Abstract
Chromosome segregation during mitosis requires the assembly of a large proteinaceous structure termed the kinetochore. In Caenorhabditis elegans, KNL-1 is required to target multiple outer kinetochore proteins. Here, we demonstrate that the vertebrate KNL1 counterpart is essential for chromosome segregation and is required to localize a subset of outer kinetochore proteins. However, unlike in C. elegans, depletion of vertebrate KNL1 does not abolish kinetochore localization of the microtubule-binding Ndc80 complex. Instead, we show that KNL1 and CENP-K, a subunit of a constitutively centromere-associated complex that is missing from C. elegans, coordinately direct Ndc80 complex localization. Simultaneously reducing both hKNL1 and CENP-K function abolishes all aspects of kinetochore assembly downstream of centromeric chromatin and causes catastrophic chromosome segregation defects. These findings explain discrepancies in kinetochore assembly pathways between different organisms and reveal a surprising plasticity in the assembly mechanism of an essential cell division organelle.
INTRODUCTION
The kinetochore is a large proteinaceous assembly that associates with both centromeric DNA and spindle microtubule polymers and is required for chromosome segregation in all eukaryotes. Constitutively present centromere-specific nucleosomes containing the histone H3 variant CENP-A form the platform for kinetochore assembly. In vertebrates, a group of 14 proteins (CENP-C, -H, -I, and -K to -U), which we will refer to as the constitutive centromere-associated network (CCAN), associates with CENP-A throughout the cell cycle (Foltz et al., 2006; Okada et al., 2006). The CCAN includes a subgroup of three proteins (CENP-H, -I, and -K) that are interdependent for localization and play essential roles in kinetochore structure and function (Okada et al., 2006). Additional proteins begin to associate with the CENP-A/CCAN platform during mitotic entry to generate the mature mitotic kinetochore structure that forms a dynamic interface with spindle microtubules and serves as a platform for signaling pathways controlling the fidelity of chromosome segregation.
Previous work in the nematode Caenorhabditis elegans has examined pairwise localization dependency relationships for the vast majority of kinetochore proteins (Oegema et al., 2001; Desai et al., 2003; Cheeseman et al., 2004). These studies have revealed a largely linear hierarchy of kinetochore assembly in this organism. A key player in kinetochore assembly in C. elegans is KNL-1, which functions downstream of the inner kinetochore components such as CENP-A, but upstream of all known outer kinetochore proteins (Desai et al., 2003; Cheeseman et al., 2004). KNL-1 belongs to a conserved protein family that includes budding yeast Spc105 (Nekrasov et al., 2003), fission yeast Spc7 (Kerres et al., 2004), Drosophila Spc105R (Przewloka et al., 2007), and a novel protein previously identified in human cells known as AF15q14, D40, CASC5, or Blinkin (Cheeseman et al., 2004; Obuse et al., 2004; Kiyomitsu et al., 2007). AF15q14 is present in all vertebrates including chicken (Gallus gallus). Here we will refer to the human and chicken proteins as hKNL1 and ggKNL1, respectively. The C. elegans, human, Drosophila, and fission yeast KNL-1 proteins directly interact with the Ndc80 and Mis12 complexes (Cheeseman et al., 2004; Obuse et al., 2004; Liu et al., 2005; Przewloka et al., 2007). We have termed this interacting protein set that is conserved throughout eukaryotes the KNL-1/Mis12 complex/Ndc80 complex (KMN) network (Cheeseman et al., 2006). The KMN network is a key structural component of the kinetochore, but also directly binds to microtubules via the Ndc80 subunit of the Ndc80 complex and KNL-1 (Cheeseman et al., 2006).
Here, we focus on the contribution of KNL1 to kinetochore assembly in vertebrates. We demonstrate that KNL1 localizes to kinetochores throughout mitosis and is required for the localization of a subset of outer kinetochore proteins. However, in contrast to C. elegans KNL-1, depletion of hKNL1 or ggKNL1 does not abolish Ndc80 complex localization. This apparent difference was explained by analysis of the CCAN subunit CENP-K. KNL1 and CENP-K appear to work coordinately to target the Ndc80 complex to kinetochores. Simultaneously reducing the levels of both hKNL1 and the CENP-K results in a failure of kinetochore assembly downstream of CENP-A and -C and severe defects in chromosome segregation. These results reveal that the KMN network and the CENP-K subgroup of the CCAN coordinately direct kinetochore assembly in vertebrates. The system is evolutionarily plastic, because the CCAN pathway appears to have been lost in C. elegans.
MATERIALS AND METHODS
Cell Culture and siRNA Transfection
HeLa cells and clonal lines derived from HeLa cells stably expressing GFP-Mis12 (Cheeseman et al., 2004), YFP-CENP-A (Kops et al., 2005), and GFP-CENP-H, and GFP-CENP-Q (Okada et al., 2006) were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), penicillin/streptomycin, and l-glutamine (Invitrogen, Carlsbad, CA) at 37°C in a humidified atmosphere with 5% CO2. RNA interference (RNAi) experiments were conducted as described previously (Kline et al., 2006). CENP-K (Okada et al., 2006), hDsn1 (Kline et al., 2006), and hNuf2 (DeLuca et al., 2002) small interfering RNAs (siRNAs) have been previously described. For hKNL1, a combination of four pooled sequences were predesigned by Ambion (Austin, Tx). These include GGAAUCCAAUGCUUUGAGAtt, GAAAUAAGAAAAACUCUCGtt, GAUACUAUAAAGGUAUUCCtt, and GGAAGCUUCAGAUAAUUGUtt.
The chicken KNL1 target disruption construct was generated such that exons 8 and 9 were replaced with a histidinol- (hisD) or puromycin- (puro) resistance cassette under control of the β-actin promoter resulting in a severely truncated ggKNL1. Target constructs were transfected with a Gene Pulser II electroporator (Bio-Rad, Richmond, CA). Chicken DT40 cells were cultured and transfected as described previously (Fukagawa et al., 2001, 2004). All DT40 cells were cultured at 38°C in Dulbecco's modified medium supplemented with 10% fetal calf serum, 1% chicken serum, and penicillin/streptomycin. To repress the expression of the tetracycline-responsive transgenes, tetracycline (tet; Sigma, St. Louis, MO) was added to the culture medium to a final concentration of 2 μg/ml.
Immunofluorescence
Immunofluorescence in human cells was conducted as described previously (Kline et al., 2006). For fluorescence of microtubules, DM1α (Sigma) was used at 1:500. For visualization of kinetochore proteins, mouse anti-HEC1 (9G3; Abcam, Cambridge, MA) was used at 1:1000, mouse anti-CENP-C (a generous gift of Lars Jensen and Don Cleveland, Ludwig Institute for Cancer Research, La Jolla, CA) was used at 1:200, and human anti-centromere antibodies (ACA; Antibodies, Davis, CA) were used at 1:100. YFP-CENP-A–, GFP-CENP-H–, or GFP-CENP-Q–expressing HeLas were counterstained with goat anti-green fluorescent protein (GFP; a generous gift of Andres Ladurner, European Molecular Biology Laboratory, Heidelberg, Germany) at 1:1000. Affinity-purified rabbit polyclonal antibodies were generated against hKNL1 (residues 1413-1624) as described previously (Desai et al., 2003). Polyclonal antibodies against hDsn1 and hKNL1 were used at 1 μg/ml. Cy2-, Cy3-, and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used at 1/100, although in some cases directly labeled antibodies against hDsn1 and hKNL1 were used without secondaries. DNA was visualized using 10 μg/ml Hoechst in Tris-buffered saline-TX.
Immunofluorescent staining of DT40 cells was performed as described previously (Okada et al., 2006). DT40 cells were collected onto slides with a cyto-centrifuge and fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature or 100% methanol for 15 min at −20°C, permeabilized in 0.5% NP-40 in PBS for 15 min at room temperature, rinsed three times in 0.5% bovine serum albumin (BSA) in PBS, and incubated for 1 h at 37°C with primary antibody. Binding of primary antibody was then detected with fluorescein isothiocyanate–conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) diluted to an appropriate concentration in 0.5% BSA in PBS. Affinity-purified rabbit polyclonal antibodies were generated against recombinant chicken CENP-C (Fukagawa et al., 1999), chicken CENP-I (Nishihashi et al., 2002), chicken CENP-O (Okada et al., 2006), chicken Mis12 (Kline et al., 2006), and chicken Ndc80 (Hori et al., 2003). To examine localization of ggKNL1-FLAG, anti-FLAG antibody (Sigma) was used. Chromosomes and nuclei were counterstained with DAPI at 0.2 μg/ml in Vectashield Antifade (Vector Labs, Burlingame, CA).
Microscopy and Image Acquisition
Images of fixed cells were acquired on a DeltaVision deconvolution microscope (Applied Precision Instruments, Issaquah, WA) equipped with a CoolSnap CCD camera (Roper Scientific, Tucson, AZ). Z-sections (n = 30–40) were acquired at 0.2-μm steps using a 100×, 1.3 NA Olympus U-PlanApo objective with 1 × 1 binning (Melville, NY). For analysis of microtubules and spatial localization within the kinetochore, images were deconvolved using the DeltaVision software. Measurements of the intensity of kinetochore localization were conducted on nondeconvolved images. All images for a specific experiment used identical exposure settings and scaling. Quantitative analysis of kinetochore localization intensities were performed using MetaMorph software as described previously (Universal Imaging, West Chester, PA; Kline et al., 2006). Images of DT40 cells were collected with a cooled CCD camera (Cool Snap HQ, Photometrics Image Point; Roper Scientific, Woburn, MA) mounted on an Olympus IX71 inverted microscope with a 60× objective lens (PlanApo 60×, NA 1.40) together with a filter wheel. Images were analyzed with an IPLab software (Signal Analytics, Vienna, VA) or DeltaVision deconvolution system (Applied Precision).
RESULTS
Vertebrate KNL1 Localizes to Kinetochores During Mitosis and Is Required for Chromosome Alignment
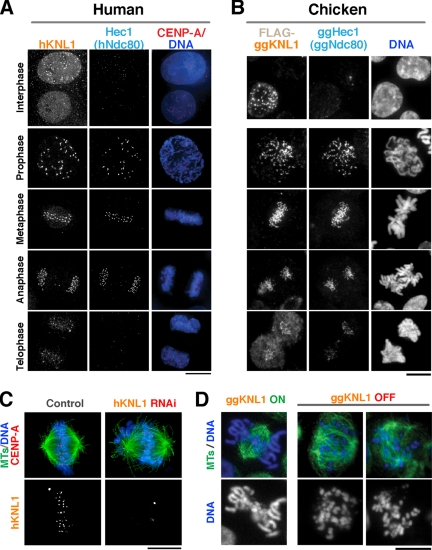
We previously demonstrated that transiently transfected GFP-hKNL1 localizes to human kinetochores during mitosis (Cheeseman et al., 2004). To monitor localization of hKNL1 and ggKNL1 throughout the cell cycle, we generated affinity-purified antibodies against amino acids 1413-1624 of hKNL1 and a DT40 chicken cell line stably expressing FLAG-ggKNL1. The FLAG-ggKNL1 fusion is fully functional based on its ability to rescue the depletion of untagged ggKNL1 (not shown). Immunofluorescence analysis indicated that hKNL1 and ggKNL1 localize to kinetochores throughout mitosis, but begin to disappear from kinetochores during telophase (Figure 1, A and B). Localization was also observed in a subset of interphase cells, which likely correspond to cells in G2. KNL1 exhibited similar temporal localization to subunits of the human Mis12 complex (not shown; (Goshima et al., 2003; Kline et al., 2006); both KNL1 and the Mis12 complex localize to kinetochores before the Ndc80 complex subunit Ndc80HEC1 (Figure 1, A and B; top row).
Figure 1.
Vertebrate KNL1 localizes to kinetochores during mitosis and is required for chromosome segregation. hKNL1 and ggKNL1 localize to kinetochores during mitosis. (A) Images showing hKNL1, Hec1 (hNdc80), stably expressed GFP-CENP-A (red), and DNA (blue) throughout the cell cycle in human cells. (B) Images showing FLAG-ggKNL1, ggHEC1 (ggNdc80), and DNA throughout the cell cycle. (C) hKNL1 depletion results in chromosome misalignment. Immunofluorescence images of control siRNA-transfected or hKNL1 RNAi-transfected cells stained for microtubules (green), GFP-CENP-A (red), DNA (blue), and hKNL1. (D) ggKNL1 depletion results in chromosome misalignment. Immunofluorescence images of conditional ggKNL1 DT40 cell lines either in the absence of tetracycline (ON) or 36 h after the addition of tetracycline (OFF) showing microtubules and DNA. See Supplementary Figure 1 for details on the cell line and extent of depletion. Scale bars, 10 μm.
To assess the role of vertebrate KNL1 in chromosome segregation, we depleted hKNL1 from HeLa cells using RNAi. Depletion was heterogeneous within the population, and thus we restricted our analysis to cells that were visibly depleted on the basis of immunofluorescence. In all hKNL1-depleted cells analyzed in this article, there was no detectable localization of hKNL1 above background; control cells processed in parallel and imaged at identical exposure showed high signal (60–90% of camera pixel saturation) at kinetochores. As a complementary approach to RNAi, we also generated a conditional knockout for ggKNL1 in chicken DT40 cells (Supplementary Figure 1). ggKNL1 is essential for viability. The inviable ggKNL1 deletion was maintained by a cDNA expressed from a tetracycline repressible promoter. After addition of tetracycline, ggKNL1 was depleted to undetectable levels on immunoblots within 18 h (Supplementary Figure 1).
Analysis of hKNL1-depleted cells indicated that many chromosomes were able to align in the middle of the cell. However, misaligned chromosomes were always detected at spindle poles or in an inappropriate orientation relative to the spindle axis (Figure 1C). This phenotype is reminiscent of that previously described after depletion of components of the hMis12 complex (Obuse et al., 2004; Kline et al., 2006), or human CENP-H/I/K (Okada et al., 2006), but is much less severe than depletion of the hNdc80 complex (DeLuca et al., 2002). Depletion of ggKNL1 also resulted in severe consequences in chromosome alignment and penetrant cell lethality (Figure 1D; Supplementary Figure 1). These results indicate that KNL1 is essential for cell viability and proper chromosome segregation in vertebrates.
KNL1 Does not Contribute to Localization of Constitutive Centromere Proteins But Is Required to Localize CENP-F and Zwint
We next examined the localization of a diverse collection of kinetochore proteins after hKNL1 RNAi. Although the CCAN has not been identified in C. elegans, previous work has demonstrated an important role for its subunits, especially of the CENP-H/I/K subgroup, in kinetochore assembly in budding yeast, fission yeast, and vertebrates (De Wulf et al., 2003; Kerres et al., 2006; Liu et al., 2006; Okada et al., 2006). Thus, we examined the same range of kinetochore proteins in cells where CENP-K levels were reduced using previously established RNAi conditions (Okada et al., 2006). We also analyzed conditional DT40 knockout cell lines of ggKNL1, ggCENP-K, and ggCENP-H. To detect the localization of different kinetochore proteins, we used a combination of affinity-purified antibodies and clonal cell lines stably expressing GFP fusions. The monitored fusion proteins are at least partially functional based on their similar localization to the endogenous protein and their copurification in multisubunit kinetochore subcomplexes (Cheeseman et al., 2004; Kops et al., 2005; Kline et al., 2006; Okada et al., 2006).
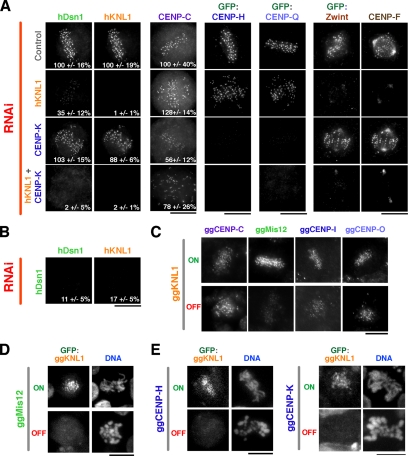
Depletion of hKNL1 abolished the localization of the outer kinetochore proteins Zwint and CENP-F and reduced the localization of hDsn1, a component of the Mis12 complex (Figure 2A). However, there was no significant effect observed on the localization of CENP-A, -C, -H, or -Q. CENP-Q is in a distinct subgroup of the CCAN from CENP-H/I/K (Okada et al., 2006). Similarly, ggKNL1 depletion reduced the localization of ggMis12, but did not strongly affect ggCENP-C, -I, or -O (Figure 2C). CENP-I and -O are present in the same subgroups of the CCAN as CENP-H and -Q, respectively. Finally, both hKNL1 and ggKNL1 require subunits of the Mis12 complex (hDsn1 or ggMis12, respectively) for their localization to kinetochores (Figure 2, B and D), similar to what is observed in C. elegans (Cheeseman et al., 2004).
Figure 2.
Localization of kinetochore proteins after KNL1/CENP-H/IK inhibitions. (A) Immunofluorescence images acquired using antibodies to hDsn1, hKNL1, CENP-C, and CENP-F, or using anti-GFP antibody in cells stably expressing GFP-CENP-H, GFP-CENP-Q, and GFP-Zwint. Control cells, hKNL1-depleted cells, CENP-K–depleted cells and cells doubly depleted for CENP-K and hKNL1 are shown. (B) hDsn1-depleted cells costained for hDsn1 and hKNL1 as in A. In A and B, when partial defects in localization were evident, kinetochore fluorescence intensity was measured relative to control cells. The number in bottom right represents the average kinetochore fluorescence intensity ± SD expressed as a percentage relative to control cells. Intensities were measured for between four and six cells, with 30–80 kinetochores per cell for each condition. (C) Immunofluorescence images of conditional ggKNL1 DT40 cell lines either in the absence of tetracycline (ON) or 36 h after the addition of tetracycline (OFF), showing ggCENP-C, ggMis12, ggCENP-I, or ggCENP-O localization detected using specific antibodies. (D) Immunofluorescence images of conditional ggMis12 DT40 cell lines either in the absence of tetracycline (ON) or 36 h after the addition of tetracycline (OFF), showing localization of a stably transfected GFP-ggKNL1 fusion protein. (E). Immunofluorescence images of conditional ggCENP-H and ggCENP-K DT40 cell lines either in the absence of tetracycline (ON) or 36 h after the addition of tetracycline (OFF), showing localization of a stably transfected GFP-ggKNL1 fusion protein. Scale bars, 10 μm.
CENP-K RNAi in human cells resulted in the complementary pattern to hKNL1 depletion; localization of CENP-H and -Q was abolished, whereas localization of hKNL1, CENP-F or Zwint was unaffected (Figure 2A). CENP-K depletion reduced the kinetochore levels of CENP-C in agreement with prior analysis of the effect of CENP-H and -K depletions on CENP-C localization in interphase cells (Fukagawa et al., 2001; Kwon et al., 2007) and with the close association of CENP-H/I/K with CENP-C (Foltz et al., 2006; Izuta et al., 2006; Figure 2A). Because CENP-H and CENP-Q localization is strongly affected after CENP-K RNAi (Figure 2A; also see quantitative analysis in Okada et al., 2006), this suggests that the RNAi conditions used here do significantly reduce CENP-K levels. However, we do not currently have the means to directly examine the depletion of CENP-K in human cells. In contrast to what we observed here, a previous study found that RNAi-mediated depletion of CENP-I did affect CENP-F localization (Liu et al., 2003). In addition, in chicken cells CENP-K or -H depletion reduced the localization of a GFP-ggKNL1 fusion protein (Figure 2E). Preliminary results in chicken cells also indicate that the localization of ggMis12 to mitotic kinetochores is reduced after CENP-H or -K depletion (Hori and Fukagawa., unpublished observations), in contrast to the normal hDsn1 localization observed after CENP-K RNAi in human cells (Figure 2A). In light of these observations, we acknowledge the possibility that the CENP-K knockdown in HeLa cells in this study may be partial. All the experiments including CENP-K RNAi are interpreted taking this point into consideration.
In the human cell RNAi experiments, hKNL1 appears to be significantly depleted because both endogenous hKNL1 and endogenous CENP-F signals were eliminated. CENP-K is at least partially depleted with clear consequences on localization of its closely associated CCAN subunits, but no significant effect on hKNL1, CENP-F, or Zwint localization. Under these conditions, largely nonoverlapping protein groups downstream of CENP-A and -C depend on hKNL1 or CENP-K for their kinetochore localization. This finding predicts that codepletion of hKNL1 and CENP-K should prevent localization of all of the components tested above except for CENP-A and -C. This was indeed the case: no localization of hDsn1, CENP-H, CENP-Q, Zwint, or CENP-F was observed after the double RNAi against hKNL1 and CENP-K (Figure 2A). The dependency relationships for kinetochore localization observed in this work, together with the relationships defined by other recent studies, are summarized in Figure 5.
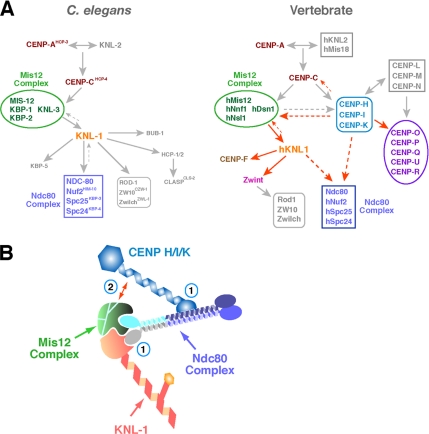
Figure 5.
Models for kinetochore assembly and synergistic contribution of KNL-1 and CENP-H/I/K to Ndc80 complex localization. (A) Diagram of kinetochore assembly hierarchies in C. elegans embryos and vertebrate cells. Red arrows indicate dependencies defined in this study. These schematics are not comprehensive and are focused primarily on stably associated proteins/complexes that do exhibit rapid turnover at kinetochores. (B) Schematic of potential mechanisms explaining the synergistic defect in Ndc80 complex localization in hKNL1/CENP-K double-depleted cells. The “two hand” model (1) is based on distinct sites for association of KNL1 and CENP-H/I/K on the four-subunit Ndc80 complex. Loss of a single association site would weaken, but not abolish, localization. Biochemical and two-hybrid studies provide some support for this idea. Partial localization interdependence between CENP-H/I/K and the Mis12 complex (2), which is known to affect Ndc80 complex localization in both C. elegans and vertebrates, may also indirectly contribute to the observed synergistic role of CENP-K and hKNL1 in Ndc80 complex localization. These two proposals are not mutually exclusive, and both are likely to contribute to some degree in vertebrates to ensure proper Ndc80 complex localization and function.
hKNL1 and CENP-K Act Coordinately to Recruit the Ndc80 Complex to the Outer Kinetochore
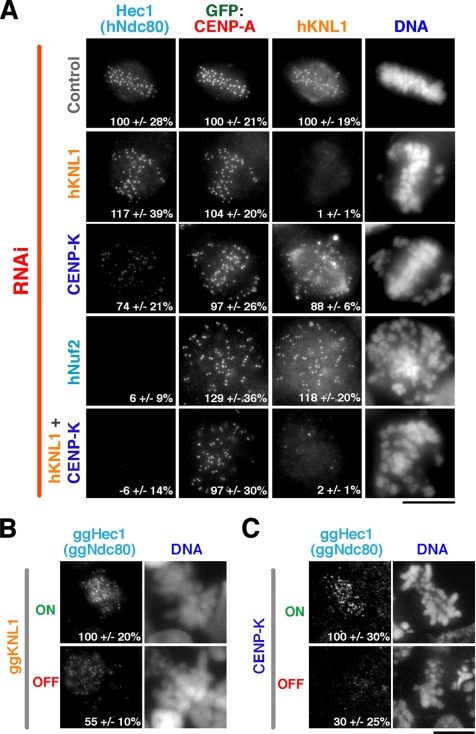
The Ndc80 complex plays a critical role in interactions with spindle microtubules at kinetochores (Cheeseman et al., 2006; DeLuca et al., 2006; Wei et al., 2007). Thus, it is of fundamental importance to properly recruit the Ndc80 complex during kinetochore assembly. The Ndc80 complex localizes external to KNL1 and CENP-K within the kinetochore for a reduced window of the cell cycle (Figure 1, A and B; Okada et al., 2006). In addition, the Ndc80 complex physically associates with hKNL1 and the Mis12 complex within the KMN network (Cheeseman et al., 2004; Obuse et al., 2004) and has been reported to associate with the CENP-H subunit of the CCAN (Mikami et al., 2005; Okada et al., 2006). In C. elegans, where the equivalent of the CENP-H/I/K protein group is likely missing, depletion of KNL-1 abolishes the localization of the Ndc80 complex. These prior results left open the question as to how KNL1 influences kinetochore localization of the Ndc80 complex in vertebrates. We found that hNdc80 localized at normal levels to kinetochores in human cells throughout mitosis after hKNL1 depletion (Figure 3A; data not shown). CENP-K RNAi reduced, but did not abolish, hNdc80 localization (Figure 3A; Okada et al., 2006). A reduction in ggNdc80 localization was observed in both ggKNL1 and ggCENP-K depleted chicken cells (Figure 3, B and C; Okada et al., 2006), although a more severe reduction was observed after ggCENP-K depletion, consistent with prior work (Liu et al., 2006). Thus, these results demonstrate that, unlike in C. elegans, depletion of KNL1 in vertebrates does not abolish kinetochore localization of the Ndc80 complex.
Figure 3.
KNL1 and CENP-K act coordinately to recruit the Ndc80 complex to kinetochores. (A) Immunofluorescence images acquired using antibodies against Hec1 (hNdc80), hKNL1, and an anti-GFP antibody in cells stably expressing GFP-CENP-A. Control cells, hKNL1-depleted cells, CENP-K–depleted cells, hNuf2-depleted cells, and cells doubly depleted for CENP-K and hKNL1 are shown. Kinetochore fluorescence intensities were quantified in all channels and the mean ± SD, expressed as a percentage relative to control cells, is indicated in the bottom right. The fluorescence intensity of 30–80 kinetochores per cell was measured for between 12 and 23 cells per condition. (B and C) Immunofluorescence images showing ggHec1 (ggNdc80) localization in conditional ggKNL1 (B) or ggCENP-K (C) DT40 cell lines either in the absence of tetracycline (ON) or 36 h after the addition of tetracycline (OFF). Numbers in panels indicate mean ± SD in kinetochore fluorescence intensity of ggHec1 (ggNdc80) relative to the control ON cells. Scale bars, 10 μm.
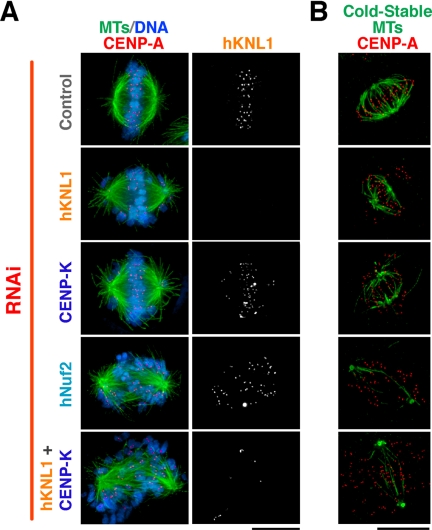
The results above prompted us to test the consequences of hKNL1 and CENP-K double RNAi on hNdc80 localization. Simultaneously reducing levels of both proteins abolished Ndc80 localization to the same extent as depletion of Nuf2, a core subunit of the Ndc80 complex (Figure 3A). Consistent with the lack of detectable hNdc80 complex localization in the double RNAi cells, severe defects in chromosome segregation were observed that were significantly worse than the individual RNAi treatments (Figure 4A). Indeed, the pattern of chromosome distribution, kinetochore orientation, and the absence of cold stable kinetochore fibers were similar to that observed after depletion of the Ndc80 complex subunit Nuf2 (Figure 4, A and B). We additionally observed significant cell lethality after codepletion of hKNL1 and CENP-K, with the majority of cells lifting from the coverslip as corpses 48 h after RNAi treatment, and interphase cells with multiple micronuclei were also evident (not shown). These defects were not the consequence of the RNAi treatment procedure, because codepletion of hKNL1 and CENP-Q (which is present in a different subgroup of the CCAN and does not affect Ndc80 complex localization) resulted in defects that were largely identical to the hKNL1 depletion alone (not shown).
Figure 4.
Simultaneous depletion of hKNL1 and CENP-K results in catastrophic defects in chromosome segregation. (A) Microtubules (green), DNA (blue), CENP-A (red), and hKNL1 in control cells, hKNL1-depleted cells, CENP-K–depleted cells, hNuf2-depleted cells, and cells doubly depleted for CENP-K and hKNL1. (B) Microtubules (green) and CENP-A (red) staining after the indicated cells were incubated at 0°C for 10 min to selectively visualize cold stable kinetochore microtubule fibers. Scale bars, 10 μm.
In total, these results indicate that hKNL1 and CENP-K act coordinately during vertebrate kinetochore assembly. Simultaneously reducing the function of both results in catastrophic defects in kinetochore assembly with no detectable localization of any of the seven tested components downstream of CENP-A and -C. This defect in kinetochore assembly resulted in severe chromosome segregation defects.
DISCUSSION
In this article, we show that vertebrate KNL1 makes important contributions to kinetochore assembly and chromosome segregation. We note that other recent work on human KNL1 also indicates an important role for KNL1 in connecting to the mitotic checkpoint (Kiyomitsu et al., 2007). Our results reveal that KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrate cells. Combined with other recent work (Kline et al., 2006; Liu et al., 2006; Okada et al., 2006), these results lead to a model for assembly of the vertebrate kinetochore depicted in Figure 5A. For comparison, the kinetochore assembly hierarchy established in C. elegans is also depicted.
On the basis of previous work in C. elegans (Desai et al., 2003; Cheeseman et al., 2004) and the close biochemical association between KNL-1 and the Ndc80 complex that is observed in C. elegans, fission yeast, Drosophila, and humans, we expected vertebrate KNL1 to function upstream of the Ndc80 complex. In agreement with this expectation, in the DT40 chicken cell conditional knockout analysis, we observed a twofold reduction in Ndc80 levels at the kinetochore after KNL1 depletion. However, in both the chicken cell analysis and in the human RNAi experiments significant amounts of Ndc80 were still able to localize to kinetochores after KNL1 depletion. Our work suggests that at least one explanation for this is a contribution from the CENP-H/I/K complex to the targeting of the Ndc80 complex. The CENP-H/I/K complex is likely to be upstream of KNL1 in vertebrate kinetochore assembly and the analysis in chicken cells with genetic CENP-H or -K deletions is consistent with this notion. However, Ndc80 complex targeting downstream of the CENP-H/I/K complex does not appear to work exclusively via KNL1 in vertebrates. This finding is supported not only by the localization analysis in human and chicken cells but also by the dramatic synergy in phenotypic defects in the KNL1/CENP-K double RNAi experiments. Work in fission yeast is also consistent with this conclusion as discussed further below.
Thus far, despite genome-wide RNAi screening and extensive biochemical analysis, we have been unable to identify C. elegans counterparts to any of the subunits of the CCAN, with the exception of CENP-C. In light of the results presented here, the apparent absence of the CENP-H/I/K group proteins is consistent with KNL-1 being absolutely required for Ndc80 complex localization in C. elegans. Because C. elegans chromosomes are holocentric, with diffuse kinetochores assembling along the length of each sister chromatid, one possibility is that loss of the CCAN is a necessary prerequisite for holocentricity. However, with the exception of CENP-C, no counterparts to the CCAN have been found to date in Drosophila, which has monocentric chromosomes. The reported dependency of Ndc80 complex localization on the KNL1 ortholog in Drosophila (Przewloka et al., 2007) suggests that, as in C. elegans, the CCAN protein group may have been lost in this lineage, further work is needed to test if this indeed the case.
The CENP-H/I/K class of proteins and other components of the CCAN are present in fungi, albeit weakly conserved, suggesting that they arose early in eukaryotic evolution and have been lost in specific lineages. Recent work in fission yeast demonstrated that the KNL-1 homologue Spc7 is not required for Ndc80 localization, but also reported synthetic lethal genetic interactions between spc7 and mis6 (CENP-I) or sim4 (CENP-K) mutants (Kerres et al., 2004), consistent with the synergistic defects we describe here for hKNL1/CENP-K double inhibitions. Overall, these results suggest evolutionary plasticity in kinetochore assembly, specifically in the recruitment of the critical Ndc80 complex. Because orthologues of KNL1, the Ndc80 complex and the Mis12 complex—the three parts of the KMN network—are found throughout eukaryotes and are essential in all systems where they have been tested, whereas the CCAN appears to have been lost in specific lineages, the future challenge will be to elucidate the mechanism of CCAN action and the reason for its dispensability in specific organisms.
There are several possibilities that could explain the synergistic defect in Ndc80 complex localization after hKNL1 and CENP-K inhibitions in human cells. KNL1 and CENP-H/I/K proteins may act in parallel pathways to direct Ndc80 localization or may function coordinately in intersecting pathways. The reduction in KNL1 and Mis12 complex localization after CENP-K depletion in chicken cells as well as the reduction in CENP-H/I/K and KNL1 localization after Mis12 complex inhibition is more consistent with the latter. In addition to providing a structural scaffold for the association of the Ndc80 complex, these proteins sets could also function via a regulatory mechanism directing Ndc80 complex localization to kinetochores. On the basis of the physical associations that have been previously reported for these proteins, we speculate that one possible explanation for our results is that hKNL1 and the CENP-H/I/K subgroup of the CCAN directly associate with distinct regions of the four-subunit Ndc80 complex (Figure 5B). KNL1 is likely to associate with the Spc24/Spc25 dimer that is oriented toward the inner kinetochore, as suggested by reconstitution experiments with C. elegans proteins (Cheeseman et al., 2006), whereas CENP-H/I/K may associate with the Ndc80/Nuf2 dimer that is projecting out toward the spindle microtubules, as suggested by two-hybrid analysis in chickens (Mikami et al., 2005). In this scenario, loss of either hKNL1 or CENP-K would partially compromise Ndc80 complex localization, whereas loss of both would dramatically inhibit localization. Such a “two-hand” mechanism is evolutionarily plastic, in the sense that one interaction may be compensated for by a change in the affinity of the other. We note that interactions with KNL-1 modulate the microtubule-binding activity of the C. elegans Ndc80 complex (Cheeseman et al., 2006). Thus, although a substantial amount of Ndc80 complex persists at kinetochores after depletion of KNL1, its function in generating interactions with microtubules may be compromised. Such a “two-hand” mechanism should be testable in future work using reconstituted complexes in vitro.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Reto Gassman, Paul Maddox, and Defne Yarar for helpful discussions. This work was supported by grants from the National Institutes of Health to A.D. (R01-GM074215), funding from the Ludwig Institute for Cancer Research to A.D., and by Grants-in-Aid for Scientific Research on Priority Areas “Cancer Cell Biology,” “Cell Cycle,” and “Nuclear Dynamics” from the Ministry of Education, Science, Sports, and Culture of Japan to T.F. I.M.C. was a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. A.D. is the Connie and Bob Lurie Scholar of the Damon Runyon Cancer Research Foundation (DRS 38-04).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1051) on November 28, 2007.
REFERENCES
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., III, Oegema K., Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A., Salmon E. D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Moree B., Hickey J. M., Kilmartin J. V., Salmon E. D. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Rybina S., Muller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L.E.T., Black B. E., Bailey A. O., Yates J. R., Cleveland D. W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Pendon C., Morris J., Brown W. CENP-C is necessary but not sufficient to induce formation of a functional centromere. EMBO J. 1999;18:4196–4209. doi: 10.1093/emboj/18.15.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu T., Yoda K., Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- Kerres A., Jakopec V., Beuter C., Karig I., Pohlmann J., Pidoux A., Allshire R., Fleig U. Fta2, an essential fission yeast kinetochore component, interacts closely with the conserved Mal2 protein. Mol. Biol. Cell. 2006;17:4167–4178. doi: 10.1091/mbc.E06-04-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerres A., Vietmeier-Decker C., Ortiz J., Karig I., Beuter C., Hegemann J., Lechner J., Fleig U. The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore-spindle association. Mol. Biol. Cell. 2004;15:5255–5267. doi: 10.1091/mbc.E04-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. Human Blinkin/AF15q14 is Required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Kline S. L., Cheeseman I. M., Hori T., Fukagawa T., Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G. J., Kim Y., Weaver B. A., Mao Y., McLeod I., Yates J. R., 3rd, Tagaya M., Cleveland D. W. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M. S., Hori T., Okada M., Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-T., Rattner J. B., Jablonski S. A., Yen T. J. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. T., Hittle J. C., Jablonski S. A., Campbell M. S., Yoda K., Yen T. J. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 2003;5:341–345. doi: 10.1038/ncb953. [DOI] [PubMed] [Google Scholar]
- Liu X., McLeod I., Anderson S., Yates J. R., 3rd, He X. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami Y., Hori T., Kimura H., Fukagawa T. The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol. Cell. Biol. 2005;25:1958–1970. doi: 10.1128/MCB.25.5.1958-1970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V. S., Smith M. A., Peak-Chew S., Kilmartin J. V. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:4931–4946. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi T., Hiraoka Y., Ikemura T., Regnier V., Dodson H., Earnshaw W. C., Fukagawa T. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2002;2:463–476. doi: 10.1016/s1534-5807(02)00144-2. [DOI] [PubMed] [Google Scholar]
- Obuse C., Iwasaki O., Kiyomitsu T., Goshima G., Toyoda Y., Yanagida M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 2004;6:1135–1141. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1225. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Przewloka M. R., Zhang W., Costa P., Archambault V., D'Avino P. P., Lilley K. S., Laue E. D., McAinsh A. D., Glover D. M. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE. 2007;2:e478. doi: 10.1371/journal.pone.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R. R., Al-Bassam J., Harrison S. C. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.