kinetics of changes in GFP-OSH2-PH2x localization after AngII addition was indistinguishable regardless whether cells were preincubated with 10 mM Li+ or not (data not shown). Because Ins(1,4)P2 levels accumulate to very high levels and remain elevated in Li+-treated cells (e.g., Balla et al., 1988) due to the inhibition of the phosphatase that dephosphorylates Ins(1,4)P2 (Majerus et al., 1999), these findings suggest that the AngII-induced translocation of the GFP-OSH2-PH2x protein from the membrane is caused by the changes in membrane PtdIns4P levels rather than increases in Ins(1,4)P2 levels in the cytosol. The slight delay in the response of GFP-OSH2-PH2x compared with that of PLCδ1PH-mRFP may reflect the slower (probably Ca2+-dependent) activation of the PLC isoform that hydrolyzes PtdIns4P, or a slight delay in the conversion of PtdIns4P to PtdIns(4,5)P2, but the contribution of the rapid Ins(1,4,5)P3 increase to PLCδ1PH-mRFP translocation can also be responsible for its faster kinetics. Nevertheless, these data together were consistent with the conclusion that the GFP-OSH2-PH2x reporter can monitor plasma membrane PtdIns4P changes during agonist activation.
Figure 6.
Angiotensin II-induced changes in membrane phosphoinositides simultaneously monitored by OSH2-PH2x-GFP and PLCδ1PH-mRFP. HEK-293 cells stably transfected with the AT1a angiotensin receptors were cotransfected with the PLCδ1PH-mRFP and OSH2-PH2x-GFP constructs for 24 h. Live cells were examined in a confocal microscope at 35°C during stimulation with AngII (10−7 M), and pictures were taken every 5 s. Note the faster response observed with the PLCδ1PH-mRFP. Quantification of membrane/cytosolic fluorescence intensities as function of time calculated from line-intensity histograms from five to seven cells obtained in two separate experiments (means ± SEM).
Pharmacological Manipulations of the Membrane Localization of OSH2–2x-PH
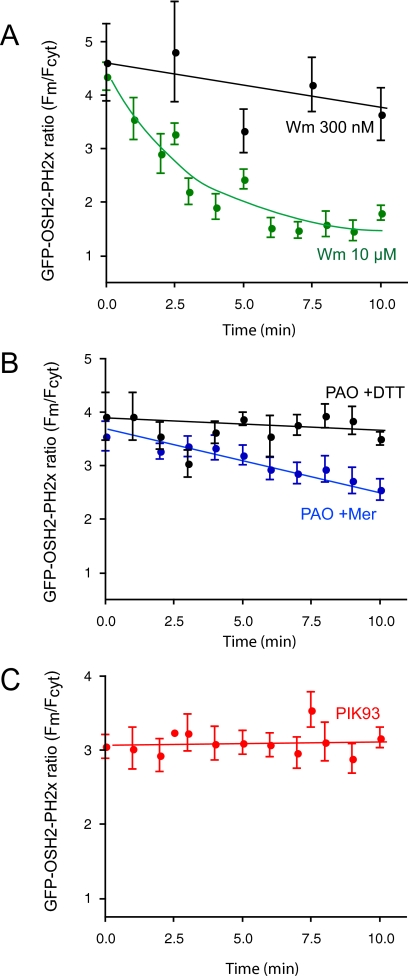
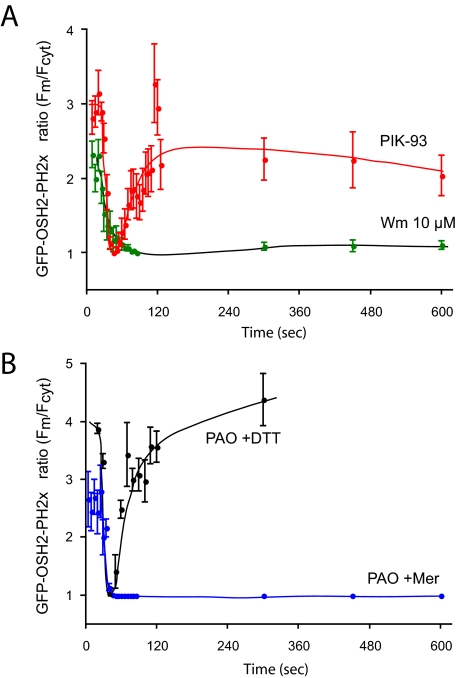
In response to the PI 4-kinase inhibitors, the GFP-OSH2-PH2x domain showed changes in good agreement with the [32P]phosphate and myo-[3H]inositol labeling results: the localization of the GFP-OSH2-PH2x was monitored as a change in the membrane to cytosol ratio of fluorescence. (We also attempted to use FRET for analysis of OSH2-PH2x. However, the FRET signal was significantly smaller than with the PLCδ1PH domain, which may reflect a lower abundance of PtdIns4P in the membrane or a lower density of the lipid in the membrane microdomains where it is found). The membrane-bound fluorescence of GFP-OSH2-PH2x already started to decrease during the 10-min Wm (10 μM) preincubation, and ∼70–80% of the membrane localization was eliminated by the end of this period. Lower concentrations of Wm (300 nM) did not show this effect (Figure 7A). Subsequent addition of AngII rapidly eliminated the remaining localization and no relocalization of the probe was observed throughout the 10-min stimulation (Figure 8A). It is important to note that the effects of Wm became almost undetectable when prolonged rapid sampling of the confocal pictures was used to follow localization of the PH domains. This was again attributed to the photolability of Wm inhibition discussed above and also observed in Warashina (1999). Ten micromolar PAO (with 1 mM β-mercaptoethanol) had a similar inhibitory effect on the basal localization, although this treatment did not evoke the same decrease as Wm did before AngII addition (Figure 7B). The effect of PAO was completely reversed by simultaneous addition of 1 mM dithiothreitol (DTT) (Figure 7B). PAO also prevented the relocalization of the GFP-OSH2-PH2x during AngII stimulation (Figure 8B). In contrast, PIK93 (250 nM) had no effect on either the basal localization of the GFP-OSH2-PH2x probe (Figure 7C) nor did it affect the response to AngII stimulation (Figure 8A).
Figure 7.
Effects of PI4K inhibitors on the membrane localization of the OSH2-PH2x-GFP. HEK-293 cells stably transfected with the AT1a angiotensin receptors were cotransfected with the OSH2-PH2x-GFP constructs for 24 h. Live cells were examined in a confocal microscope at 35°C. Calculation of membrane/cytosolic fluorescence intensities as function of time was performed from line-intensity histograms from images taken at every minute during a 10-min incubation with the inhibitors. The concentrations of the inhibitors were 250 nM PIK93, 10 μM PAO with 1 mM Mer, 10 μM PAO with 1 mM DTT, and 10 μM or 300 nM Wm. Means ± SEM from three to 25 cells from two to four separate experiments are shown.
Figure 8.
Effects of AngII on the membrane localization of the OSH2-PH2x-GFP after treatment with PI4K inhibitors. HEK-293 cells stably transfected with the AT1a angiotensin receptors were cotransfected with the OSH2-PH2x-GFP constructs for 24 h. Live cells were examined in a confocal microscope at 35°C. Calculation of membrane/cytosolic fluorescence intensities as function of time was performed from line-intensity histograms from images taken at the indicated times after AngII addition. The concentrations of the inhibitors applied for 10 min before AngII were 250 nM PIK93, 10 μM PAO with 1 mM Mer, and 10 μM Wm. Means ± SEM from three to 25 cells from two to four separate experiments are shown. The initial ratio values of Wm-treated cells are higher in these experiments than those shown in Figure 7 at the end of Wm treatment, because cells with still measurable OSH2-PH2x-GFP localization after Wm treatment were selected for this analysis.
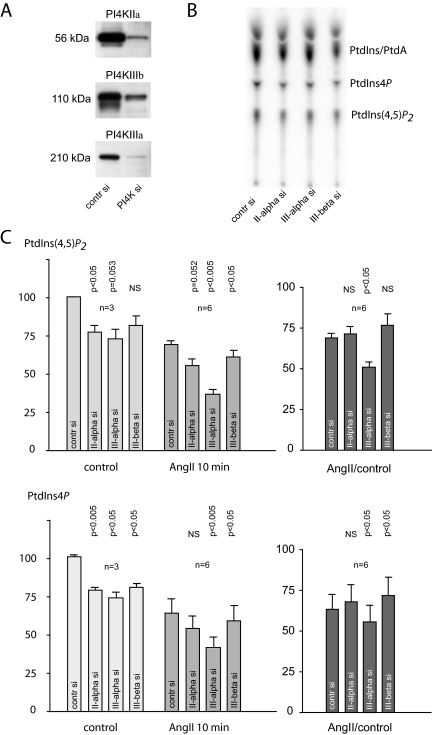
Effects of RNAi-mediated Knockdown on Phosphoinositides and Ca2+ Signaling
The results of the pharmacological analysis pointed to PI4KIIIα as the enzyme important in the maintenance of the agonist-sensitive phosphoinositide pools of the plasma membrane. We wanted to confirm these data with RNAi-mediated gene silencing. All PI4Ks were individually knocked down, and the basal and AngII-induced changes in 32P-labeled phosphoinositides and cytoplasmic Ca2+ were determined. These experiments showed that knockdown of either PI4Ks (confirmed in each experiments by Western blot analysis) (Figure 9A) resulted in no appreciable change in the Ca2+ response of the cells in response to AngII regardless of whether these measurements were performed in cells suspension or in single attached cells (data not shown). 32P-labeling of cells depleted in the respective PI4Ks showed variations due to the loss of cells in the PI4K down-regulated groups (Figure 9B). To normalize for this difference, in each experiments the ratio of PtdIns4P and PtdIns(4,5)P2 to the PtdIns/phosphatidic acid (PtdA) spots from unstimulated cells were calculated and compared between the control siRNA or PI4K siRNA-treated groups. After such corrections, the labeling of both PtdIns4P and PtdIns(4,5)P2 showed an ∼25% decrease upon depletion of either of PI4KIIα, PI4KIIIα, or PI4KIIIβ. This difference was statistically significant (p < 0.05) for all enzyme knockdowns in PtdIns4P but only for PI4KIIα in PtdIns(4,5)P2 (Figure 9C). To assess PtdIns4P and PtdIns(4,5)P2 synthesis during stimulation, the level of these lipids were measured after 10 min of AngII exposure, because by this time impaired resynthesis of these lipids was expected to be detectable (see Figure 2 for full kinetics). In these experiments, we also included treatment with 250 nM PIK93 before AngII treatment for each group to determine whether compensatory changes in PI4KIIIβ (although not apparent by Western analysis) could mask the effects of PI4KIIα or PI4KIIIα knockdown. Because there was absolutely no difference between the PIK93-AngII–treated group and those only treated with AngII alone (data not shown), these measurements have been combined for statistical analysis.
Figure 9.
Effects of knockdown of the individual PI 4-kinases on [32P]phosphate labeling of phosphoinositides and their response to AngII stimulation in HEK-293-AT1 cells. HEK-293 cells stably transfected with the AT1a receptors were treated with siRNA for 3 d as described under Materials and Methods. The extent of knockdown was determined by Western blot analysis (A). On the fourth day, cells were labeled with [32P]phosphate for 3 h in phosphate-free medium. Cells were then either treated with AngII (10−7 M) or saline and incubated for an additional 10 min. Reactions were terminated by PCA, and lipids were extracted from the cell pellets, separated by TLC, and analyzed by a PhosphorImager (B). Due to cell loss in the PI4K-depleted cells, the individual PtdIns4P and PtdIns(4,5)P2 spots were normalized to the PtdA/PtdIns values measured in unstimulated samples of the respective groups. These ratios were then compared between the PI4K down-regulated or control siRNA-treated cells in each of the three experiments performed in duplicates (C). In each experiments, the effect of 250 nM PIK93 was also determined in the AngII group. Because PIK93-treated cells showed no difference compared with those treated with AngII only, these values were pooled in the statistical analysis. Means ± SEM are shown, and the p values obtained in one-sample t tests compared with control siRNA-treated cells are shown above the bars.
As shown in Figure 9C, by 10 min of AngII treatment, PtdIns(4,5)P2 and PtdIns4P levels returned to ∼60–70% of their prestimulatory values (also see Figure 2) in control cells1. However, in cells depleted in PI4KIIIα, this value was significantly (p < 0.05) lower (∼50%) for both lipids, the difference being more pronounced in PtdIns(4,5)P2, which could be measured with better accuracy. These differences were relatively minor and required a fine balance between siRNA treatment that still preserved enough cells for analysis yet showed sufficient knockdown to capture even these small changes.
In cells labeled with myo-[3H]inositol, the picture was further complicated because we observed an increased labeling of PtdIns4P and PtdIns(4,5)P2 (and the inositol phosphates) in cells in which PI4KIIIα was depleted. This paradoxical change was attributed to a significant decrease in the free inositol level of PI4KIIIα knockdown cells and hence an increased specific activity of the labeled inositol pool (data not shown). The reason for the consistent decrease in the level of inositol in cells depleted in PI4KIIIα is not known, and it is currently under further investigation.
DISCUSSION
In the present study, we asked the question of which of the PI4K enzymes is important for the generation of the plasma membrane pool of PtdIns4P, the precursor of PtdIns(4,5)P2 during PLC activation in agonist-stimulated cells. This question has not been thoroughly addressed before for lack of appropriate tools to investigate it properly. In a previous study, we showed that Wm-sensitive type III PI4Ks are required for the maintenance of agonist-sensitive phosphoinositide pools and hence Ins(1,4,5)P3 and calcium signaling (Nakanishi et al., 1995). These studies have been confirmed in other laboratories using different cell lines and different Ca2+-mobilizing receptors and agonists (Willars et al., 1998), but the identity of the enzyme(s) responsible for supplying PtdIns4P for the plasma membrane so far has eluded identification. The recent discovery of a PI 3-kinase inhibitor that discriminates between the type III PI4Ks has proven to be an invaluable tool to reinvestigate this question. Based on studies using 32P-labeled phosphoinositides and myo-[3H]inositol-labeled inositol phosphates and cytoplasmic Ca2+ measurements, we were able to show that the effects of Wm are reproduced by inhibitors of PI4KIIIα but not PI4KIIIβ. It was also important to demonstrate that the PtdIns4P pool that is affected by the PI4KIIIα enzyme is in fact in the plasma membrane. This question was especially relevant, because none of the type III PI4Ks can be found in detectable amounts in the plasma membrane by immunofluorescence analysis of COS-7 or HEK-293 cells. Based on enzymatic characterization of the plasma membrane-associated PI4K activity performed two decades ago, it was concluded that the smaller and wortmannin-insensitive type II PI4Ks are present in the plasma membrane. This is supported by more recent immunocytochemical analysis that finds only the type II enzymes partially in the plasma membrane. Of the type III enzymes, PI4KIIIβ is found in the Golgi, and PI4KIIIα in the ER/Golgi compartment (see Balla and Balla, 2006 for citation of the original studies). So, it was of particular interest that the PH domain of the OSH2 protein, one of the yeast homologues of oxysterol binding proteins, is capable of reporting on changes in the plasma membrane that are consistent with changes of PtdIns4P levels (see also Yeung et al., 2006).
These studies using PH domains of PLCδ1 and OSH2-PH2x to monitor PtdIns(4,5)P2 and PtdIns4P, respectively, confirmed the conclusions of the labeling and Ca2+ experiments that PI4KIIIα is the enzyme necessary for the maintenance of the plasma membrane pool of these lipids. The exact mechanism how this is achieved by the enzyme that is primarily ER/Golgi localized remains to be answered, but its is quite possible that a small fraction of the enzyme is in fact found at the plasma membrane regulated by a unique mechanism, whereas the larger fraction of the protein has additional functions in the ER/Golgi compartments. It is interesting to note that in S. cerevisiae the Stt4p PI4K, a homologue of PI4KIIIα, was reported to generate the plasma membrane PtdIns4P pool, but unlike in mammalian cells, the yeast enzyme is primarily found in the plasma membrane or a tightly associated compartment (Audhya and Emr, 2002). Paradoxically, there are well-documented ER and vacuolar functions of Stt4p in yeast; yet, the protein has not been detected in those locations (Trotter et al., 1998; Audhya et al., 2000).
RNAi-mediated depletion of the individual PI4Ks has produced only small effects on 32P-labeled PtdIns4P and PtdIns(4,5)P2 but confirming the identity of the PI4KIIIα enzyme as one responsible for PtdIns4P and PtdIns(4,5)P2 production at the plasma membrane during AngII stimulation. Although the levels of the enzymes can be knocked down to 15–30% of their original values, this level of knockdown produced only small effects in the labeled lipids and none on AngII-induced Ca2+ signaling. This probably reflects the fact that small amounts of these enzymes are sufficient to support the production of this signaling pool of the lipids and cells with more severe knockdown could simply be eliminated or developed adaptive mechanisms to cope with the lack of the protein. This highlights the difficulties with knockdown studies of enzymes of crucial importance and the value of inhibitors that can be tested in short-term experiments.
In recent studies, we were able to show the role of PI4Ks in other aspects of cellular PtdIns4P production where the knockdown data gave as clear results as the pharmacological approach. These included the recruitment of the FAPP1PH domain to the Golgi being mediated by both the PI4KIIIβ and PI4KIIα enzymes (Balla et al., 2005), and the role of PI4KIIIβ in the ER-to-Golgi transport of ceramide (Toth et al., 2006). In fact, in the former study we found that PI4KIIIα knockdown was able to decrease the plasma membrane recruitment of the FAPP1PH domain during recovery from a large Ca2+-mediated PLC activation (Balla et al., 2005). These studies collectively suggest that PI4Ks probably have multiple functions within the cells that are affected at different level of enzyme depletion.
Last, the experiments using the FAPP1PH, OSBP-PH, and OSH2-PH domains highlight the values and the limitations of PH domain reporters. They clearly demonstrate that specific pools of PtdIns4P are detected by the individual probes and that one reporter may not detect PtdIns4P in all locations within the cells. The FAPP1 and OSBP PH domains detect primarily the Golgi pool, although under special circumstances such as during recovery from massive Ca2+ induced PLC activation they can also detect PtdIns4P in the plasma membrane (Balla et al., 2005). In contrast, the OSH2–2x-PH-GFP reporter detects PtdIns4P in the plasma membrane but not in the Golgi compartment in mammalian cells. Only the yeast OSH1-PH decorates both the Golgi and the plasma membrane. Importantly, the plasma membrane localization of all of the PtdIns4P reporters depends on the activity of Wm-sensitive, type III PI4Ks, and based on the current studies, specifically of the PI4KIIIα enzyme.
In summary, the present results establish PI4KIIIα as an important component of the generation of the plasma membrane pool of phosphoinositides. This conclusion is based on pharmacological studies and confirmed by RNAi-mediated gene silencing, although the latter could not decrease the enzyme levels sufficiently to appreciably impair Ca2+ signaling. These results also establish the OSH2-PH2x domain as a useful reporter to follow PtdIns4P changes in the plasma membrane. Further studies are in progress to identify the mechanism by which the apparently ER/Golgi localized PI4KIIIα can affect the supply of plasma membrane PtdIns4P.
ACKNOWLEDGMENTS
We are grateful to Dr. Roger Y. Tsien for the monomeric red fluorescent protein, Dr. Philip W. Majerus for the human type-IV 5-ptase clone, and Drs. Alberto-Jesus Olivares-Reyes and Kevin J. Catt for the HEK-293-AT1 cells. The confocal imaging was performed at the Microscopy and Imaging Core of the National Institute of Child Health and Human Development, National Institutes of Health (NIH), with the kind assistance of Drs. Vincent Schram and James T. Russell. This research was supported in part by the Intramural Research Program of the National Institute of Child Health and Human Development of the National Institutes of Health, and by an appointment of P.V. to the Senior Fellowship Program at the NIH. This latter program is administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the National Institutes of Health. A.B and P.V. are also Bolyai Fellows of the Hungarian Academy of Science and were supported by the Hungarian Scientific Research fund (OTKA NF-68563 and PF 63893) and the Medical Research Council (ETT 440/2006).
Abbreviations used:
- 5-ptase
type-IV phosphoinositide 5-phosphatase
- AngII
angiotensin II
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- FAPP
four phosphate adaptor protein
- FRET
fluorescence resonance energy transfer
- GFP
enhanced green fluorescent protein
- InsP3
inositol trisphosphate
- Mer
β-mercaptoethanol
- mRFP
monomeric red fluorescent protein
- OSBP
oxysterol binding protein
- PAO
phenylarsine oxide
- PI4K
phosphatidylinositol 4-kinase
- PtdIns4P
phosphatidylinositol 4-phosphate
- PtdIns(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PtdA
phosphatidic acid
- PLC
phospholipase C
- PH
pleckstrin homology
- siRNA
small interfering RNA
- Wm
wortmannin.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0713) on December 12, 2007.
Because AngII treatment increases the labeling of PtdIns/PtdA, and the knockdown of the individual enzymes could also affect this response, the correction for cell loss due to knockdown of the enzymes was based on the PtdIns/PtdA values obtained in the unstimulated cells also for the in AngII-stimulated samples. We assumed that AngII treatment does not cause cell loss.
REFERENCES
- Audhya A., Emr S. D. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Audhya A., Foti M., Emr S. D. Distinct roles for the yeast phosphatidylinositol 4-kinases, stt4p and pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A., Balla T. Phosphatidylinositol 4-kinases; old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Balla A., Tuymetova G., Barshishat M., Geiszt M., Balla T. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J. Biol. Chem. 2002;277:20041–22050. doi: 10.1074/jbc.M111807200. [DOI] [PubMed] [Google Scholar]
- Balla A., Tuymetova G., Tsiomenko A., Varnai P., Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T., Baukal A. J., Guillemette G., Catt K. J. Multiple pathways of inositol polyphosphate metabolism in angiotensin-stimulated adrenal glomerulosa cells. J. Biol. Chem. 1988;263:4083–4091. [PubMed] [Google Scholar]
- Balla T., Downing G. J., Jaffe H., Kim S., Zolyomi A., Catt K. J. Isolation and molecular cloning of wortmannin-sensitive bovine type-III phosphatidylinositol 4-kinases. J. Biol. Chem. 1997;272:18358–18366. doi: 10.1074/jbc.272.29.18358. [DOI] [PubMed] [Google Scholar]
- Barylko B., Wlodarski P., Binns D. D., Gerber S. H., Earnest S., Sudhof T. C., Grichine N., Albanesi J. P. Analysis of the catalytic domain of phosphatidylinositol 4-kinase type II. J. Biol. Chem. 2002;277:44366–44375. doi: 10.1074/jbc.M203241200. [DOI] [PubMed] [Google Scholar]
- Berridge M. J., Irvine R. F. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- Creba J. A., Downes C. P., Hawkins P. T., Brewster G., Michell R. H., Kirk C. J. Rapid breakdown of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate in rat hepatocytes stimulated by vasopressin and other Ca2+-mobilizing hormones. Biochem. J. 1983;212:733–747. doi: 10.1042/bj2120733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughman R. L., Firestone A. J., Anderson R. A. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J. Membr. Biol. 2003;194:77–89. doi: 10.1007/s00232-003-2027-7. [DOI] [PubMed] [Google Scholar]
- Godi A., Di Campi A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Kisseleva M. V., Wilson M. P., Majerus P. W. The isolation and characterization of a cDNA encoding phospholipid-specific inositol polyphosphate 5-phosphatase. J. Biol. Chem. 2000;275:20110–20116. doi: 10.1074/jbc.M910119199. [DOI] [PubMed] [Google Scholar]
- Knight Z. A., et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T. P., Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Majerus P. W., Kisseleva M. V., Norris F. A. The role of phosphatases in inositol signaling reactions. J. Biol. Chem. 1999;274:10669–10672. doi: 10.1074/jbc.274.16.10669. [DOI] [PubMed] [Google Scholar]
- Minogue S., Waugh M. G., De Matteis M. A., Stephens D. J., Berditchevski F., Hsuan J. J. Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J. Cell Sci. 2006;119:571–581. doi: 10.1242/jcs.02752. [DOI] [PubMed] [Google Scholar]
- Nakanishi S., Catt K. J., Balla T. A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositolphospholipids. Proc. Natl. Acad. Sci. USA. 1995;92:5317–5321. doi: 10.1073/pnas.92.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Levine T. P. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- Toth B., Balla A., Ma H., Knight Z. A., Shokat K. M., Balla T. Phosphatidylinositol 4-kinase IIIbeta regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J. Biol. Chem. 2006;281:36369–36377. doi: 10.1074/jbc.M604935200. [DOI] [PubMed] [Google Scholar]
- Trotter P. J., Wu W. I., Pedretti J., Yates R., Voelker D. R. A genetic screen for aminophospholipid transport mutants identifies the phosphatidylinositol 4-kinase, Stt4p, as an essential component in phosphatidylserine metabolism. J. Biol. Chem. 1998;273:13189–13196. doi: 10.1074/jbc.273.21.13189. [DOI] [PubMed] [Google Scholar]
- van Der Wal J., Habets R., Varnai P., Balla T., Jalink K. Monitoring phospholipase C activation kinetics in live cells by FRET. J. Biol. Chem. 2001;276:15337–15344. doi: 10.1074/jbc.M007194200. [DOI] [PubMed] [Google Scholar]
- Varnai P., Balla A., Hunyady L., Balla T. Targeted expression of the inositol 1,4,5-triphosphate receptor (IP3R) ligand-binding domain releases Ca2+ via endogenous IP3R channels. Proc. Natl. Acad. Sci. USA. 2005;102:7859–7864. doi: 10.1073/pnas.0407535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai P., Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium-and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P., Lin X., Lee S. B., Tuymetova G., Bondeva T., Spat A., Rhee S. G., Hajnoczky G., Balla T. Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J. Biol. Chem. 2002;277:27412–27422. doi: 10.1074/jbc.M109672200. [DOI] [PubMed] [Google Scholar]
- Varnai P., Thyagarajan B., Rohacs T., Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J. Cell Biol. 2006;175:377–382. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sun H. Q., Macia E., Kirchhausen T., Watson H., Bonifacino J. S., Yin H. L. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol. Biol. Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. J., Li W. H., Wang J., Xu K., Dong P., Luo X., Yin H. L. Critical role of PIP5KIgamma 87 in InsP3-mediated Ca(2+) signaling. J. Cell Biol. 2004;167:1005–1010. doi: 10.1083/jcb.200408008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirschhausen T., Albanesi J. P., Roth M. G., Yin H. L. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- Warashina A. Light-evoked recovery from wortmannin-induced inhibition of catecholamine secretion and synaptic transmission. Arch. Biochem. Biophys. 1999;367:303–310. doi: 10.1006/abbi.1999.1273. [DOI] [PubMed] [Google Scholar]
- Wiedemann C., Schäfer T., Burger M. M. Chromaffin granule-associated phosphatidylinositol 4-kinase activity is required for stimulated secretion. EMBO J. 1996;15:2094–2101. [PMC free article] [PubMed] [Google Scholar]
- Willars G. B., Nahorski S. R., Challiss R. A. Differential regulation of muscarinic acetylcholine receptor-sensitive polyphosphoinositide pools and consequences for signaling in human neuroblastoma cells. J. Biol. Chem. 1998;273:5037–5046. doi: 10.1074/jbc.273.9.5037. [DOI] [PubMed] [Google Scholar]
- Yeung T., Terebiznik M., Yu L., Silvius J., Abidi W. M., Philip M., Levine T., Kapus A., Grinstein S. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., Lemmon M. A. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]