Abstract
Autophagy is the major degradative process for recycling cytoplasmic constituents and eliminating unnecessary organelles in eukaryotic cells. Most autophagy-related (Atg) proteins are recruited to the phagophore assembly site (PAS), a proposed site for vesicle formation during either nonspecific or specific types of autophagy. Therefore, appropriate recruitment of Atg proteins to this site is critical for their function in autophagy. Atg11 facilitates PAS recruitment for the cytoplasm-to-vacuole targeting pathway, which is a specific, autophagy-like process that occurs under vegetative conditions. In contrast, it is not known how Atg proteins are recruited to the PAS, nor which components are involved in PAS formation under nonspecific autophagy-inducing, starvation conditions. Here, we studied PAS assembly during nonspecific autophagy, using an atg11Δ mutant background to eliminate the PAS formation that occurs during vegetative growth. We found that protein complexes containing the Atg1 kinase have two roles for PAS formation during nonspecific autophagy. The Atg1 C terminus mediates an interaction with Atg13 and Atg17, facilitating a structural role of Atg1 that is needed to efficiently organize an initial step of PAS assembly, whereas Atg1 kinase activity affects the dynamics of protein movement at the PAS involved in Atg protein cycling.
INTRODUCTION
Macroautophagy (hereafter autophagy) is a major degradative pathway that allows cells to respond to various types of environmental stress (Klionsky, 2005; Yorimitsu and Klionsky, 2007). Autophagy is an evolutionarily conserved pathway in all eukaryotic cells (Reggiori and Klionsky, 2002). After certain environmental cues such as nutrient deprivation or hormonal stimuli, cells dynamically sequester portions of the cytoplasm within double-membrane cytosolic vesicles, called autophagosomes (Klionsky and Ohsumi, 1999; Klionsky and Emr, 2000). The completed vesicles subsequently fuse with lysosomes/vacuoles, allowing the degradation of the sequestered components and the recycling of the resulting macromolecules. Autophagy has critical roles in cellular remodeling through degradation, thus acting at various stages of development (Klionsky, 2004; Levine and Klionsky, 2004), and it is involved in protection against a range of diseases such as pathogen infection, tumor formation and certain types of neurodegeneration (Shintani and Klionsky, 2004a; Rubinsztein et al., 2005).
Autophagy is usually considered a nonselective process for the bulk degradation of cytoplasmic components; however, there are many types of specific autophagy. For example, peroxisomes in methylotrophic yeasts are specifically degraded when cells are shifted from conditions where these organelles are required for metabolism to ones where they are no longer necessary (Dunn et al., 2005). In addition to its role in adapting to changing metabolic demands, selective autophagy is involved in removing damaged organelles such as mitochondria (Kissova et al., 2007; Zhang et al., 2007). Specific types of autophagy also occur in higher eukaryotes as seen, for example, with the targeting of pathogenic microbes (Birmingham and Brumell, 2006; Colombo et al., 2006; Talloczy et al., 2006; Vergne et al., 2006; Webster, 2006; Yoshimori, 2006). A particularly unique case of selective autophagy is the cytoplasm-to-vacuole targeting (Cvt) pathway, which is a biosynthetic autophagy-like pathway that occurs in growing conditions in yeast (Yorimitsu and Klionsky, 2005b; Farre et al., 2007). The precursor form of the vacuolar hydrolase aminopeptidase I (Ape1), and α-mannosidase, are recognized and sequestered within double-membrane vesicles and transported into the vacuole through this alternative targeting route that overlaps extensively with bulk autophagy (Klionsky et al., 1992; Hutchins and Klionsky, 2001). There are clear morphological and mechanistic differences between the Cvt pathway and autophagy, even though both share most of the autophagy-related (Atg) proteins, the machinery that drives these processes. One of the key proteins required for both pathways, the serine-threonine kinase Atg1, plays a critical role in autophagy induction and is regulated by Tor-dependent signaling (Kamada et al., 2000). Recently, it was reported that the Atg1-interacting partners Atg13 and Atg17 participate in regulating the conversion between the Cvt pathway and autophagy through modulating the autophagic response (Kamada et al., 2000; Cheong et al., 2005; Kabeya et al., 2005).
The phagophore assembly site (also called the preautophagosomal structure; PAS) is thought to be an organizing center for formation of the sequestering vesicles, autophagosomes and Cvt vesicles, which form during bulk and specific autophagy, respectively. Although the PAS is presumed to play a prominent role in these processes, the nature of this structure is poorly understood. For example, through elegant circular reasoning the site where most Atg proteins reside is defined as the PAS, and the PAS is defined in part as the site where most Atg proteins reside. The crux of this logic is that the PAS is primarily detected through fluorescence microscopy as a perivacuolar punctum in yeast cells expressing fluorophore-tagged Atg proteins. Even though this site is poorly defined, the correct PAS targeting of Atg proteins seems to be required for their function (Suzuki et al., 2001; Kim et al., 2002; Nice et al., 2002). Although the role of the PAS is not fully understood, one model is that this site serves to facilitate the nucleation and/or expansion of the phagophore, the precursor to the autophagosome, through the recruitment of Atg proteins (Mizushima et al., 2001; Suzuki and Ohsumi, 2007). Accordingly, to understand the initial steps of sequestering vesicle formation, it is important to understand how Atg proteins are recruited to the PAS, which proteins interact at this site, and how they function.
Recently, we reported that Cvt cargo complexes, composed of the cargo protein precursor Ape1 (prApe1), the receptor Atg19 and the adaptor Atg11, have a critical role in PAS organization under vegetative conditions (Shintani and Klionsky, 2004b). In contrast, PAS assembly in the absence of these proteins occurs normally during starvation, which suggests that Cvt cargo complexes are most likely involved in the formation of a Cvt-specific PAS, but they are not needed for PAS formation during nonspecific autophagy. These results raise questions about targeting of Atg proteins to the PAS during specific and nonspecific types of autophagy, and in particular about the mechanism of Atg protein recruitment under autophagy-inducing conditions. In this study, we examined the role of proteins that comprise the Atg1 kinase complex in PAS assembly. Our results suggest that Atg1, Atg13, and Atg17 have a critical role in PAS organization during nonspecific autophagy and that the interaction among these proteins is essential for PAS recruitment of Atg proteins.
MATERIALS AND METHODS
Media
Yeast were grown in YPD medium (1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal medium (SMD; 0.67% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as required). Starvation medium was SD-N (0.17% yeast nitrogen base without ammonium sulfate or amino acids, containing 2% glucose).
Strains and Plasmids
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Tagging of proteins by the integration of modified genes at the corresponding chromosomal loci, and gene deletions were performed by a polymerase chain reaction (PCR)-based procedure (Longtine et al., 1998). For gene disruption, the entire coding region was replaced with the Escherichia coli kanr, Schizosaccharomyces pombe HIS5, Kluyveromyces lactis URA3, Saccharomyces kluyveri HIS3, or S. cerevisiae TRP1, LEU2, or URA3 gene by using PCR primers containing ∼40 bases of identity to the regions flanking the open reading frame. Western blotting, PCR, or both verified putative gene knockout and tagged strains. The functionality of tagged constructs was confirmed by examining the processing of prApe1, green fluorescent protein (GFP)-Atg8, or both.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| AHY001 | SEY6210 atg11Δ:: HIS5 S.p. | Kim et al., 2001b |
| BY4742 | MATα his3Δ leu2Δ lys2Δ ura3Δ | ResGen/Invitrogen |
| CWY233 | SEY6210 atg13Δ::KAN | This study |
| CWY235 | AHY001 atg13Δ::KAN | Yorimitsu and Klionsky, 2005a |
| CWY239 | SEY6210 atg17Δ::KAN | Cheong et al., 2005 |
| FRY143 | SEY6210 vps4Δ::TRP1 pep4Δ::LEU2 | Cheong et al., 2005 |
| HCY31 | FRY143 atg17Δ::Kan | This study |
| HCY37 | FRY143 atg11Δ:: HIS3 S.k. | This study |
| HCY38 | HCY31 atg11Δ:: HIS3 S.k. | This study |
| HCY66 | CWY239 atg11Δ:: LEU2 | This study |
| HCY76 | SEY6210 vps4Δ::TRP1 pep4Δ::Kan | |
| atg1Δ::URA3 | This study | |
| HCY114 | SEY6210 GFP-ATG8::URA3 | This study |
| HCY116 | TYY158 GFP-ATG8::URA3 | This study |
| HCY118 | SEY6210 ATG17-3GFP::URA3 | This study |
| HCY119 | TYY158 ATG17-3GFP::URA3 | This study |
| HCY131 | AHY001 ATG16-GFP::TRP1 | This study |
| HCY132 | CWY233 ATG16-GFP::TRP1 | This study |
| HCY133 | HCY66 ATG16-GFP::TRP1 | This study |
| HCY134 | CWY235 ATG16-GFP::TRP1 | This study |
| HCY135 | TYY158 ATG16-GFP::TRP1 | This study |
| HCY136 | SEY6210 ATG14-GFP::HIS5 | This study |
| HCY137 | WHY001 ATG14-GFP::TRP1 | This study |
| HCY138 | CWY233 ATG14-GFP:: HIS5 | This study |
| HCY139 | CWY239 ATG14-GFP::TRP1 | This study |
| HCY140 | AHY001 ATG14-GFP::TRP1 | This study |
| HCY141 | HCY66 ATG14-GFP::TRP1 | This study |
| HCY142 | CWY235 ATG14-GFP::TRP1 | This study |
| HCY143 | TYY158 ATG14-GFP::TRP1 | This study |
| KTY148 | SEY6210 ATG14-GFP::TRP1 | This study |
| KTY149 | atg1Δ:: URA3 ATG16-GFP::TRP1 | This study |
| KTY161 | atg17Δ::LEU2 ATG16-GFP::TRP1 | This study |
| PJ69-4A | MATa trp1-Δ901 leu2-3,112 ura3-52 his3-Δ200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al., 1996 |
| SEY6210 | MATα leu2-3,112 ura3-52 his3-200 trp1-901 lys2-801 suc2-9 GAL | Robinson et al., 1988 |
| TYY158 | AHY001 atg1Δ::KAN | This study |
| UNY1 | YTS159 atg1Δ::HIS5 S.p. | This study |
| UNY3 | TN121 atg1Δ | This study |
| UNY27 | CWY233 atg1Δ::HIS5 S.p. | This study |
| WHY001 | SEY6210 atg1Δ::HIS5 S.p. | Shintani et al., 2002 |
| YTS157 | SEY6210 atg1Δ::HIS5 S.p. atg11Δ::LEU2 | Yorimitsu and Klionsky, 2005a |
| YTS159 | BY4742 pho8::pho8Δ60 pho13::Kan | This study |
The pRS316 GFP-APG1 plasmid was a kind gift from Dr. Yoshinori Ohsumi (National Institute for Basic Biology, Okazaki, Japan). A plasmid expressing GFP-Atg8 [pGFP-Aut7(416)] was described previously (Huang et al., 2000). To integrate GFP-Atg8, we used a pRS306-based plasmid containing the GFP-ATG8 gene with the endogenous ATG8 promoter (Suzuki et al., 2001); integration was done at the URA3 locus. For constructing an ATG17-3xGFP plasmid containing three tandem copies of the gene encoding GFP, the full-length ATG17 gene was PCR amplified and ligated into the ClaI–AgeI sites of pRS306-3xGFP, which was described previously (Rossanese et al., 2001). The resulting plasmid was used for yeast transformation to integrate ATG17-3xGFP at the ATG17 chromosomal locus. The atg1K54A and atg1M102A mutants were described previously (Abeliovich et al., 2003; Kamada et al., 2000). The plasmid containing the double point mutation, atg1K54A,M102A was generated by PCR-based site-directed mutagenesis. DNA sequencing was used to verify the mutation.
For yeast two-hybrid screening, we used an N-terminally truncated version of Atg1, lacking a kinase domain (Atg1ΔN), that was introduced into the yeast two-hybrid bait vector pGBDU-C1 by using BamHI and SalI sites, as described previously (Yorimitsu and Klionsky, 2005a). The C-terminal deletion mutant (ΔC20) of this version of Atg1 was generated by PCR and cloned into pGBDU-C1 by using the same restriction enzyme sites. The pGBDU-C1 plasmids expressing point mutants that substituted alanine for tyrosine and arginine at positions 878 and 885, respectively (Atg1Y878A,R885A) and glutamate for arginine and lysine at positions 885 and 892, respectively (Atg1R885E,K892E) were generated by PCR-based site-directed mutagenesis using the pGBD-Atg1ΔN construct. Full-length Atg13 (pAD-Atg13), described previously (Scott et al., 2000), was used as prey. Each set of bait and prey plasmids were transformed into strain PJ69-4A, and interactions were assayed by streaking transformants on SD plates lacking adenine, and examining growth.
To generate mutant versions of native Atg1, we started with the wild-type ATG1 gene containing its endogenous promoter and terminator that was cloned into pRS315 [pATG1(315); Abeliovich et al., 2003], in which an NheI site was introduced after the stop codon. The Atg1ΔC20, Atg1Y878A,R885A and Atg1R885E,K892E mutations were each cloned from the pGBDU-C1 vector containing these respective mutants, into pATG1(315) as SphI–NheI fragments. The mutations were verified by DNA sequencing. These constructs were used for the Pho8Δ60 alkaline phosphatase assay and the fluorescence and electron microscopy analyses.
For protein A (PA) affinity isolation, the Atg1 open reading frame with a BamHI site engineered just after the ATG start codon, was amplified by PCR and cloned as a SpeI–SalI fragment into the pCu416 vector (Labbe and Thiele, 1999). A PA tag on a BamHI fragment from plasmid pTS480 was used to create an N-terminal PA-tagged version of this construct, and the functionality was determined by monitoring prApe1 maturation in an atg1Δ strain expressing this protein. DNA fragments containing the Atg1ΔC20 deletion or point mutations were excised as SphI–SalI fragments from the pGBDU-C1 vector, and they were used to replace the wild-type Atg1 sequence in the PA-tagged construct after digestion with the same set of enzymes.
Full-length ATG13, containing its endogenous promoter and terminator sequences, was cloned into pRS315 (Kamada et al., 2000). An AvrII site was introduced by site-directed mutagenesis, immediately after the start codon of ATG13. DNA containing three tandem copies of the hemagglutinin (HA) coding sequence was amplified from 3HA-Apg12 (a kind gift from Dr. Noboru Mizushima, Tokyo Medical and Dental University) by primers containing an XbaI site and ligated into the AvrII site to create an N-terminal fusion of 3xHA with Atg13. Western blotting and PCR were used for verifying this construct, and the functionality was assayed by examining prApe1 maturation. HA-tagged Atg11 has been described previously (Yorimitsu and Klionsky, 2005a).
Protein A Affinity Isolation
Cells were grown in 50 ml of SMD lacking the appropriate auxotrophic amino acids to OD600 = 0.6. Rapamycin (0.2 μg/ml; Fermentek, Jerusalem, Israel) was added, and cells were cultured for an additional 2 h. PA-affinity isolation was performed as described previously (He et al., 2006), with only one exception: the cell extracts were incubated with immunoglobulin (Ig)G-Sepharose beads for 2 h at 4°C. The eluates were resolved by 6% SDS-polyacrylamide gel electrophoresis (PAGE), transferred to Immobilon polyvinylidene difluoride (IPVH00010; Millipore, Billerica, MA) and immunoblotted with monoclonal anti-HA antibody (Santa Cruz Biotechnology, CA).
Enzyme Assays
The alkaline phosphatase assay using Pho8Δ60, and an in vitro phosphorylation assay using HA-tagged Atg1 were performed as described previously (Kamada et al., 1995; Noda et al., 1995). β-Galactosidase assays with AD-Atg13 and BD-Atg1 variants in strain PJ69-4A were performed as described previously (Rose et al., 1990).
Microscopy
For fluorescence microscopy, cells were grown to mid-log phase in YPD or SMD medium lacking the appropriate auxotrophic amino acids to maintain plasmids. For starvation, cells were shifted to SD-N medium for 2 h. The cells were viewed using a DeltaVision Spectris (Applied Precision, Issaquah, WA) microscope fitted with differential interference contrast (DIC) optics and an Olympus camera IX-HLSH100 with softWoRx software. For image quantification of fluorescence intensity, a stack of 12 z-sections at intervals of 0.5 μm was collected, which completely covered the whole cell along the z-axis. The stack of 12 z-sections was then projected into a two-dimensional image by softWoRx software. A circle was drawn around the PAS dot and the intensity within this area was measured. An area of the same size encompassing the adjacent cytosol was subtracted as background. For the time-lapse experiment, cells were immobilized on SD-N medium containing 2% agar on concavity slides (Fisher Scientific) and pictures were taken every minute.
Electron microscopy was performed as described previously (Kaiser and Schekman, 1990). To quantify size, the diameter of autophagic bodies with clear membrane boundaries was measured.
RESULTS
Atg17 Is Important for PAS Organization under Starvation Conditions
Atg17 and Atg11 were identified as interacting proteins of Atg1, which are required for either nonspecific autophagy or the specific Cvt pathway, respectively (Kamada et al., 2000; Kim et al., 2001b). Atg11 plays a role in cargo packaging and transport in the Cvt pathway, but it is not required for nonspecific autophagy (Shintani et al., 2002; Yorimitsu and Klionsky, 2005a). Conversely, Atg17 is not needed for the Cvt pathway, but it modulates the magnitude of the autophagic response; the atg17Δ mutant has fewer, and smaller, autophagosomes (Cheong et al., 2005; Kabeya et al., 2005). Except for this role, however, the physiological function of Atg17 in autophagy is not understood.
Recently, we reported that the Cvt-specific proteins Atg11 and Atg19 (the prApe1 receptor), along with the prApe1 cargo protein, recruit Atg proteins to the PAS to facilitate Cvt vesicle formation (Shintani and Klionsky, 2004). In contrast, these proteins are not needed for PAS formation during starvation conditions, which suggests that other proteins are involved in PAS formation during nonspecific autophagy. Accordingly, we hypothesized that one of the autophagy-specific Atg proteins has a similar role to Atg11 in PAS formation under starvation conditions. Atg17 is one promising candidate because of the unique autophagy-specific phenotype of the null mutant. In addition, some Atg proteins do not display a normal PAS localization in the absence of Atg17 under starvation conditions; for example, GFP-Atg13 does not localize at the PAS in the atg17Δ strain (our unpublished data; Suzuki et al., 2007). Accordingly, we decided to examine the role of Atg17 in PAS formation during nonspecific autophagy.
We first used GFP-tagged Atg1 and Atg8 as independent PAS markers and examined PAS formation under rich and starvation conditions. Cells expressing GFP-Atg8 or GFP-Atg1 were grown to mid-log phase and imaged by fluorescence microscopy. In wild-type cells, both Atg proteins usually displayed a prominent punctate signal and also a more faint cytosolic diffuse pattern (Figure 1). The punctate structure was located in a perivacuolar region and represents the phagophore assembly site (Suzuki et al., 2001; Kim et al., 2002). Approximately 30–40% of the cells contained a GFP-Atg8 or GFP-Atg1 punctate signal in vegetative (SMD medium) conditions. Under starvation conditions (SD-N medium), we could usually detect more than one punctum for GFP-Atg1, and many of the GFP-Atg8 cells displayed some level of vacuolar fluorescence, reflecting autophagic delivery of this chimera that remains associated with the completed autophagosome (Kim et al., 2001a). More than 50% of the cells still had clear punctate signals corresponding to the PAS in starvation conditions (Figure 1).
Figure 1.
Atg17 is important for PAS organization under starvation conditions. Wild-type (SEY6210), atg11Δ (AHY001), atg17Δ (CWY239), and atg11Δ atg17Δ (HCY66) strains expressing GFP-Atg8 or GFP-Atg1 from centromeric plasmids were grown in SMD lacking auxotrophic amino acids and shifted to SD-N for 2 h before microscopy. The cells were examined by fluorescence microscopy as described in Materials and Methods.
The atg17Δ mutant showed comparable fluorescence patterns to the wild type, even under starvation conditions (Figure 1); this is particularly evident for GFP-Atg8. In contrast, the atg11Δ mutant showed primarily a diffuse fluorescent signal of GFP-Atg8 and GFP-Atg1 in the cytosol rather than a punctate signal at the PAS under vegetative conditions, in agreement with previous data (Shintani and Klionsky, 2004b); however, this mutant showed a normal punctate signal for both marker proteins during starvation (Figure 1). This result suggested that Atg11 plays a role in recruiting Atg8 and Atg1 to the PAS only for the Cvt pathway; however, the Atg11-dependent PAS that forms during vegetative growth may persist in the cell after the induction of autophagy, complicating the analysis of the atg17Δ mutant. Thus, to examine the role of Atg17 in PAS formation during nonspecific autophagy, we used an atg11Δ mutant background. We performed a similar fluorescence microscopy analysis by observing GFP-Atg8 or GFP-Atg1 in an atg11Δ atg17Δ double mutant. The double mutant showed a diffuse fluorescent signal of GFP-Atg8 and GFP-Atg1 in the cytosol under both growing and starvation conditions (Figure 1), suggesting that Atg17 has an important role in PAS formation during nonspecific autophagy.
Next, to test whether the defect in PAS recruitment of Atg1 and Atg8, and potentially other Atg proteins, in the atg11Δ atg17Δ strain affects autophagosome formation, we carried out a morphological examination by electron microscopy. One of the unique features of Cvt vesicles and autophagosomes is the presence of double membranes; after fusion of the autophagosome outer membrane with the vacuole, single-membrane autophagic bodies are released into the vacuole lumen (Takeshige et al., 1992). The autophagic bodies are subsequently degraded in a Pep4-dependent manner; pep4Δ mutants accumulate autophagic bodies, which can be observed by electron microscopy. Additionally, we deleted the VPS4 gene to eliminate the vesicles derived from the multivesicular body pathway (Reggiori et al., 2004). Cells in the pep4Δ vps4Δ background were grown in YPD, shifted to SD-N for 4 h to induce autophagy and observed by electron microscopy, as described in Materials and Methods.
The wild-type (pep4Δ vps4Δ) vacuole was filled with a large number of autophagic bodies that accumulated after starvation (Figure 2). In contrast, the atg17Δ strain accumulated a reduced number and size of autophagic bodies in agreement with our previous results (Cheong et al., 2005). We then focused on two strains, atg11Δ and atg11Δ atg17Δ, which showed vegetative-specific or complete (i.e., including starvation conditions) defects in PAS formation (Figure 1). Under starvation conditions, the atg11Δ vacuole accumulated a comparable number and size of autophagic bodies as seen in the wild type. In contrast, the atg11Δ atg17Δ strain essentially did not contain any detectable autophagic bodies, having at most <5% the number seen in the atg11Δ mutant (Figure 2). This result indicates that PAS formation in autophagy conditions is closely linked to normal autophagosome formation. Furthermore, Atg17 plays an important role in the PAS localization of Atg proteins, which presumably facilitates assembly of the autophagosome.
Figure 2.
The atg11Δ atg17Δ double mutant is defective for autophagosome formation. (A) The wild-type (FRY143; pep4Δ vps4Δ), atg17Δ (HCY31; pep4Δ vps4Δ), atg11Δ (HCY37; pep4Δ vps4Δ), and atg11Δ atg17Δ (HCY38; pep4Δ vps4Δ) strains were grown to mid-log stage in YPD and transferred to SD-N medium for 4 h. Cells were fixed with permanganate and examined by electron microscopy as described in Materials and Methods. (B) Quantification of autophagic body accumulation. 50 sections for each strain were scored for autophagic body accumulation.
Atg1 and Atg13 Are Also Required for PAS Recruitment of Atg Proteins under Starvation Conditions
To confirm that the defect in PAS recruitment in the atg11Δ atg17Δ background was due to loss of Atg17 function, we transformed this strain with plasmids expressing Atg17 or Atg17C24R; the mutation of cysteine to arginine at position 24 of Atg17 causes a loss of interaction with Atg1 and Atg13 (Kabeya et al., 2005). The presence of wild-type Atg17 restored the ability of the atg11Δ atg17Δ cells to assemble a PAS in starvation conditions, based on the recruitment of GFP-tagged Atg1 or Atg8 (our unpublished data). In contrast, Atg17C24R did not complement the defect in PAS assembly (our unpublished data). Atg17 is part of a complex that includes the Atg1 kinase and other interacting proteins, including Atg13 (Kamada et al., 2000; Nice et al., 2002). Accordingly, we investigated whether Atg1 and/or Atg13 have a role in PAS assembly during nonspecific autophagy similar to Atg17. To address this question, we first carried out the fluorescence microscopy analysis using PAS markers in atg11Δ atg13Δ or atg1Δ atg11Δ mutants.
Similar to the results with the atg11Δ atg17Δ strain, the PAS localization of Atg8 or Atg1 was defective in the atg11Δ atg13Δ double mutant under starvation conditions, whereas PAS localization of these proteins was essentially normal in the atg13Δ single-deletion strain, presumably due to the presence of the Atg11-dependent PAS formed during vegetative growth (Figures 3 and 4, A and B). Similarly, the atg1Δ atg11Δ mutant showed a diffuse fluorescent signal of GFP-Atg8 in the cytosol (Figures 3A and 4A); we could not use the GFP-Atg1 marker in this strain, because the hybrid protein expressed from the plasmid complemented the chromosomal deletion of ATG1 (our unpublished data). Therefore, we examined Atg17-GFP as another PAS marker in the atg1Δ atg11Δ double mutant strain. In both vegetative and starvation conditions, the Atg17-GFP signal was diffuse in the cytosol (Figure 3C). Overall, these results suggest that Atg1 and its modulators, Atg13 and Atg17, have essential roles in PAS formation during nonspecific autophagy.
Figure 3.
Atg13 and Atg1 are required for PAS recruitment of Atg proteins under starvation conditions. Localization of GFP-Atg8 (A) or GFP-Atg1 (B) in atg11Δ (AHY001), atg13Δ (CWY233), atg1Δ atg11Δ (TYY158), and atg11Δ atg13Δ (CWY235) strains, and GFP-Atg17 (C) in the atg1Δ atg11Δ (TYY158) strain was examined by fluorescence microscopy as described in Figure 1.
Figure 4.
Atg1 and its modulators Atg13 and Atg17 are required for the PAS recruitment of various Atg proteins under starvation conditions. The localization of Atg proteins was examined by fluorescence microscopy as described in Figure 1. The number of cells that contained PAS puncta was quantified for GFP-Atg8 (A), GFP-Atg1 (B), Atg14-GFP (C), and Atg16-GFP (D), after shifting to SD-N for 2 h. Approximately 150–250 cells for each strain were analyzed for scoring the percentage of cells with fluorescent PAS puncta. ND, not determined.
Even though Atg8 is frequently used as a representative marker for the PAS, most of the Atg proteins are also recruited to this site. Therefore, we extended our analysis by examining additional proteins that are parts of functional groups acting at different stages of autophagy. GFP-Atg8 is a ubiquitin-like protein and its localization represents the functionality of Atg3, Atg4 and Atg7, which are all required for the proper posttranslational processing of Atg8. Similarly, Atg1 localization in part reflects the proteins that are part of the Atg1 kinase complex, including Atg13 and Atg17. We next examined the localization of Atg14 and Atg16 under starvation conditions. Atg14 is part of the phosphatidylinositol 3-kinase complex I, which functions along with Atg6/Vps30, Vps15 and Vps34 in autophagy (Kihara et al., 2001). Atg16 is involved in the Atg12—Atg5 conjugation system, which is required for autophagosome formation (Kuma et al., 2002). Both Atg14-GFP and Atg16-GFP displayed reduced punctate signals compared with Atg8-GFP; under vegetative conditions, only ∼20–30% of the cells showed weak PAS puncta (our unpublished data). In both cases, the signals were stronger under starvation conditions and localization at the PAS was evident in the single deletion strains (Figure 4, C and D). When we examined localization in the atg11Δ atg17Δ, atg11Δ atg13Δ, or atg1Δ atg11Δ double mutants under starvation conditions, we observed a clear reduction in PAS puncta and a concomitant increase in the diffuse cytosolic staining (Figure 4, C and D) similar to the results with Atg8-GFP and GFP-Atg1 (Figure 4, A and B). Together, these results further suggest that Atg1 and its modulators, Atg13 and Atg17, are required in PAS organization during nonspecific autophagy.
Atg1 Kinase Activity Is Not Necessary for PAS Recruitment of Atg Proteins but Is Needed for Their Normal Localization
Atg13 and Atg17 modulate Atg1 kinase activity (Kamada et al., 2000), but the role of Atg1 kinase activity is not clear in either autophagy or the Cvt pathway (Nair and Klionsky, 2005). Conflicting reports suggest that higher kinase activity plays a more important role in autophagy (Kamada et al., 2000) or the Cvt pathway (Abeliovich et al., 2003; Nair and Klionsky, 2005). To examine the potential role of Atg1 kinase activity in PAS formation, we decided to use a mutant form of Atg1 with reduced kinase activity, which combined two previously characterized mutant alleles. The Atg1K54A mutant has an alteration in the kinase active site, but still displays a low level of function based on the Pho8Δ60 assay (Kamada et al., 2000; Figure 5A). When combined with a mutation of methionine to alanine at position 102, the Atg1K54A,M102A double mutant displays a level of autophagic activity similar to an atg1Δ strain (Figure 5A). To confirm that the loss of autophagic activity corresponded with a reduction in kinase activity we used an in vitro kinase assay. Cells expressing HA-tagged Atg1, Atg1K54A, and Atg1K54A,M102A were treated with 0.2 μg/ml rapamycin for 1.5 h. A protein extract from each mutant was subjected to immunoprecipitation with anti-HA antibody under native conditions, and then the immunocomplex was analyzed for in vitro kinase activity by using myelin basic protein as a substrate (Kamada et al., 1995). Compared with the wild-type Atg1 control, the Atg1K54A and Atg1K54A,M102A mutants showed nearly a complete loss of in vitro kinase activity (Figure 5B). The Pho8Δ60, but not the kinase assay, revealed a difference in the activity of the two Atg1 mutants; we suspect this reflects the nature of the assays, as the Pho8Δ60 assay is more quantitative.
Figure 5.
Atg1 kinase activity is required for autophagy. (A) Atg1 mutants are defective for autophagy. The alkaline phosphatase activity from Pho8Δ60, a marker for nonspecific autophagy, was monitored to determine the level of autophagy activity. The wild-type (YTS159) and atg1Δ (UNY1) strains harboring plasmids expressing Atg1, Atg1K54A (Atg1K), Atg1K54A,M102A (Atg1KM), or empty vector (pRS415) were grown in SMD lacking auxotrophic amino acids and shifted to SD-N for 4 h. Samples were collected and protein extracts assayed for alkaline phosphatase activity as described in Materials and Methods. The results represent the mean and SD of three experiments. (B) Atg1 mutants are defective for kinase activity. The atg1Δ mutant (WHY001) cells expressing HA-Atg1, HA-Atg1K54A, or HA-Atg1K54A,M102A were treated with 0.2 μg/ml rapamycin for 2 h. The extract of each mutant was immunoprecipitated with anti-HA antibody, and then immunocomplexes were assayed for Atg1 kinase activity as described in Materials and Methods.
To extend our examination of the effect of Atg1 kinase activity on autophagy, we again decided to perform an electron microscopy analysis to obtain morphological data. Cells harboring the atg1Δ pep4Δ vps4Δ mutations were transformed with plasmids expressing Atg1, Atg1K54A, or Atg1K54A,M102A. Each strain was grown to mid-log stage in selective media, shifted to SD-N for 4 h to induce autophagy, and examined by electron microscopy, as described in Materials and Methods. The cells harboring wild-type Atg1 showed extensive accumulation of autophagic bodies within the vacuole after starvation, whereas cells transformed with an empty vector displayed an essentially empty vacuole due to the deletion of ATG1 (Figure 6). In the cells harboring the Atg1 kinase mutants, the overall accumulation of autophagic bodies was drastically reduced compared with the wild type. In agreement with the results from the Pho8Δ60 assay, a small number of autophagic bodies with reduced size were detected in cells expressing the Atg1K54A mutation, whereas almost no autophagic bodies were found in cells expressing Atg1K54A,M102A. Overall, these results suggest that Atg1 kinase activity has an essential role in autophagy, although higher levels of kinase activity are needed for the Cvt pathway (Abeliovich et al., 2003).
Figure 6.
Atg1 kinase activity is required for autophagosome formation. (A) Atg1 mutants are defective for autophagic body accumulation. The atg1Δ (HCY76; pep4Δ vps4Δ) strains harboring plasmids expressing Atg1, Atg1K54A (Atg1K), Atg1K54A,M102A (Atg1KM), or the empty vector (pRS415) were grown in selective medium and shifted to SD-N for 4 h. Cells were fixed with permanganate and examined by electron microscopy as described in Materials and Methods. (B) Quantification of autophagic body accumulation. Fifty sections for each strain were scored for autophagic body accumulation in the vacuole.
Having established the phenotype of the Atg1 kinase mutants, we could now ask the question of whether Atg1 kinase activity is involved in the initiation of autophagy, and in particular in the assembly of the PAS and/or recruitment of Atg proteins. We used the atg1Δ atg11Δ strain with GFP-Atg8 or Atg17-GFP integrated at their respective chromosomal loci, and expressing Atg1 or Atg1K54A,M102A from a plasmid, and performed a fluorescence microscopy analysis. As we showed in Figure 2, GFP-Atg8 or Atg17-GFP showed a diffuse cytosolic signal in the atg1Δ atg11Δ double mutant strain under starvation conditions (Figure 7A, vector). Wild-type Atg1 fully complemented the PAS localization defect of GFP-Atg8 or Atg17-GFP in this strain (Figure 7A). The same cells harboring a plasmid expressing Atg1K54A,M102A also showed a strong punctum corresponding to GFP-Atg8 or Atg17-GFP at the PAS (Figure 7A). These data suggest that Atg1 kinase activity is not absolutely required for the PAS recruitment of Atg8 or Atg17 in starvation conditions that induce nonspecific autophagy.
Figure 7.
Atg1 kinase activity is not essential for PAS localization of Atg8 and Atg17, but it is required for normal localization of these proteins under starvation conditions. (A) Cells from the atg1Δ atg11Δ strain expressing GFP-Atg8 (HCY116) and Atg17-3xGFP (HCY119) from the respective chromosomal loci were transformed with plasmids encoding Atg1, empty vector, or Atg1K54A,M102A. The cells were grown in SMD and shifted to SD-N for 2 h before fluorescence microscopy, which was performed as described in Materials and Methods. (B) For quantification of fluorescence intensity at the PAS, 30 individual cells for each strain in A were observed by fluorescence microscopy. The relative percentage of fluorescence intensity was calculated as described in Materials and Methods and normalized to the value for wild-type Atg1 cells, which were set at 100%. Error bars indicate the SD of three independent experiments. (C) Redistribution of Atg8 from the PAS is affected by Atg1 kinase activity under starvation conditions. The atg1Δ atg11Δ strain expressing GFP-Atg8 from the chromosomal locus (HCY116) was cotransformed with plasmids encoding ATG1ts and Atg1K54A,M102A or ATG1ts and empty vector. The cells were grown in SMD medium at 24°C and transferred to SD-N for 1 h before the temperature shift. For inactivation of Atg1ts, cells were shifted to 37°C for 2 h, and then shifted back to 24°C for 2 h. The cells were examined by fluorescence microscopy as described in Materials and Methods. (D) For quantification of fluorescence intensity at the PAS, 30 individual cells from each strain in C were observed by fluorescence microscopy. The relative percentage of fluorescence intensity was calculated and normalized based on the value for GFP-Atg8 in cells expressing Atg1ts and the empty vector, which was set at 100% at the permissive temperature. Error bars indicate the SD of three independent experiments. (E) Quantification of the number of cells that contained GFP-Atg8 puncta as a PAS marker from C. Approximately 150–200 cells for each strain were analyzed for scoring the percentage of cells containing fluorescent dots.
The localization pattern of these two proteins, however, was not exactly the same as that seen in the presence of wild-type Atg1. GFP-Atg8 and Atg17-GFP displayed only strong punctate dots essentially without cytosolic staining in the strains expressing Atg1K54A,M102A, whereas these proteins had weaker PAS signals and obvious cytosolic diffuse signals in the presence of wild-type Atg1 (Figure 7A). These results suggested that Atg1 kinase activity is required for normal localization of Atg8 or Atg17, rather than assembly of the PAS. To explore this further we decided to perform quantitative fluorescence microscopy to carefully examine the effect of Atg1 kinase activity on PAS formation under conditions of nonspecific autophagy. The atg1Δ atg11Δ strains with GFP-Atg8 or Atg17-GFP integrated at the chromosomal loci, and harboring plasmids expressing Atg1 or Atg1K54A,M102A were prepared as described above, and the fluorescence intensity of each chimeric protein at the PAS was quantified as described in Materials and Methods. The relative fluorescence intensity of GFP-Atg8 at the PAS in the Atg1K54A,M102A strain was ∼2 times higher than in cells containing wild-type Atg1 (Figure 7B). Similarly, Atg17-GFP also showed a substantially stronger PAS intensity in the strain expressing Atg1K54A,M102A being ∼1.5 times that of the wild type.
To demonstrate that the accumulation of Atg17 or Atg8 at the PAS in the presence of Atg1 mutants is not due to a chronic defect associated with the Atg1 kinase mutation itself, we took advantage of a temperature-sensitive allele of Atg1 (Atg1ts) (Suzuki et al., 2001) to generate the equivalent of a conditional Atg1 kinase mutation. We cotransformed plasmids expressing Atg1ts and Atg1K54A,M102A into the atg1Δ atg11Δ strain that expressed either GFP-Atg8 or Atg17-GFP integrated at their respective chromosomal loci. As a control, we used an empty vector instead of the plasmid expressing Atg1K54A,M102A. First, we analyzed GFP-Atg8 processing to monitor the rate of Atg1ts inactivation under starvation conditions. GFP-Atg8 is delivered to the vacuole inside the autophagic body and after lysis in the vacuole lumen the Atg8 portion of the chimera is degraded, whereas GFP is relatively stable; the appearance of free GFP can then be used to monitor autophagy (Cheong et al., 2005). Cells expressing only Atg1ts in the atg1Δ atg11Δ background were grown in SMD at 24°C and transferred to SD-N for 1 h before the temperature shift. The accumulation of free GFP resulting from autophagy-dependent cleavage of GFP-Atg8 was significantly reduced within 2 h, which suggested inactivation of Atg1 occurred within this time frame after the temperature shift to 37°C (our unpublished data). Next, we used this conditional Atg1ts background to examine how the Atg1K54A,M102A kinase mutant affected the dynamics of GFP-Atg8 association/dissociation with the PAS.
In vegetative conditions, GFP-Atg8 showed a diffuse cytosolic signal in the atg1Δ atg11Δ double mutant at either 24°C or 37°C due to the absence of Atg11 (our unpublished data). Under starvation conditions GFP-Atg8 displayed a clear signal at the PAS in addition to an obvious cytosolic diffuse pattern in strains coexpressing Atg1ts and Atg1K54A,M102A, or Atg1ts and empty vector at 24°C (Figure 7C). This pattern was essentially the same as that of the wild-type strain, but distinct from the Atg1K54A,M102A mutant with regard to the level of cytosolic staining (Figure 7A), suggesting that the Atg1ts protein at the permissive temperature complemented the atg1Δ mutation and was dominant to the Atg1K54A,M102A phenotype. After a shift to the nonpermissive temperature of 37°C, GFP-Atg8 was restricted to the PAS without cytosolic staining within 2 h, only in the presence of Atg1K54A,M102A. In contrast, the strain expressing Atg1ts alone, displayed an increase in the diffuse cytosolic signal and a decrease in the PAS puncta when Atg1 was inactivated. Therefore, the Atg1K54A,M102A phenotype that resulted rapidly after the Atg1ts mutant was inactivated was the accumulation of a strong GFP-Atg8 signal at the PAS. When the cells were shifted back to the permissive temperature, GFP-Atg8 in both strains regained the dual PAS and cytosolic localization originally seen, indicating that the accumulation at the PAS was not a terminal phenotype (Figure 7C).
Quantification of the data indicated that the relative fluorescence intensity of GFP-Atg8 at the PAS was ∼2.5 times higher at the nonpermissive relative to the permissive temperature for the Atg1K54A,M102A strain (Figure 7D). Similarly, Atg17-GFP also showed a stronger PAS intensity in the strain expressing Atg1K54A,M102A at the nonpermissive temperature, increasing ∼1.5-fold (our unpublished results). In addition we counted the number of cells containing a GFP-Atg8 PAS dot in these conditions to examine whether Atg1 kinase activity affected the total number of PAS that were assembled (Figure 7E). The number of GFP-Atg8 puncta in the strain expressing Atg1K54A,M102A was comparable with that of the wild-type Atg1 strain at both permissive and nonpermissive conditions. In contrast, the atg1Δ atg11Δ mutant expressing Atg1ts alone showed a clear decrease in the number of PAS dots at the nonpermissive temperature, whereas the intensity of GFP-Atg8 at the few detectable PAS was unaffected (the latter presumably correspond to PAS sites that have not yet disassembled). These results support the idea that autophagy-specific PAS recruitment is a dynamic process depending on the Atg1 protein and that there may be two separable Atg1-dependent functions; recruitment of Atg proteins needed for assembly of the PAS (and ultimately for autophagosome formation), and subsequent dissociation from this site, steps that are independent of and dependent on Atg1 kinase activity, respectively. Although Atg1 kinase activity is not needed for PAS recruitment of the Atg proteins that we analyzed, in the absence of normal kinase activity the Atg proteins are not able to undergo normal dissociation from this site, which presumably reflects a defect in function that translates into a reduced ability to form autophagosomes.
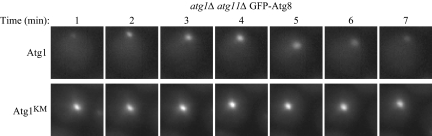
During this quantitative analysis, we noticed that the GFP-Atg8 puncta displayed a variable intensity, increasing and decreasing over time. To examine this dynamic effect carefully, we performed time-lapse fluorescence microscopy as described in Materials and Methods. As shown in Figure 7A, GFP-Atg8 in the atg1Δ atg11Δ strain expressing Atg1 displayed normal recruitment at the PAS upon the shift to starvation conditions. The GFP-Atg8 fluorescence reached a peak at ∼4 min, followed by a loss of signal after 7 min (Figure 8). In contrast, the same strain harboring the plasmid expressing Atg1K54A,M102A showed a strong punctate signal of GFP-Atg8 without any substantial change in the fluorescence pattern at the PAS over the 7-min time course (Figure 8). These results support the hypothesis that Atg1 kinase activity affects the dynamics of Atg8 retention at, or dissociation from, the PAS.
Figure 8.
Atg1 kinase activity is important for Atg8 dynamics during starvation. Cells from the atg1Δ atg11Δ strain expressing GFP-Atg8 (HCY116) were transformed with plasmids encoding Atg1 and Atg1K54A,M102A (Atg1KM). The cells were grown in SMD medium and shifted to SD-N for 2 h before fluorescence microscopy. For time-lapse experiments, the cells were immobilized on SD-N medium containing 2% agar on concavity slides as described in Materials and Methods, and pictures were taken every minute. Images are shown over the 7-min time course.
The Interaction of Atg1 with Atg13 Is Important for Autophagy
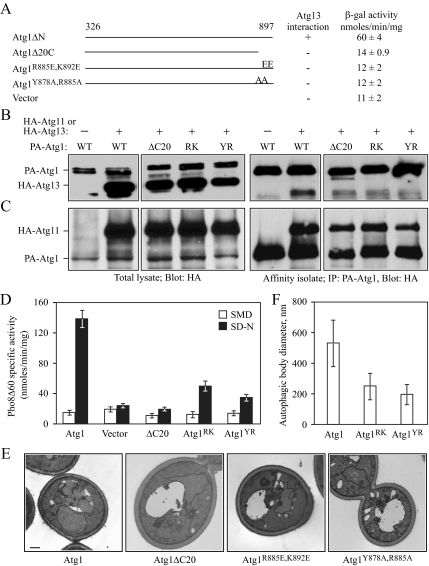
Atg1 interacts with the Cvt-specific protein Atg11, and with Atg13 and Atg17 (Kamada et al., 2000; Yorimitsu and Klionsky, 2005a). Because of our results with the Atg1 kinase mutants, we decided to examine whether the interaction between Atg1 and Atg13 is necessary for initial PAS formation under conditions of nonspecific autophagy. Previous data suggest that the C terminus of Atg1 may have a role in formation of a complex with its interacting partners (Abeliovich et al., 2003). Based on this information, we decided to investigate whether C-terminally truncated mutants of Atg1 lost their ability to interact with either of its known-binding partners, Atg11 or Atg13, by a yeast two-hybrid analysis. Accordingly, we generated a series of C-terminally truncated Atg1 fragments by incrementally deleting 20 amino acids. It was necessary to carry out this analysis with an Atg1 construct lacking the kinase domain, which otherwise results in autoactivation in this assay (our unpublished data). Two-hybrid analysis with cells containing plasmids expressing Atg13 and an Atg1 fragment lacking only the C-terminal 20 amino acids in addition to the absence of the N-terminal kinase domain (Atg1ΔC20) completely lost the ability to grow on selective plates lacking adenine, whereas a mutant lacking only the Atg1 kinase domain (Atg1ΔN) displayed clear growth (Figure 9A).
Figure 9.
The interaction between Atg1 and Atg13 is important for autophagy. (A) Mutations in the C terminus of Atg1 disrupt its interaction with Atg13 based on a yeast two-hybrid assay. PJ69-4A cells expressing Atg13 (AD-Atg13) and either Atg1 lacking the N-terminal kinase domain (BD-Atg1ΔN), Atg1ΔN deleted for the C-terminal 20 amino acids (BD-Atg1ΔC20), Atg1ΔN with substitutions of R885E and K892E (BD-Atg1R885E,K892E), Atg1ΔN with substitutions of Y878A and R885A (BD-Atg1Y878A,R885A), or the BD-empty vector were grown for 5 d on plates lacking adenine. Growth or lack of growth on these plates was scored as + or −, respectively. The strength of the interaction for each set of proteins was quantified by measuring β-galactosidase activity from three independent experiments. (B and C) Atg1 C-terminal mutants are defective for interacting with Atg13 but not Atg11. The B, atg1Δ atg13Δ (UNY27) or C, atg1Δ atg11Δ (YTS157) cells were transformed with a plasmid expressing 3xHA-Atg13 under the control of its endogenous promoter or 3xHA-Atg11 under the control of the CUP1 promoter as indicated, and CUP1 promoter-driven protein A (PA)-tagged fusions of either wild-type Atg1 (WT), Atg1ΔC20 (ΔC20), Atg1R885E,K892E (RK), or Atg1Y878A,R885A (YR). As negative controls UNY27 and YTS157 cells were transformed with a plasmid expressing CUP1 promoter-driven wild-type PA-Atg1 and an empty pRS315 or pRS414 plasmid, respectively. Protein extracts were subjected to affinity isolation, eluted proteins were separated on a 6% SDS-PAGE gel and detected with anti-HA antibody as described in Materials and Methods. IP, immunoaffinity-purified isolate. (D) The Atg1 C-terminal mutants are defective in autophagy. The Pho8Δ60 assay was used to monitor autophagy activity for the atg1Δ (UNY3) strain expressing the indicated wild-type Atg1, empty vector (pRS315), or Atg1 mutant plasmids expressing Atg1 under the control of the endogenous promoter, as described in Materials and Methods. Alkaline phosphatase activity was monitored from protein extracts prepared from cells grown in SMD or after a 4 h shift to SD-N. The results represent the mean and SD of three independent experiments. (E) The Atg1ΔC20 mutant is defective in forming autophagosomes (detected as autophagic bodies), whereas small autophagosomes are formed in cells expressing C-terminal point mutants. An atg1Δ pep4Δ vps4Δ strain (HCY76) was transformed individually with empty vector, or plasmids expressing wild-type Atg1 or the indicated Atg1 mutant. Cells were grown to mid-log phase in SMD medium, shifted to SD-N medium to induce starvation, fixed with potassium permanganate, and examined by electron microscopy as described in Materials and Methods. No autophagic bodies were detected in cells transformed with the empty vector. (F) Quantification of the diameters of the autophagic bodies was carried out from 50 sections for each strain. Essentially no autophagic bodies were detected in the strain expressing Atg1ΔC20.
We then measured β-galactosidase activity to quantitatively evaluate the strength of these interactions (Rose et al., 1990). The Atg1ΔN construct in the presence of Atg13 generated ∼60 U of β-galactosidase activity, whereas the Atg1ΔC20 protein exhibited only background levels of activity (Figure 9A). We decided to extend our analysis by isolating point mutations within the C-terminal domain of Atg1 that disrupt interaction with Atg13. This region of Atg1 is highly conserved among the Atg1 homologues of different Saccharomyces species. We carried out site specific mutagenesis and generated two sets of mutants within this region; in the first, the tyrosine and arginine residues at positions 878 and 885, respectively, were substituted with alanine (Y878A,R885A), and in the second, an arginine residue at position 885 and lysine at position 892, were substituted with glutamic acid (R885E,K892E) (Figure 9A). Both pairs of mutations resulted in a loss of Atg1–Atg13 interaction based on the yeast two-hybrid assay (Figure 9A). To confirm that these Atg1 point mutants were defective in interacting with Atg13, we examined the interaction between these two proteins under physiological conditions.
Cells that contain protein-A tagged wild-type Atg1 or mutant Atg1 derivatives, and Atg13 tagged with the HA epitope, were treated with rapamycin to induce autophagy and examined by affinity isolation as described in Materials and Methods. The wild-type and mutant protein A-tagged Atg1 were isolated with IgG-Sepharose and the presence of HA-Atg13 was analyzed by western blot. HA-Atg13 was coisolated with wild-type Atg1 (Figure 9B). By comparison, the amount of HA-Atg13 coisolated with the Atg1 mutants was substantially reduced; the point mutants showed essentially an equivalent loss of interaction with Atg13 as seen with the ΔC20 truncation. As a control, we examined a strain expressing PA-Atg1 without HA-Atg13 and were not able to detect any coisolating bands. These results indicate that the mutations at the C terminus of Atg1 caused a substantial reduction in the ability of this protein to interact with Atg13. We also examined the ability of the different Atg1 proteins to interact with Atg11. Under starvation conditions, wild-type Atg1 and all of the Atg1 C-terminal mutants interacted strongly with Atg11 (Figure 9C). Thus, the extreme C terminus of Atg1 is important for forming a complex with Atg13, but apparently not with Atg11. Recently, it was reported that the interaction between Atg17 and Atg1 is mediated through Atg13 (Cheong et al., 2005; Kabeya et al., 2005); thus, it is possible that these Atg1 mutants might also show altered interactions with Atg17.
The function of Atg1 is required for both autophagy and the Cvt pathway. In contrast, the function of Atg13 seems to be more significant for autophagy rather than the Cvt pathway; an atg13Δ mutant exhibits a partial maturation of prApe1, but it is completely blocked in autophagy (Abeliovich et al., 2003; Kamada et al., 2000). To determine the physiological function of disrupting the Atg1–Atg13 interaction, we tested whether the Atg1 mutants were capable of prApe1 maturation via the Cvt pathway and/or autophagic activity. Under vegetative conditions, prApe1 maturation was completely blocked in the Atg1ΔC20 mutant, indicating a defect in the Cvt pathway (our unpublished data). Conversely, an atg1Δ strain harboring each of the point mutants, Atg1Y878A,R885A and Atg1R885E,K892E, exhibited wild-type prApe1 maturation patterns (our unpublished data). Thus, in contrast to an atg13Δ mutant, which only partially affects the Cvt pathway, the Atg1ΔC20 mutant shows a complete block in the Cvt pathway. Because the Atg1ΔC20 mutant shows a greater Cvt defect compared with an atg13Δ mutant, we conclude that the last 20 amino acids are not merely important for interacting with Atg13 but also are required for other Atg1-specific functions.
We quantitatively monitored bulk autophagy by measuring the activity of a nonspecific autophagy marker, Pho8Δ60, which encodes an altered form of alkaline phosphatase; Pho8Δ60 remains in the cytosol and can only be delivered to the vacuole via autophagy (Noda et al., 1995; Klionsky et al., 2007). In the vacuole, Pho8Δ60 is processed to its mature, active form. Thus, measuring Pho8Δ60 activity under starvation conditions provides a quantitative analysis of autophagy. The alkaline phosphatase activity was measured in atg1Δ cells transformed with a plasmid expressing wild-type Atg1, an empty vector, Atg1ΔC20, Atg1Y878A,R885A, or Atg1R885E,K892E, in vegetative and starvation conditions (Figure 9D). The atg1Δ cells transformed with an empty vector were defective in bulk autophagy, and showed only a basal level of Pho8Δ60 activity after 4 h starvation. In contrast, the Pho8Δ60 activity in atg1Δ cells expressing wild-type Atg1 increased after starvation, indicating that the wild-type Atg1 protein complemented the autophagy defect in atg1Δ cells. The Atg1ΔC20 mutant was completely blocked for bulk autophagy, displaying a level of activity after starvation that was essentially the same as cells transformed with an empty vector. The atg1Δ cells expressing the Atg1R885E,K892E and Atg1Y878A,R885A mutations exhibited ∼35 and 30%, respectively, of the Pho8Δ60 activity seen with wild-type cells, indicating a substantial defect in bulk autophagy. Together, these results show that the C terminus of Atg1 is essential for both the Cvt and autophagy pathways. Furthermore, the reduced autophagic capacity in strains harboring each set of point mutants indicates that the interaction between Atg1 and Atg13 that is mediated through the Atg1 C terminus is important for efficient nonspecific autophagy.
We decided to complete our analysis of the Atg1 mutants by examining their effect on autophagosome formation. Deletion of Atg17 results in cells with fewer and smaller autophagosomes (Cheong et al., 2005). Therefore, we examined the morphology of autophagosomes produced in cells expressing either the Atg1ΔC20, Atg1Y878A,R885A or Atg1R885E,K892E mutants by electron microscopy. We transformed atg1Δ pep4Δ vps4Δ mutant cells with plasmids expressing wild-type or mutant Atg1. The transformants were grown in selective media, shifted to SD-N for 4 h to induce autophagy, fixed with potassium permanganate, and embedded in Spurr's resin as described in Materials and Methods. Similar to the results shown in Figure 6, after 4-h starvation, cells expressing wild-type Atg1 showed an accumulation of ∼10–14 autophagic bodies within the vacuole (Figure 9E). Consistent with the basal level of pho8Δ60 activity seen in cells expressing Atg1ΔC20, most cells harboring this mutant version of Atg1 exhibited an empty vacuole, whereas a few cells exhibited vacuoles with a very small number of autophagic bodies (Figure 9E). Also in agreement with the reduced bulk autophagy seen in cells expressing the Atg1 point mutants, electron microscopy analysis revealed that the Atg1Y878A,R885A or Atg1R885E,K892E mutants produced smaller autophagic bodies (i.e., having a reduced diameter), with presumably reduced autophagic capacity compared with those in the wild-type strain (Figure 9, E and F). However, the total number of autophagic bodies produced in cells expressing these Atg1 point mutants (12.87 ± 4.83 for Atg1Y878A,R885A and 11.23 ± 4.04 for Atg1R885E,K892E) was not significantly different from the wild type (12.28 ± 3.63). Together, these data suggest that the interaction between Atg1 and Atg13 regulates the size of the autophagosomes (and the resulting autophagic bodies), and thereby the optimal magnitude of the autophagic response.
The Interaction of Atg1 with Atg13-Atg17 Is Needed for Starvation-induced PAS Organization
Finally, we used the various Atg1 C-terminal mutants to examine the effect of the loss of interaction between Atg1 and Atg13-Atg17 on PAS formation. GFP-Atg8 and Atg17-GFP were monitored in the atg1Δ atg11Δ strain expressing Atg1, Atg1ΔC20, Atg1Y878A,R885A, or Atg1R885E,K892E. Under starvation conditions, wild-type Atg1 complemented the PAS localization defect of GFP-Atg8 and Atg17-GFP in this strain, and once again we detected puncta at the PAS and weak cytosolic signals (Figure 10, A and B). GFP-Atg8 also displayed some level of vacuolar fluorescence under these conditions. In contrast, the Atg1ΔC20 or Atg1Y878A,R885A mutants showed only a diffuse cytosolic fluorescent signal for GFP-Atg8 and Atg17-GFP, with no evidence for PAS recruitment (Figure 10, A and B). The Atg1R885E,K892E mutant in this strain occasionally showed weak puncta for both chimeras but the predominant pattern was a diffuse cytosolic signal. To quantify the results for PAS recruitment of GFP-Atg8 and Atg17-GFP, we counted the number of cells containing a PAS dot in the strains that harbored the various Atg1 mutants (Figure 10C). The atg1Δ atg11Δ strain expressing Atg1ΔC20, Atg1Y878AR885A or Atg1R885E,K892E showed a significant reduction in the number of GFP-Atg8 and Atg17-GFP dots at the PAS compared with the same strains expressing wild-type Atg1. These results support the hypothesis that the interaction among Atg1, Atg13, and Atg17 plays an important role in PAS assembly/organization for nonspecific autophagy, a function that is distinct from Atg1 kinase activity.
Figure 10.
The interaction of Atg1 with Atg13 is required for PAS formation under starvation conditions. Cells from the atg1Δ atg11Δ strain expressing GFP-Atg8 (HCY116) (A) or Atg17-GFP (HCY119) (B) from the chromosomal locus were transformed with plasmids encoding wild-type Atg1, or the Atg1ΔC20 (ΔC20), Atg1R885E,K892E (RK), or Atg1 Y878A,R885A (YR) mutants. The cells were grown in SMD and shifted to SD-N for 2 h before fluorescence microscopy, which was carried out as described in Materials and Methods. (C) Quantification of the number of cells that contained GFP-Atg8 or Atg17-GFP puncta as a PAS marker from A and B. Approximately 150–200 cells for each strain were analyzed for scoring the percentage of cells with fluorescent dots.
DISCUSSION
One of the major unresolved issues in the autophagy field concerns the nature of the phagophore assembly site. Although this site is thought to play a critical role in autophagosome formation, and it is the site where the majority of Atg proteins reside at least transiently, little is known about PAS assembly or the recruitment process. Previously, we showed that Atg11 plays an important role in formation of the PAS under vegetative conditions (Shintani and Klionsky, 2004b). Accordingly, we propose that Atg11 is one of the first factors to target to the PAS and that it is required for the subsequent recruitment of additional Atg proteins. However, under starvation conditions, PAS assembly is apparently normal in the absence of Atg11, which is a Cvt-specific Atg protein. Therefore, we hypothesized that an additional component(s) would play a similar role to Atg11 under starvation conditions.
Atg17 is characterized as a protein that is only needed for nonspecific autophagy, and it thus seemed like a reasonable candidate for playing an equivalent role to Atg11 under starvation conditions. We examined the role of Atg17 in strains deleted for ATG11 to eliminate the PAS that would otherwise form under vegetative conditions. The atg11Δ atg17Δ double mutant displayed a strong block in PAS formation in starvation conditions based on the localization of GFP-Atg8 and GFP-Atg1 (Figure 1), supporting this hypothesis. Similarly, the double mutant showed a much greater defect in autophagosome/autophagic body formation compared with the atg17Δ mutant (Figure 2). Thus, Atg11 is sufficient in the absence of Atg17 for dictating PAS assembly in vegetative conditions, whereas Atg17 is necessary for allowing assembly in starvation conditions; the absence of both proteins prevents PAS recruitment of GFP-Atg8, GFP-Atg1, and other Atg marker proteins including Atg14 and Atg16 under either condition.
During the course of our analysis, a report was published suggesting that Atg11 and Atg17 are the two initial factors that establish the assembly sequence for Atg proteins at the PAS (Suzuki et al., 2007). Atg17 is proposed to act as a scaffold for recruiting other Atg proteins, similar to the function we proposed for Atg11 (Yorimitsu and Klionsky, 2005a; He et al., 2006). However, in our present analysis, we found that PAS assembly under starvation conditions requires more than Atg17; cells deleted for ATG11 and either ATG1 or ATG13 displayed essentially the same phenotype as the atg11Δ atg17Δ double mutant (Figure 3). The analysis by Suzuki et al., 2007) relied primarily on an examination of strains disrupted for individual ATG genes in the presence of rapamycin, whereas the present analysis includes double deletion strains, and autophagy was induced by starvation. Both studies note the role of Atg17 as a scaffold that functions during autophagy-inducing conditions in a similar manner to that of Atg11 during vegetative growth. However, single deletions of ATG1, ATG13, or ATG17 did not result in defective PAS localization phenotypes, especially for GFP-Atg1 or GFP-Atg8 (Figure 4). Thus, the role of Atg1 and Atg13 was not examined in the previous analysis, which only noted the effect of the atg11Δ atg17Δ double mutant (Suzuki et al., 2007).
Atg1, Atg13, and Atg17 associate with each other regardless of Atg1 kinase activity, and the affinity of these interactions seems to increase upon starvation (Kamada et al., 2000; Kabeya et al., 2005). Accordingly we examined whether the interaction among Atg1, Atg13, and Atg17 affects PAS formation under starvation conditions. We mapped an interaction site for Atg13 in the Atg1 C terminus (Figure 9). Deletion of the C-terminal 20 amino acids, or point mutations within this region strongly, but not completely, abolished interaction with Atg13; thus, there may be additional sites in Atg1 that mediate this interaction. Similar to the results with the atg1Δ and atg13Δ strains, we found that Atg1 mutants that are defective in their interaction with Atg13 also display defects in PAS assembly/recruitment of Atg proteins under starvation conditions in the atg11Δ background (Figure 10).
Atg13 and Atg17 are key modulators of Atg1 kinase activity, although the function of the latter is not yet clear. In a previous analysis (Abeliovich et al., 2003), we reported that Atg1 kinase activity is more important for the Cvt pathway and, based on analysis of the Atg1K54A mutant, that it was not needed for autophagy. It is now apparent that this conclusion is partially incorrect in that kinase activity is needed for both autophagy and the Cvt pathway. Part of the reason for this discrepancy is that the Atg1K54A mutant is clearly not completely lacking kinase activity (Kamada et al., 2000; Figure 6). Therefore, we decided to examine the Atg1K54A,M102A kinase mutant, which displays a greater defect in kinase-related function than the previously described Atg1K54A mutant. Surprisingly, we found that Atg1 kinase activity was not essential for PAS recruitment of GFP-Atg8 or Atg17-GFP (Figure 7). Rather, mutants that were defective in Atg1 kinase activity accumulated higher levels of PAS marker proteins. Therefore, kinase activity seems to play a role in disassembly of the PAS or the dissociation kinetics of Atg proteins, which may be part of the autophagosome formation cycle (Figures 7 and 8). In addition, our electron microscopy results revealed a significant reduction in the size and number of autophagosomes/autophagic bodies in cells that expressed the Atg1K54A mutant, which is partly defective for kinase activity (Figure 6). Thus, Atg1 kinase activity may affect the rate of autophagosome-expansion and formation, although we cannot conclude at this time that the defect in autophagosome formation is due to the block in Atg8 or Atg17 release from the PAS. Because they did not detect a requirement for Atg1 in the initial steps of PAS assembly, the study by Suzuki et al. (2007), did not examine the role of Atg1 kinase activity.
Significantly, the results with the Atg1 kinase mutants contrast with those of the Atg1 C-terminal mutants, even though loss of interaction with Atg13 in the latter should also reduce kinase activity. Nonetheless, Atg1 mutants that are not able to interact with Atg13, similar to strains deleted for ATG1, ATG13, or ATG17 lose the ability to efficiently assemble the PAS under starvation conditions, whereas the Atg1 kinase mutants can assemble a PAS, but they are defective in the kinetics of Atg protein dissociation and display reduced autophagosome formation. These results suggest that Atg1 has a nonkinase, perhaps structural, role in PAS assembly that is mediated though its C terminus, and that it apparently involves interactions with its binding partners. Furthermore, this function is epistatic to the Atg1 kinase function with regard to PAS assembly. Additional work will be necessary to determine the mechanism of Atg1 action during PAS assembly and autophagosome formation, but these studies suggest that one possibility involves changes in Atg1 conformation that are mediated via its interactions in the Atg1–Atg13–Atg17 complex.
ACKNOWLEDGMENTS
We thank Drs. Takahiro Shintani (Tohoku University) and Fulvio Reggiori (University Medical Center Utrecht) for helpful discussions and technical advice. This work was supported by National Institutes of Health Public Health Service grant GM53396 (to D.J.K.).
Abbreviations used:
- Atg
autophagy-related
- Cvt
cytoplasm-to-vacuole targeting
- GFP
green fluorescent protein
- PAS
phagophore assembly site
- prApe1
precursor aminopeptidase I.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0826) on December 12, 2007.
REFERENCES
- Abeliovich H., Zhang C., Dunn W. A., Jr, Shokat K. M., Klionsky D. J. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham C. L., Brumell J. H. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy. 2006;2:156–158. doi: 10.4161/auto.2825. [DOI] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C.-W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M. I., Gutierrez M. G., Romano P. S. The two faces of autophagy: Coxiella and Mycobacterium. Autophagy. 2006;2:162–164. doi: 10.4161/auto.2827. [DOI] [PubMed] [Google Scholar]
- Dunn W. A., Jr, Cregg J. M., Kiel J.A.K.W., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Farre J. C., Vidal J., Subramani S. A cytoplasm to vacuole targeting pathway in P. pastoris. Autophagy. 2007;3:230–234. doi: 10.4161/auto.3905. [DOI] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W.-L., Legakis J. E., Klionsky D. J. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.-P., Scott S. V., Kim J., Klionsky D. J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Hutchins M. U., Klionsky D. J. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Kamada Y., Baba M., Takikawa H., Sasaki M., Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C. A., Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Jung U. S., Piotrowski J., Levin D. E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Klionsky D. J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 2001a;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Stromhaug P. E., Klionsky D. J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001b;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I., Salin B., Schaeffer J., Bhatia S., Manon S., Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. Cell biology: regulated self-cannibalism. Nature. 2004;431:31–32. doi: 10.1038/431031a. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Cuervo A. M., Seglen P. O. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Cueva R., Yaver D. S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Emr S. D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- Kuma A., Mizushima N., Ishihara N., Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- Labbé S., Thiele D. J. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U., Klionsky D. J. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J. Biol. Chem. 2005;280:41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- Nice D. C., Sato T. K., Stromhaug P. E., Emr S. D., Klionsky D. J. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D. J. Autophagy in the eukaryotic cell. Eukaryot. Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Wang C.-W., Nair U., Shintani T., Abeliovich H., Klionsky D. J. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. Assay of β-galactosidase in yeast; pp. 155–159. [Google Scholar]
- Rossanese O. W., Reinke C. A., Bevis B. J., Hammond A. T., Sears I. B., O'Connor J., Glick B. S. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 2001;153:47–62. doi: 10.1083/jcb.153.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D. C., Ravikumar B., Acevedo-Arozena A., Imarisio S., O'Kane C. J., Brown S. D. Dyneins, autophagy, aggregation and neurodegeneration. Autophagy. 2005;1:177–178. doi: 10.4161/auto.1.3.2050. [DOI] [PubMed] [Google Scholar]
- Scott S. V., et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P. E., Klionsky D. J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Autophagy in health and disease: a double-edged sword. Science. 2004a;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004b;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallóczy Z., Virgin H. W., IV, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- Vergne I., et al. Autophagy in immune defense against Mycobacterium tuberculosis. Autophagy. 2006;2:175–178. doi: 10.4161/auto.2830. [DOI] [PubMed] [Google Scholar]
- Webster P. Cytoplasmic bacteria and the autophagic pathway. Autophagy. 2006;2:159–161. doi: 10.4161/auto.2826. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 2005a;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005b;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Eating the endoplasmic reticulum: quality control by autophagy. Trends Cell Biol. 2007;17:279–285. doi: 10.1016/j.tcb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Yoshimori T. Autophagy vs. Group A Streptococcus. Autophagy. 2006;2:154–155. doi: 10.4161/auto.2822. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Qi H., Taylor R., Xu W., Liu L. F., Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]