Abstract
The ESCRT protein complexes are recruited from the cytoplasm and assemble on the endosomal membrane into a protein network that functions in sorting of ubiquitinated transmembrane proteins into the multivesicular body (MVB) pathway. This transport pathway packages cargo proteins into vesicles that bud from the MVB limiting membrane into the lumen of the compartment and delivers these vesicles to the lysosome/vacuole for degradation. The dissociation of ESCRT machinery by the AAA-type ATPase Vps4 is a necessary late step in the formation of MVB vesicles. This ATP-consuming step is regulated by several Vps4-interacting proteins, including the newly identified regulator Ist1. Our data suggest that Ist1 has a dual role in the regulation of Vps4 activity: it localizes to the ESCRT machinery via Did2 where it positively regulates recruitment of Vps4 and it negatively regulates Vps4 by forming an Ist1-Vps4 heterodimer, in which Vps4 cannot bind to the ESCRT machinery. The activity of the MVB pathway might be in part determined by outcome of these two competing activities.
INTRODUCTION
Multivesicular bodies (MVBs) are endosomal structures that function in the transport of proteins between trans-Golgi, plasma membrane, and lysosomes/vacuoles. One route of traffic mediated by MVBs is the delivery of transmembrane proteins to the lumen of lysosomes/vacuoles for degradation (for review, see Gruenberg and Stenmark, 2004; Piper and Katzmann, 2007). This route requires the formation of cargo-containing vesicles that bud into the lumen of MVBs. On fusion with the lysosome/vacuole the MVB vesicles are exposed to hydrolytic enzymes that degrade both lipids and proteins. The MVB pathway represents the major protein degradation pathway for plasma membrane proteins and therefore plays an important role in regulating numerous cell surface–mediated processes, including cell signaling, nutrient uptake, and adaptation to environmental conditions.
Cargo sorting and vesicle formation in the MVB pathway are thought to be executed by a group of protein complexes called ESCRT-I, -II, and -III (endosomal sorting complex required for transport). These protein complexes are transiently recruited from the cytoplasm to the endosomal membrane where they bind transmembrane proteins previously marked for degradation by monoubiquitination. The current working model predicts that the ESCRTs concentrate the ubiquitinated proteins and package this cargo into forming vesicles that bud into the lumen of the compartment (for review, see Babst, 2005; Hurley and Emr, 2006; Williams and Urbe, 2007). The mechanism of membrane deformation and vesicle budding remains unclear. One hypothesis suggests that MVB vesicle formation is intimately linked to the assembly of ESCRT-III, a complex composed of at least four subunits (Vps2, Vps24, Vps20, Snf7). The ESCRT-III subunits appear to assemble on the membrane into a large protein network that restricts cargo diffusion and might deform the membrane (Babst et al., 2002; Muziol et al., 2006; Shim et al., 2007).
The disassembly of ESCRT-III is an essential step in the MVB vesicle formation and is executed by the AAA-type ATPase Vps4 (Babst et al., 1998, 2002). Without Vps4 function the ESCRT machinery remains associated with the endosome and MVB formation is blocked. The ATPase activity of Vps4 is regulated by its oligomeric state (Babst et al., 1998; Scott et al., 2005a). As a dimer, Vps4 is inactive and localizes mainly to the cytoplasm. ESCRT-III recruits Vps4 dimers by binding to the N-terminal microtubule interacting and trafficking microtubule-interacting and transport (MIT) domain of Vps4. ESCRT-III associated Vps4 oligomerizes into a large ring structure of 12 subunits. This large Vps4 oligomer hydrolyzes the bound ATP, resulting in the disassembly of ESCRT-III and the dissociation of Vps4 into ADP-bound dimers that recycle back to the cytoplasm.
Two MVB-associated proteins, Vta1 and Did2 (also known as Fti1/Vps46), play distinct roles in regulating Vps4 activity. Vta1 interactions with Vps4 have been demonstrated both in vitro and in vivo (Yeo et al., 2003; Shiflett et al., 2004; Azmi et al., 2006; Lottridge et al., 2006). Biochemical studies indicate a positive regulatory role for Vta1 in promoting the oligomeric assembly of Vps4 and stimulating its ATPase activity (Azmi et al., 2006). Did2 associates with ESCRT-III and binds to both Vps4 and Vta1 (Howard et al., 2001; Lottridge et al., 2006; Nickerson et al., 2006). Loss of Did2 results in the accumulation of ESCRT-III components on the endosomal membrane (Nickerson et al., 2006). These data point toward a role for Did2 in coordinating the disassembly of ESCRT-III from endosomal membranes via Vps4.
In this study we provide evidence for an additional regulator of Vps4 activity, Ist1. By interacting with Did2 and Vps4, Ist1 appears to regulate the recruitment and oligomerization of Vps4. We propose that Ist1, Did2, and Vta1 form a network of interconnected regulatory proteins that modulate Vps4 activity, thereby regulating the flow of cargo through the MVB pathway.
MATERIALS AND METHODS
Materials
HA-tag (hemagglutinin-tag) specific antibody was purchased from Covance (Princeton, NJ). The antisera against Snf7 and Vps24 have been characterized previously (Babst et al., 1998).
Strains and Media
Table 1 lists the Saccharomyces cerevisiae strains used in this work. Yeast strains were grown in standard yeast extract-peptone-dextrose (YPD) or synthetic medium supplemented with essential amino acids as required for maintenance of plasmids (YNB; Sherman et al., 1979). To construct deletion strains, yeast cells were transformed with DNA fragments containing the HIS3 or URA3 gene flanked by 50 base pairs specific for the 5′ and 3′ region of the corresponding gene. The cells were selected for the presence of the marker gene and the deletions were confirmed by PCR analysis of the chromosomal DNA.
Table 1.
S. cerevisiae strains and plasmids used in this study
| Strain or plasmid | Descriptive name | Genotype or description | Reference or source |
|---|---|---|---|
| Strain | |||
| MBY41 | vps4Δvps2Δ | MBY3, VPS2::HIS3 | Babst et al. (2002) |
| EEY26-1 | vps4Δdid2Δ | MBY3, DID2::HIS3 | This study |
| MBY12 | vps4Δvps24Δ | MBY3, VPS24::HIS3 | Babst et al. (2002) |
| EEY17 | did2Δ | SEY6210, DID2::HIS3 | This study |
| JPY47 | vta1Δ | SEY6210.1, VTA1::HIS3 | Azmi et al. (2006) |
| EEY12 | vps4Δdid2Δ | MBY3, SNF7::HIS3 | Babst et al. (2002) |
| MBY63 | ist1Δ | SEY6210, IST1::HIS3 | This study |
| MCY2 | vta1Δist1Δ | SEY6210, VTA1::HIS3, IST1::HIS3 | This study |
| MCY3 | vps4Δist1Δ | MBY3, IST1::HIS3 | This study |
| CAY2 | did2Δist1Δ | EEY17, IST1:URA3 | This study |
| MBY3 | vps4Δ | SEY6210, VPS4::TRP1 | Babst et al. (1997) |
| MCY31 | did2Δ | W303, DID2::HIS3 | This study |
| W303 | WT | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | Thomas and Rothstein (1989) |
| SEY6210 | WT | MATα leu2-3,112ura3-52his3-Δ200trp1-Δ901lys2-801suc2-Δ9 | Robinson et al. (1988) |
| Plasmids | |||
| pGO45 | GFP-CPS | URA3 ApR (pRS426) GFP-CPS1 | Odorizzi et al. (1998) |
| pCS24 | Ste2-GFP | URA3 ApR (pRS426) STE2-GFP | Odorizzi et al. (1998) |
| pMB241 | IST1-HA | URA3 ApR (pRS416) IST1-HA | This study |
| pMB243 | IST1-GFP | URA3 ApR (pRS416) IST1-GFP | This study |
| pMB287 | GST-IST1(CC) | ApR (pGEX-KG) GST-IST1(CC) | This study |
| pCD2 | GST-IST1 | ApR (pGEX-KG) GST-IST1 | This study |
| pCJ7 | IST1(ΔN)-GFP | URA3 ApR (pRS416) IST1(ΔN)-GFP | This study |
| pAH26 | 2 μDID2-HA | URA3 ApR (pRS425) DID2-HA | This study |
| pMB149 | vps4(E233Q)-HA | URA3 ApR (pRS416) vps4(E233Q)-HA | Babst et al. (2002) |
| pMB283 | VPS4-HA | URA3 ApR (pRS416) VPS4-HA | This study |
| pAH27 | DID2(ΔC58)-HA | LEU2 ApR (pRS425) DID2(ΔC58)-HA | This study |
| pAH25 | DID2(ΔC29)-HA | LEU2 ApR (pRS415) DID2(ΔC29)-HA | This study |
| pMB65 | vps4-ts | LEU2 ApR (pRS415) vps4-ts2–29 | This study |
| pMB286 | P(GAL1)-IST1 | TRP1 ApR (pRS414) P(GAL1)-IST1 | This study |
| pMB277 | P(GAL1)-IST1-HA | URA3 ApR (pRS416) P(GAL1)-IST1-HA | This study |
DNA Manipulations
Plasmids used in this study are listed in Table 1. The pRS shuttle vectors used in this study have been described previously (Christianson et al., 1992). The IST1 gene including the promoter region was amplified by PCR from SEY6210 chromosomal DNA and fused either to a 3-HA tag or to the green fluorescent protein (GFP) containing NheI/SalI fragment of pEGFP-C1 (Clontech Laboratories, Palo Alto, CA), resulting in the gene fusions IST1-HA (pMB241) and IST1-GFP (pMB243). The IST1 gene was amplified by PCR and inserted into the BamHI/SalI digested Escherichia coli expression vector pGEX-KG, resulting in the plasmid pCD2. A SacI/HindIII fragment of pMB241 encoding the C-terminal coiled-coil domain of Ist1 fused to 3-HA was inserted into the SacI/HindIII sites of pGEX-KG, resulting in the plasmid pMB287 [GST-IST1(CC)-HA]. pCJ7 was constructed by fusing a PCR product containing the 3′ region of IST1 with NheI/SalI fragment of pEGFP-C1 [IST1(ΔN)-GFP, see Figure 1]. The DID2 gene was amplified by PCR from genomic DNA, fused to the 3-HA tag and inserted into SacI/SalI of pRS426, resulting in the plasmid pAH26 (2μ DID2-HA). A SacI/SalI fragment of pMB76 (Babst et al., 1998) was inserted into pRS416 to obtain plasmid pMB283 (VPS4-HA). PCR products encoding the N-terminal 146 or 175 amino acids of Did2 were fused to 3-HA tags, resulting in pAH27 [DID2(ΔC58)-HA] and pAH25 [DID2(ΔC29)-HA], respectively. pMB65 was obtained by inserting the NotI/SalI fragment of pMB59 (Babst et al., 1997) into pRS415. PCR products containing either IST1 or IST1-HA were fused to the GAL1 promoter and inserted into pRS414 or pRS416 to obtain pMB286 [P(GAL1)-IST1] and pMB277 [P(GAL1)-IST1-HA].
Figure 1.
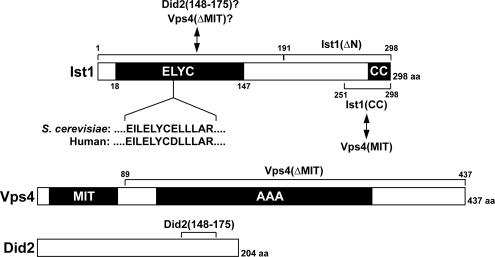
Domain structure of Ist1, Vps4, and Did2. The borders of the ELYC domain were determined based on domain prediction by Swissprot (DUF292). Arrows indicate interactions between Ist1 and Vps4 and between Ist1 and Did2. Interactions with the N-terminal region containing the ELYC domain are predicted based on the observation that deletion of this domain results in loss of Ist1 binding to Did2 and Vps4(ΔMIT), which are labeled with question marks.
Experimental Procedures
Immunofluorescence microscopy was performed on fixed spheroplasted cells as described (Babst et al., 1998). Fluorescence microscopy was performed on a deconvolution microscope (DeltaVision, Applied Precision, Issaquah, WA). Sephacryl S300 (16/60 column, GE Healthcare, Waukesha, WI) gel filtration analysis of yeast cell extract was performed as described in Curtiss et al. (2007). Superose 6 (3.2/30 column, GE Healthcare) gel filtration analysis of purified proteins was performed in presence of 1 mM ATP, 150 mM KAc, 20 mM HEPES, pH 7.4, and 5 mM MgAc2 at a flow rate of 40 μl/min. The method used for the ATPase activity tests was described previously (Babst et al., 1998). For subcellular fractionation experiments yeast was grown in 8 ml minimal medium to a density of OD600 = 0.6. The cells were harvested, washed with water, treated with 100 mM Tris, pH 9.4, 10 mM dithiothreitol for 10 min, and spheroplasted with oxolyticase in minimal medium containing 1 M sorbitol for 45 min. The spheroplasts were lysed by osmotic stress in 0.5 ml of 100 mM KCl, 50 mM KAc, 20 mM PIPES, pH 6.8, 5 mM MgAc2, 100 mM sorbitol, 0.1 mM AEBSF, and protease inhibitor cocktail (Complete, Roche Molecular Biochemicals, Indianapolis, IN). The cell extract was separated by centrifugation at 5000 × g for 10 min into a soluble and a pellet fraction. The proteins of the fractions were precipitated by the addition of 10% trichloroacetic acid (TCA). The resulting pellet was washed with acetone and resuspended in 0.25 ml SDS-PAGE sample buffer (2% SDS, 0.1 M Tris, pH 6.8, 10% glycerol, 0.01% bromophenol blue, 5% β-mercaptoethanol). Five microliters of each sample were used for Western blot analysis.
RESULTS
Identification of IST1
We identified the Ist1 gene product in a two-hybrid screen as a potential Vps4-interacting protein. Three different IST1 clones were isolated in this screen encoding either the full-length Ist1 protein or N-terminal truncated derivatives of it (data not shown). The shortest clone encoded the final 96 amino acids of Ist1, suggesting that this C-terminal region was responsible for the interaction between Vps4 and Ist1. IST1 (SGD: YNL265c) was previously identified in a study where increased sodium tolerance was observed in strains deleted for this gene (IST: Increased Sodium Tolerance; Entian et al., 1999).
The IST1 gene encodes a 298-amino acid soluble protein that is predicted to contain a C-terminal coiled-coil motif (predicted by Coiles, Lupas et al., 1991; Figure 1). Amino acid sequence comparison with other genomes indicated that Ist1 is conserved among all eukaryotes with the most conserved region within the first ∼150 amino acids of the protein. This region contains a highly conserved amino acid sequence, ELYC, which we used to name this putative domain (Figure 1).
Ist1 Binds to Vps4 and Prevents Vps4 Assembly
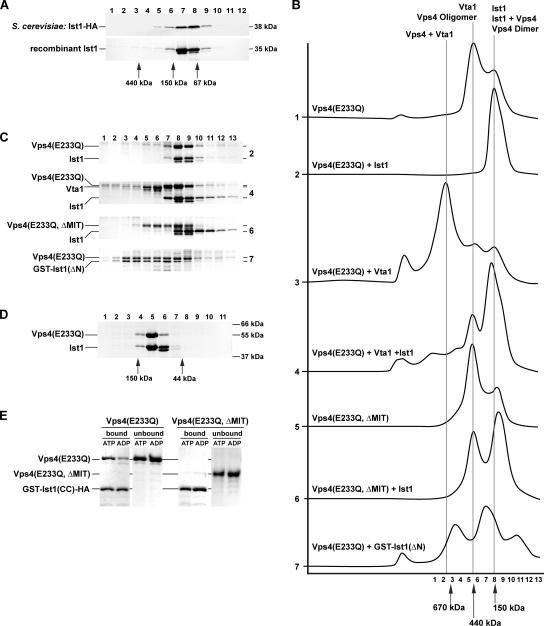
To characterize the potential interaction between Ist1 and Vps4 in vitro, we expressed IST1 in E. coli and purified the protein. The native molecular weight of recombinant Ist1 was determined by gel filtration analysis. Relative to protein standards Ist1 eluted from the column at ∼90 kDa, more than twice its predicted molecular weight of 35 kDa which suggested that Ist1 may dimerize (Figure 2A). To compare this result with endogenously expressed Ist1, we constructed a functional C-terminal HA-tagged version of Ist1 (Ist1-HA, Supplementary Figure 1), expressed the IST1-HA fusion in wild-type yeast, and analyzed its molecular weight by gel filtration. The result was consistent with the size observed with recombinant Ist1 (Figure 2A). Molecular weight determination by gel filtration is sensitive to the shape of the protein and thus the Ist1 data could be misleading. Therefore, analytical centrifugation experiments were performed using purified Ist1 at concentrations of 1, 0.5, and 0.25 mg/ml. These experiments indicated that at all three concentrations the majority of Ist1 is present as a monomer (measured MW/theoretical MW = 1.2). However, the analysis was complicated by the presence of higher molecular weight particles, suggesting that Ist1 has the tendency to oligomerize/aggregate. Together, the data from gel filtration and analytical centrifugation experiments suggested that Ist1 is a monomeric protein with a nonglobular, elongated structure.
Figure 2.
Ist1 binds to Vps4 and inhibits oligomerization of the ATPase. (A) Gel filtration analysis of cell extracts and purified recombinant protein using an S300-Sephacryl column. The resulting fractions were analyzed for the presence of Ist1-HA by Western blot and for the presence of the recombinant Ist1 by SDS-PAGE followed by Coomassie staining (top panel, extract from MBY63 pMB241; bottom panel, purified Ist1). (B) Superose S6 gel filtration analysis of purified recombinant proteins in presence of 1 mM ATP (X-axis, time; Y-axis, UV280 nm absorption). All proteins analyzed were present at 1 mg/ml in the loading sample. (C) SDS-PAGE analysis of the gel filtration fractions from the experiments in B. The sample number corresponds to the fraction number in B. The gel number indicates from which experiment in B the samples originate. (D) Gel filtration analysis using an S200-Sephacryl column of purified Ist1 and Vps4(E233Q) in presence of 1 mM ATP (1 mg/ml protein each). The fractions were analyzed by SDS-PAGE. (E) GST-Ist1(CC)-HA was immobilized on GSH-Sepharose and incubated with either Vps4(E233Q) or Vps4(E233Q, ΔMIT) in presence of 1 mM ATP or ADP. The bound and unbound fractions were analyzed by SDS-PAGE.
We studied the predicted Ist1-Vps4 interaction in vitro using recombinant Ist1 and ATPase-deficient Vps4(E233Q) protein (Figure 2B). Gel filtration analysis has shown that the ATPase-deficient mutant protein Vps4(E233Q) assembles in presence of ATP into a stable ∼500-kDa complex, representing the higher-order ATP-bound complex (no. 1 in Figure 2B; Babst et al., 1998). Interestingly, we found that the addition of equimolar amounts of Ist1 abolished Vps4 assembly and instead resulted in the formation of a Vps4-Ist1 complex that was <150 kDa (no. 2 in Figure 2, B and C). High-resolution gel filtration analysis of this complex indicated a native molecular weight of 100 kDa, suggesting that Ist1 (35 kDa) and Vps4 (48 kDa) may assemble into a hetero-dimer (Figure 2D).
Vta1 binds Vps4 as a positive regulator, promoting Vps4 assembly and ATPase activity. As previously shown, the addition of Vta1 to ATP-bound Vps4(E233Q) resulted in the formation of a large ∼1-MDa complex, (no. 3 in Figure 2B; Azmi et al., 2006). The void volume peak present in this experiment is an artifact of Vta1 aggregation. The presence of Ist1 not only inhibited the formation of the Vps4-Vta1 complex, but it also successfully competed with Vta1 for binding to Vps4. As a consequence, the gel filtration analysis showed the presence of free Vta1 in addition to the Vps4-Ist1 complex (no. 4 in Figure 2, B and C).
The N-terminal MIT domain of Vps4 interacts with ESCRT-III subunits and thus functions as the substrate-binding domain of Vps4 (Scott et al., 2005b; Vajjhala et al., 2007). Deletion of this domain does not interfere with the homotypic oligomerization or the ATPase activity of Vps4 (no. 5 in Figure 2B; Babst et al., 1998). Gel filtration analysis showed that Ist1 did not stably associate with Vps4(E233Q, ΔMIT) and thus did not block the oligomerization of the mutant ATPase (no. 6 in Figure 2, B and C). This result indicated that the MIT domain of Vps4 is required for the formation of a stable Ist1-Vps4 complex.
The two-hybrid data suggested that the C-terminal 96 amino acids of Ist1 were able to bind to Vps4. Therefore a glutathione S-transferase (GST)-fusion protein containing the C-terminal 107 amino acids of Ist1 was purified [GST-Ist1(ΔN)], mixed with Vps4(E233Q) and analyzed by gel filtration. The resulting chromatogram and SDS-PAGE data confirmed that the C-terminal region of Ist1 could bind Vps4 (no. 7 in Figure 2, B and C). However, in contrast to full-length Ist1, the binding of GST-Ist1(ΔN) to Vps4 did not inhibit oligomerization of Vps4. As a consequence, two protein peaks were observed: one at ∼250 kDa likely representing Vps4(E233Q) dimers bound to GST-Ist1(ΔN) and one at ∼600 kDa likely formed by the binding of GST-Ist1(ΔN) to oligomeric Vps4(E233Q). This result indicated that the C-terminal region of Ist1 binds to Vps4 but this interaction alone is not sufficient to inhibit Vps4 assembly.
Thus far our experiments had identified two interaction sites important for the formation of the Vps4-Ist1 complex: the MIT domain of Vps4 and the C-terminal 107 amino acids of Ist1. The MIT domain of Vps4 has been shown to bind to the C-terminal coiled-coil domains of ESCRT-III subunits (Scott et al., 2005b). Therefore we tested if the MIT domain might also bind to the C-terminal coiled-coil motif of Ist1 (see Figure 1). A GST fusion protein containing the coiled-coil domain of Ist1 [GST-Ist1(CC)-HA] was purified and immobilized on GSH (reduced glutathione)-Sepharose beads. An excess of ATPase-deficient proteins Vps4(E233Q) and Vps4(E233Q, ΔMIT) were added either in presence of ATP or ADP. The beads were washed and the bound and unbound fractions were analyzed by SDS-PAGE. The resulting gel illustrated that in presence of ATP Vps4(E233Q) bound to GST-Ist1(CC)-HA in an ∼1:1 ratio (Figure 2E). A control experiment under the same conditions demonstrated that Vps4(E233Q) did not interact with immobilized GST (data not shown). In the presence of ADP Vps4(E233Q) still interacted with GST-Ist1(CC)-HA but the affinity between the two proteins was diminished. In contrast the MIT-deleted Vps4 mutant protein did not bind to the C-terminus of Ist1 independent of the nucleotide binding state (Figure 2E). Together, the GST pulldown experiments indicated a direct interaction between the coiled-coil motif of Ist1 and the MIT domain of Vps4. This interaction was in part dependent on the nucleotide-binding state of Vps4.
In summary, the in vitro data demonstrated a direct interaction between Ist1 and Vps4, resulting in the formation of a heterodimeric Vps4-Ist1 complex. This complex was able to compete with both the oligomerization of Vps4 and the binding of the regulator Vta1. The interaction between the Vps4 MIT domain and the coiled-coil motif of Ist1 was required for the formation of a stable Vps4-Ist1 complex. However, this interaction alone was not sufficient to prevent Vps4 oligomerization, suggesting the presence of at least one more Vps4-Ist1 interaction site.
Ist1 Inhibits ATPase Activity of Vps4
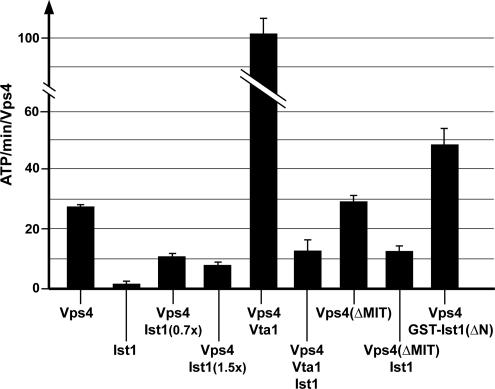
The assembly of Vps4 into the large oligomeric form is necessary for the hydrolysis of the bound ATP (Babst et al., 1998). As such, we predicted that interfering with Vps4 oligomerization by adding Ist1 would inhibit Vps4 ATP hydrolysis activity. We tested this prediction by measuring ATPase activity of Vps4 in vitro in the presence or absence of Ist1 (Figure 3). As expected, the addition of even substoichiometric amounts of Ist1 (0.7 times the amount of Vps4) strongly reduced the ATP turnover rate of Vps4. This effect on Vps4 activity only increased slightly when the concentration of Ist1 was doubled, suggesting that Ist1 reached maximal inhibition (about threefold) at approximately equimolar amounts relative to Vps4 (Figure 3). This result was consistent with the gel filtration analysis that predicted a one-to-one protein ratio of the Ist1-Vps4 complex. It is interesting to note that Ist1 is not able to completely block the ATPase activity of Vps4, suggesting that the interaction between Ist1 and Vps4 that is responsible for blocking ATPase activity might be dynamic.
Figure 3.
Ist1 inhibits ATPase activity of Vps4. Proteins were present at 1 μM concentration, except where specifically labeled (0.7× and 1.5× Ist1). The results shown represent the average activity and SEM obtained from four individual time points.
As previously demonstrated, the presence of Vta1 increased the ATPase activity of Vps4 substantially (Figure 3; Azmi et al., 2006). Consistent with the gel filtration analysis we found that Ist1 is able to compete with Vta1 and can reduce the activity of Vps4 to similar levels as observed for the Ist1-Vps4 complex alone (Figure 3). To our surprise, Ist1 reduced the ATPase activity of the MIT-deleted Vps4 protein to a similar extent as observed for wild-type Vps4 (Figure 3). The gel filtration data together with the GST pulldown experiments indicated that the MIT domain plays an important role in interactions between Ist1 and Vps4. Therefore we expected that the addition of Ist1 to Vps4(ΔMIT) would have only minor effects on the ATPase activity. The discrepancy between the two experiments could be explained by the predicted presence of a second interaction site between Ist1 and Vps4 that is dynamic and plays a key role in blocking both oligomerization and ATPase activity of Vps4. The dynamic nature of this second interaction would fit to the observation that Ist1 is not able to completely block Vps4 ATPase activity (Figure 3) and would explain why the second site alone is unable to maintain a stable Ist1-Vps4 complex during the gel filtration analysis. In support of this model we found that the presence of GST-Ist1(ΔN), which lacks the second interaction site did not inhibit but instead increased ATPase activity of Vps4 (Figure 3). This increase of ATPase activity is likely do in part to GST dimerization promoting the assembly of Vps4 bound to GST- Ist1(ΔN). However, we cannot exclude the possibility that the interaction between the C-terminus of Ist1 with the Vps4 MIT domain is responsible for the observed increase in enzymatic activity.
In summary, the ATPase activity tests and the gel filtration data supported a model of two distinct interaction sites between Ist1 and Vps4. A stable interaction between the two proteins requires the binding of the Ist1 coiled-coil motif to the Vps4 MIT domain. This interaction is nucleotide-dependent and thus resembles the binding of Vps4 to its substrate ESCRT-III. The second interaction formed between the N-terminal region of Ist1 and Vps4 is weaker but is crucial to inhibiting Vps4 assembly and ATPase activity.
Ist1 Localizes to MVBs by Binding to Did2
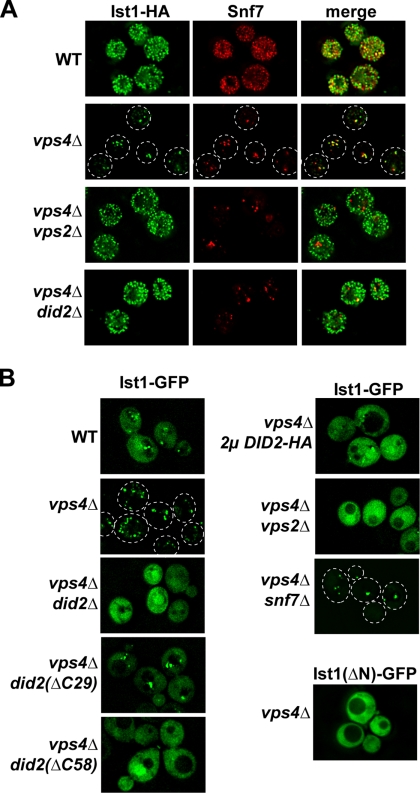
A genome-wide localization study in yeast suggested that Ist1 associates with endosomes (Huh et al., 2003). We confirmed this observation by immunofluorescence microscopy of yeast strains expressing IST1-HA. Ist1-HA localized predominantly to the cytoplasm of wild-type cells but colocalized with the ESCRT-III subunit Snf7 on enlarged endosomal structures in cells deleted for VPS4 (Figure 4A). This Vps4-dependent endosomal localization is typical for proteins of the ESCRT machinery, which have been shown to cycle on and off MVBs. We confirmed the immunofluorescence data by live cell imaging of cells expressing a C-terminal GFP-tagged version of Ist1, Ist1-GFP. This fusion protein was found to be nonfunctional (Supplementary Figure 1), but fluorescence microscopy of cells expressing Ist1-GFP showed the same Vps4-dependent endosomal localization as observed with functional Ist1-HA (Figure 4B), indicating that localization was not affected by the C-terminal GFP fusion. Live cell imaging of Ist1-GFP was necessary because fixation procedures result in a punctate distribution of soluble proteins when visualized by immunofluorescence microscopy, preventing a clear distinction between localization to the cytosol or to small compartments.
Figure 4.
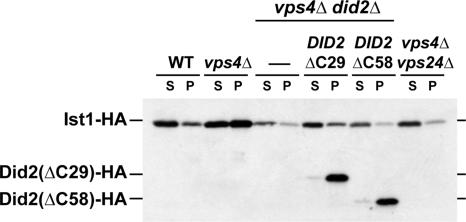
Ist1 localization to MVBs requires Did2. (A) Immunofluorescence microscopy of yeast strains expressing IST1-HA (pMB241). Fixed cells were stained using anti-HA and anti-Snf7 antibodies (WT, SEY6210; vps4Δ, MBY3; vps4Δvps2Δ, MBY41; and vps4Δdid2Δ, EEY26-1). (B) Fluorescence microscopy of cells expressing IST1-GFP (pMB243) or IST1(ΔN)-GFP (pCJ7). The following strains have been used for the experiments: MBY3 (vps4Δ), EEY26-1 (vps4Δdid2Δ), MBY3 pAH26 (vps4Δ 2μ DID2-HA), EEY26-1 pAH27 [vps4Δ did2(ΔC58)], EEY26-1 pAH25 [vps4Δ did2(ΔC29)], and EEY12 (vps4Δsnf7Δ).
We combined mutations in different ESCRTs with vps4Δ and tested which double mutant blocked endosomal localization of Ist1. These tests revealed that mutations in the Vps2-Vps24 subcomplex of ESCRT-III disrupted association of Ist1-HA and Ist1-GFP with the MVB (vps4Δvps2Δ: Figure 4, A and B; vps4Δvps24Δ: data not shown). In contrast, deletions of subunits of the ESCRT-III Snf7-Vps20 subcomplex did not affect endosomal accumulation of Ist1-GFP (vps4Δsnf7Δ: Figure 4B; vps4Δvps20Δ: data not shown). This result is consistent with the observation that localization of Vps2-Vps24 to endosomal membranes does not require the presence of Snf7 or Vps20 (Babst et al., 2002; Muziol et al., 2006; Shim et al., 2007). A recent study demonstrated that localization of the Vps4-interacting protein Did2 to the endosome was dependent on the presence of Vps2 and Vps24 (Nickerson et al., 2006). Interestingly, the vps4Δdid2Δ cells exhibited cytosolic localization of Ist1, similar to that observed in ESCRT-III mutants (Figure 4, A and B), suggesting that Ist1 associates with ESCRT-III by binding to Did2. In addition, overexpression of DID2 resulted in the redistribution of Ist1-GFP to the cytoplasm in vps4Δ, suggesting that cytoplasmic Did2 is able to interact with Ist1-GFP, thereby competing with ESCRT-III–associated Did2 for Ist1-GFP (Figure 4B). Furthermore, the mutant protein Ist1(ΔN)-GFP did not localize to endosomal structures, indicating that the N-terminal region containing the ELYC domain is required for the interaction with Did2 (Figure 4B).
Using fluorescent microscopy, we determined which region of Did2 was required for proper localization of Ist1-GFP. A previous study suggested that the N-terminal half of Did2 is involved in the binding to the Vps2-Vps24 subcomplex of ESCRT-III, whereas the C-terminus might interact with Vps4 (Nickerson et al., 2006). Our fluorescence microscopy experiments showed a partial loss of endosome-associated Ist1-GFP in cells expressing Did2(ΔC29)-HA (Did2 lacking the final 29 amino acids) and a complete redistribution of Ist1-GFP to the cytosol in cells expressing Did2(ΔC58)-HA (Figure 4B).
Additional Ist1 localization studies were performed by subcellular fractionation experiments that have been previously shown to separate cytoplasmic ESCRTs from MVB-associated ESCRT machinery (Babst et al., 1998). Yeast cells expressing Ist1-HA were spheroplasted, lysed, and separated by centrifugation at 13,000 × g into the membrane-bound, pellet fractions (P) and the cytoplasmic, soluble fractions (S). The fractions were then analyzed by Western blot for the presence of Ist1-HA. Consistent with the microscopy data Ist1-HA was found mostly in the cytoplasmic fraction of wild-type cells, accumulated in the pellet fraction in vps4Δ, and was almost completely lost from the pellet in vps4Δdid2Δ and vps4Δvps24Δ double mutants (Figure 5). The accumulation of Ist1-HA in the pellet fraction of vps4Δ was less pronounced than expected based on the microscopy data. This discrepancy between the two methods might be the result of partial dissociation of Ist1-HA from the endosomes during the fractionation procedure. As predicted from the microscopy studies, the fractionation experiments showed a partial loss of membrane-associated Ist1 in cells expressing Did2(ΔC29)-HA and a complete loss of membrane association in cells expressing Did2(ΔC58)-HA (Figure 5). However, both C-terminal deleted Did2 proteins associated with the membrane fraction, indicating that these mutant proteins are able to bind to ESCRT-III (Figure 5). In addition to changes in subcellular localization, we also observed variations in the overall amounts of Ist1 in the different yeast strains. In particular loss of Did2 resulted in reduced levels of Ist1, suggesting that Ist1 might be less stable in this mutant strain (Figure 5).
Figure 5.
Subcellular fractionation of Ist1 in different yeast strains. Yeast cell extracts were fractionated by centrifugation into soluble, cytoplasmic fractions (S) and membrane-associated pellet fractions (P). The resulting samples were analyzed for the presence of Ist1-HA (pMB241), Did2(ΔC29)-HA (pAH12), and Did2(ΔC58)-HA (pAH27) by Western blot using anti-HA antibodies (WT, SEY6210; vps4Δ, MBY3; vps4Δdid2Δ, EEY26-1; and vps4Δvps24Δ, MBY12).
In summary, the subcellular localization data indicated that Ist1 is a cytosolic protein that transiently associated with ESCRT-III by binding to Did2. This interaction was mediated by the N-terminal region of Ist1 and the C-terminal amino acids 148–175 of Did2.
Both Deletion and Overexpression of IST1 Impair MVB Sorting
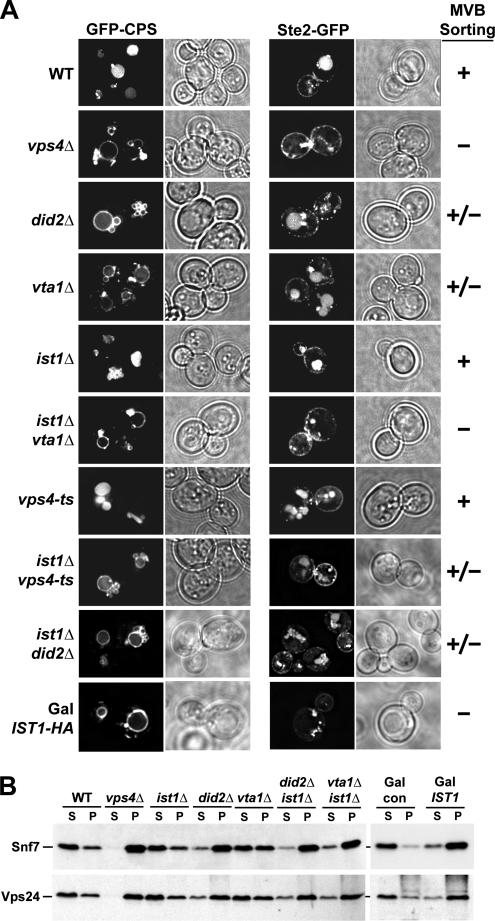
The interaction between Ist1 and Vps4 suggested that Ist1 might function in the MVB pathway. Therefore we tested the effect of an IST1 deletion on the trafficking of the MVB cargoes GFP-CPS and Ste2-GFP using fluorescence microscopy. The vacuolar peptidase CPS is synthesized as a transmembrane precursor and is delivered via the MVB sorting pathway into the lumen of the vacuole for its proper function. Ste2 is a cell-surface signaling receptor that is internalized and sorted by the MVB pathway to the vacuole for degradation (Odorizzi et al., 1998). In wild-type cells the fluorescence of both GFP-tagged cargoes are observed predominantly in the vacuole, indicating that these proteins have been efficiently sorted by the MVB machinery (Figure 6A). In contrast, mutations in the ESCRT machinery block delivery of GFP-CPS and Ste2-GFP to the vacuolar lumen causing them to accumulate in an aberrant endosomal structure called the class E compartment. Additionally, ESCRT mutants mislocalize GFP-CPS, and to a lesser extent Ste2-GFP, to the limiting membrane of the vacuole (e.g., vps4Δ in Figure 6A; Odorizzi et al., 1998).
Figure 6.
Phenotypic analysis of strains deleted for IST1 or overexpressing IST1. (A) Fluorescence microscopy of GFP-CPS (pGO45) or Ste2-GFP (pCS24) in different yeast strains (vps4-ts, MBY3 pMB65; ist1Δvps4-ts, MCY3 pMB65; other strains are listed in Table 1; for Ste2-GFP experiments the isogenic a-strains were used). Ist1-HA was overexpressed from a GAL1 promoter in wild-type cells (W303) growing in presence of galactose (Gal IST1-HA). The severity of the observed MVB trafficking phenotypes is indicated [plus (+): no phenotype; plus/minus (+/−): intermediate phenotype; minus (−): severe MVB sorting phenotype]. (B) Separation of yeast cell extracts by centrifugation into soluble (S) and membrane-associated pellet (P) fractions (see Table 1 for yeast strains). IST1 fused to a GAL1 promoter was overexpressed in wild-type cells grown in presence of galactose (Gal IST1: W303 pMB286). W303 containing an empty vector was used as a control (Gal con). The fractions were analyzed by Western blot using anti-Snf7 and anti-Vps24 antibodies.
A strain deleted for IST1 exhibited no obvious MVB trafficking defects and efficiently transported GFP-CPS and Ste2-GFP to the vacuolar lumen (Figure 6A). In comparison, mutations in two other known Vps4-interacting proteins, Vta1 and Did2, resulted in trafficking phenotypes in which sorting of GFP-CPS and Ste2-GFP was partially affected (Figure 6A; Azmi et al., 2006). This result is consistent with the model that Vta1 and Did2 are not essential factors of the ESCRT machinery but rather are regulators of Vps4 activity. Similarly, Ist1 might regulate some aspect of Vps4 activity and loss of this regulatory function might not be easily detected under optimal growth conditions present in the laboratory. Therefore we decided to test for a functional connection between IST1 and the three genes VPS4, VTA1, and DID2 by analyzing potential synthetic genetic interactions. We combined ist1Δ with a temperature-sensitive allele of VPS4, vps4-ts. This mutant allele of VPS4 encodes a protein that is active at 30°C but loses function at 37°C. Analyzing the trafficking of GFP-CPS and Ste2-GFP in the ist1Δvps4-ts strain at permissive temperature (30°C) revealed a partial MVB sorting defect demonstrating a genetic interaction between IST1 and VPS4 (Figure 6A). Similarly, deleting IST1 in vta1Δ strongly enhanced the trafficking defects, resulting in a block in vacuolar delivery of GFP-CPS and Ste2-GFP as severe as observed in vps4Δ (Figure 6A, see also Supplementary Figure 1). These results are consistent with the idea that Ist1 functions side-by-side with Vta1 in regulating Vps4 activity. In contrast, the deletion of IST1 did not change the severity of the MVB trafficking phenotype of a did2Δ strain (Figure 6A), which is consistent with the role of Did2 in localizing Ist1 to ESCRT-III. Deletion of DID2 blocks proper Ist1 function and therefore the additional deletion of IST1 cannot worsen the did2Δ phenotype. This result further suggested that the positive function of Ist1 in regulating Vps4 is dependent on Did2 and most likely occurs in association with ESCRT-III.
Vps4 functions in the disassembly of the ESCRT-III complex, resulting in the dissociation of the ESCRT-III subunits from the endosomal membrane. As a consequence, in subcellular fractionations of wild-type cells the majority of ESCRT-III subunits were found in the soluble, cytoplasmic fraction, whereas deletion of VPS4 resulted in the accumulation of ESCRT-III in the membrane-bound, pellet fraction (Figure 6B; Babst et al., 2002). Consistent with the partial MVB trafficking phenotype, mutations in VTA1 caused partial accumulation of ESCRT-III subunits Vps24 and Snf7 on endosomal membranes (Figure 6B; Azmi et al., 2006). Deleting DID2 caused a more pronounced ESCRT-III accumulation than observed in vta1Δ but in contrast to a previous publication (Nickerson et al., 2006), we reproducibly observed a less severe accumulation phenotype than in vps4Δ (Figure 6B). As expected from the lack of a trafficking phenotype, ist1Δ showed a wild-type distribution of the ESCRT-III (Figure 6B). However, deleting IST1 in a vta1Δ mutant strain resulted in a synthetic phenotype that was as severe as that observed in did2Δ (Figure 6B). In contrast, combining ist1Δ with did2Δ did not increase the ESCRT-III accumulation of the DID2 deletion strain, consistent with Did2 being necessary for Ist1 function in MVB sorting (Figure 6B).
The analysis of the MVB trafficking and ESCRT-III localization phenotypes supported the model that Ist1 functions together with Vps4 in the dissociation of ESCRT-III. However, loss of Ist1 function displayed only minor phenotypes, consistent with the idea that Ist1 functions as a regulator rather than a cofactor of Vps4.
Because the in vitro studies suggested that Ist1 functions as an inhibitor of Vps4 activity, we tested if overexpression of IST1 would have a more severe affect on Vps4 function than the deletion of IST1. For these overexpression studies we fused the IST1 gene to the GAL1 promoter, a strong, galactose-regulated promoter. Cells containing either the P(GAL1)-IST1-HA construct on a CEN plasmid or the empty vector (con) were grown in galactose medium and tested for a potential MVB trafficking phenotype by determining the localization of GFP-CPS and Ste2-GFP in these cells. Western blot analysis showed that the GAL1 promoter fusion resulted in at least 10-fold higher Ist1-HA levels then normal (Figure 7A). These high levels of Ist1 caused a strong MVB sorting defect that was as severe as observed in a VPS4 deletion strain (Figure 6A). Similarly, subcellular fractionation revealed that overexpression of IST1 caused an accumulation of ESCRT-III subunits in the membrane fraction, further indicating that high levels of Ist1 interfere with proper Vps4 function (Figure 6B).
Figure 7.
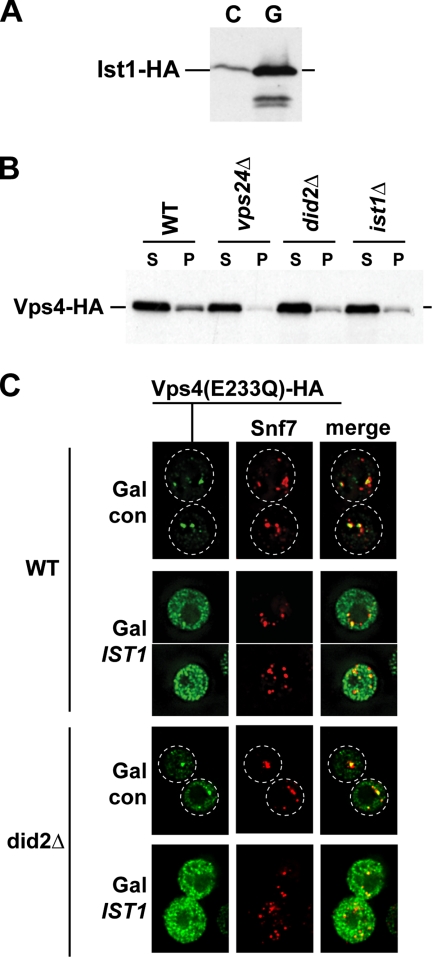
Ist1 regulates the recruitment of Vps4 to MVBs. (A) Western blot analysis of cell extracts from yeast strains grown in galactose-containing medium expressing IST1-HA either from its own promoter (C, control) or the GAL1 promoter (G). (B) Subcellular fractionation of yeast strains expressing Vps4-HA into soluble (S) and membrane-bound pellet (P) fractions (WT, MBY3 pMB283; vps24Δ, MBY12 pMB283; did2Δ, EEY26-1 pMB283; and ist1Δ, MCY3 pMB283). The fractions were analyzed by Western blot using anti- HA antibodies. (C) Localization of Vps4(E233Q)-HA (expressed from pMB149) and Snf7 in cells overexpressing IST1 from a GAL1 promoter (Gal IST1: pMB286) was determined by immunofluorescence microscopy. The strains were grown in presence of galactose (WT, W303; did2Δ, MCY31). Strains containing the empty expression vector were used as controls (Gal con).
Ist1 Regulates the Recruitment of Vps4 to ESCRT-III
The observation that Ist1 interacted both with Vps4 directly and with ESCRT-III via Did2 suggested that Ist1 might function in regulating the recruitment of Vps4 to ESCRT-III. Therefore we tested the effects of deletion or overexpression of Ist1 on the localization of Vps4 to the endosomal membrane. Subcellular fractionation demonstrated that compared with wild type, ist1Δ cells showed reduced amounts of Vps4 in the membrane-bound pellet fraction (Figure 7B). The reduction was similar to that of a DID2 deleted strain but less severe then observed in cells deleted for VPS24 (Figure 7B). These results indicated that Ist1 indeed plays a role in the recruitment of Vps4 to MVBs. The fact that Did2 is essential for the localization of Ist1 to MVBs explains why a deletion of DID2 resulted in the same Vps4 recruitment defect as found in ist1Δ. Finally, unlike Vps24 that is crucial for localizing Vps4 to MVBs (Babst et al., 2002), Did2 and Ist1 play a regulatory role in Vps4 recruitment, but are not required for the recruitment itself (Nickerson et al., 2006).
A potential effect of high levels of Ist1 on Vps4 localization was studied using immunofluorescence microscopy of cells expressing the Vps4(E233Q)-HA mutant protein. This point mutant of Vps4 is trapped in the ATP-bound state and therefore accumulates on MVBs (Babst et al., 1998). However, in cells overexpressing IST1 from a strong GAL1 promoter, Vps4(E233Q)-HA was mislocalized to the cytoplasm (Figure 7C), indicating that high cellular levels of Ist1 were able to block Vps4 recruitment to MVBs. This Vps4 mislocalization phenotype in IST1-overexpressing cells was not dependent on the presence of Did2 (Figure 7C).
Together, the phenotypic analysis suggested a role of Ist1 in regulating the recruitment of Vps4 to the MVB-localized ESCRT-III complex. Lack of Ist1 as well as high levels of Ist1 interfered with proper localization of Vps4, suggesting that cellular levels of Ist1 could play a role in regulating Vps4 function.
DISCUSSION
In the MVB pathway, the AAA-type ATPase Vps4 functions in the disassembly of the ESCRT-III complex and the subsequent recycling of all ESCRT complexes from the endosomal membrane (Babst et al., 1998, 2002). Without Vps4 function, the ESCRT machinery remains on the endosomal membrane and MVB vesicle formation is inhibited. Vps4 is the only known essential enzyme within the ESCRT machinery and therefore is ideally positioned to play a key role in regulating the MVB pathway according to cellular needs.
Previous studies suggested a model in which inactive Vps4 dimers are recruited from the cytosol to the ESCRT-III complex where they assemble into a large double-ring structure of 12 subunits (Babst et al., 1998; Scott et al., 2005a). This oligomeric form of Vps4 is the active ATPase, which disassembles the bound ESCRT-III complex using the energy from ATP hydrolysis. Vps4 activity seems to be regulated through oligomerization of the protein. One Vps4 regulator is Vta1, a dimeric protein that binds to the ATPase domain of Vps4 and promotes both Vps4 assembly as well as ATP hydrolysis (Azmi et al., 2006). In this study we identified Ist1 as a regulator of Vps4 recruitment that may function together with Vta1 in modulating Vps4 activity.
IST1 encodes a 35-kDa soluble protein that contains a conserved N-terminal domain of ∼150 amino acids, which we named the ELYC (pronounced “ellis”) domain, and a putative C-terminal coiled-coil domain of ∼30 amino acids. Size determination experiments under native conditions indicated that Ist1 is a monomeric protein with a nonglobular, elongated structure.
Yeast two-hybrid data and published proteomics studies (Krogan et al., 2006) indicated that Ist1 interacts with Vps4 in vivo. Our in vitro studies using recombinant Ist1 and Vps4 protein revealed that Ist1 binds directly to Vps4, resulting in the formation of a heterodimeric Vps4-Ist1 complex. Because Vps4 is predicted to be either a dimer or a dodecamer, the data suggested that Vps4 complexes disassemble in order to form the Vps4-Ist1 heterodimer. Moreover, this interaction requires the MIT domain of Vps4 and the coiled-coil motif of Ist1. The MIT domain has been shown to function as the substrate-binding domain Vps4 by interacting with the coiled-coil rich ESCRT-III subunits. Therefore it seems that the Ist1-Vps4 interaction mimics the binding of Vps4 to its substrate ESCRT-III.
The formation of the Vps4-Ist1 complex efficiently blocks the oligomerization of Vps4 into the dodecameric complex, resulting in a strong reduction of Vps4 ATPase activity. This inhibition of Vps4 oligomerization was observed even in presence of the assembly-promoting factor Vta1, suggesting that Ist1 is able to compete with Vta1 for interaction with Vps4. The interaction between the Vps4 MIT domain and the coiled-coil region of Ist1 is sufficient to obtain a stable Vps4-Ist1 complex, but cannot block Vps4 oligomerization, indicating the existence of at least one more interaction site, possibly via the N-terminal Ist1 ELYC domain (Figure 1). This second interaction site alone is sufficient to impair hydrolysis activity of Vps4 in ATPase assays, but is not stable enough to form an Ist1-Vps4 complex that can be observed by gel filtration analysis.
In wild-type cells Ist1 localizes to both the cytoplasm and MVBs. In contrast, in cells deleted for VPS4 Ist1 accumulates on the aberrant endosomal structures formed in these cells. Therefore Ist1 localization mimics the localization of the ESCRT complexes, suggesting that, like the ESCRTs, Ist1 cycles on and off MVBs in a Vps4-dependent manner. The recruitment of Ist1 to MVBs requires the presence of Did2, a protein homologous to the ESCRT-III subunits. Previous studies have shown that Did2 localizes to ESCRT-III by binding to the Vps24 subunit, where it functions in Vps4-dependent disassembly of the protein complex (Nickerson et al., 2006). This activity requires the binding of the Did2 C-terminal region to the Vps4 MIT domain. However, our data indicate that the same region of Did2 recruits Ist1 to ESCRT-III and that the MIT domain of Vps4 interacts with the Ist1 coiled-coil region. Further studies will be necessary to determine if Ist1 competes with the Did2-Vps4 interactions or if all three proteins are able to bind to each other simultaneously.
Deletion of IST1 caused no obvious MVB trafficking phenotypes. However, genetic studies revealed synthetic interactions between ist1Δ and mutations in VTA1 and VPS4, suggesting that Ist1 plays a positive role in Vps4 function. No synthetic genetic interactions were observed between ist1Δ and did2Δ, consistent with the idea that Ist1 requires Did2 for its positive effect on Vps4 activity. Cells deleted for IST1 or DID2 exhibited a similar reduction in the amount of membrane associated Vps4, suggesting that the Ist1-Did2 complex might function to regulate the proper recruitment of Vps4 to ESCRT-III. In contrast to the modest defects observed in ist1Δ, overexpression of IST1 resulted in a severe MVB trafficking phenotype. This phenotype was caused by a block in Vps4 recruitment to MVBs, resulting in endosomal accumulation of ESCRT-III. The overexpression data suggested that high levels of Ist1 in the cytoplasm might result in the formation of a Vps4-Ist1 complex that competes with the recruitment of Vps4 by ESCRT-III localized Did2-Ist1.
The phenotypic analysis suggests two distinct functions for Ist1 in regulating Vps4 activity. Associated with Did2, Ist1 localizes to ESCRT-III, where it seems to have a positive effect in the recruitment of Vps4. In contrast the free, cytoplasmic pool of Ist1 binds to Vps4, resulting in the formation of an Ist1-Vps4 complex that is not recruited to ESCRT-III. This latter complex seems to represent an inactive pool of Vps4. Both regulatory functions of Ist1 have mild effects on Vps4 activity. As a result of this, MVB trafficking defects are only observed either when ist1Δ is combined with other mutations in the ESCRT machinery or when Ist1 is overexpressed to artificially high levels. These observations are consistent with the idea that at normal cellular levels Ist1 acts as a modulator of the MVB pathway, affecting the kinetics of sorting but not blocking the pathway. One possible mechanism by which the cell could modulate Vps4 activity is by regulating the concentration of Ist1 in the cytoplasm.
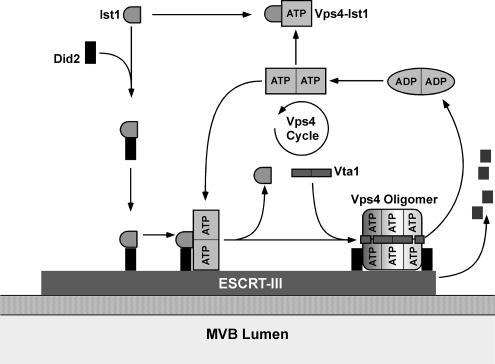
In summary, we propose the following model for the function of Ist1 in regulating Vps4 activity (Figure 8). In the cytoplasm Ist1 interacts with Did2, resulting in the formation of Ist1-Did2 complex that is recruited to ESCRT-III. On ESCRT-III Ist-Did2 seems to function in regulating the recruitment of dimeric Vps4. The Ist1-Did2 complex is not required for binding of Vps4 to ESCRT-III but the complex might affect the kinetics and the proper timing of Vps4 recruitment. Furthermore, when bound to ESCRT-III the interaction of Vps4 with Ist1-Did2 might inhibit premature oligomerization of the ATPase, ensuring that ESCRT-III has completed its function in the MVB-sorting pathway before dissociation is initiated. At the proper time Ist1 dissociates from Vps4 and Did2 and is exchanged for the assembly factor Vta1, which initiates the oligomerization of Vps4 into the active complex. Previous studies showed that Vta1 interacts with Did2, suggesting that Did2 might also function in the Vps4 assembly step (Lottridge et al., 2006). This would explain why loss of Did2 results in more severe MVB trafficking defects than observed in ist1Δ. The oligomeric form of Vps4 is the active ATPase that disassembles ESCRT-III in an ATP-dependent manner. In addition to the interaction with Did2, Ist1 can also form a heterodimeric complex with Vps4 in the cytoplasm. This Ist1-Vps4 complex represents an inactive form of Vps4 that competes with the function of Vps4 in ESCRT-III disassembly. By regulating the level of Ist1 in the cytoplasm the cell would be able to shift the equilibrium either in favor of the Ist1-Did2 or the Ist1-Vps4 complex, thereby regulating the recruitment of Vps4 and thus the efficiency of MVB sorting.
Figure 8.
Model for the regulation of Vps4 by Ist1 (see Discussion for details).
Supplementary Material
ACKNOWLEDGMENTS
We thank Tamara Darsow for critical reading of the manuscript and Debra Eckert for her assistance with the analytical ultracentrifugation experiments. This work has been supported by Grant RO1 GM074171-01 A1 from the National Institutes of Health and Grant 0530210N from the American Heart Association.
Abbreviations used:
- MVB
multivesicular body
- ESCRT
endosomal sorting complex required for transport.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0747) on November 21, 2007.
REFERENCES
- Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T. K., Banta L. M., Emr S. D. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Curtiss M., Jones C., Babst M. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol. Biol. Cell. 2007;18:636–645. doi: 10.1091/mbc.E06-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- Gruenberg J., Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Howard T. L., Stauffer D. R., Degnin C. R., Hollenberg S. M. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 2001;114:2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lottridge J. M., Flannery A. R., Vincelli J. L., Stevens T. H. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc Natl Acad Sci USA. 2006;103:6202–6207. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Muziol T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Gottlinger H., Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., West M., Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S. D. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Piper R. C., Katzmann D. J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A., et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005a;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., Sundquist W. I. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. USA. 2005b;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Lawrence L. W. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1979. [Google Scholar]
- Shiflett S. L., Ward D. M., Huynh D., Vaughn M. B., Simmons J. C., Kaplan J. Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:10982–10990. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- Shim S., Kimpler L. A., Hanson P. I. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Vajjhala P. R., Catchpoole E., Nguyen C. H., Kistler C., Munn A. L. Vps4 regulates a subset of protein interactions at the multivesicular endosome. FEBS J. 2007;274:1894–1907. doi: 10.1111/j.1742-4658.2007.05736.x. [DOI] [PubMed] [Google Scholar]
- Williams R. L., Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Yeo S. C., et al. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 2003;116:3957–3970. doi: 10.1242/jcs.00751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.