Abstract
The Golgi apparatus (GA) of mammalian cells is positioned in the vicinity of the centrosome, the major microtubule organizing center of the cell. The significance of this physical proximity for organelle function and cell cycle progression is only beginning to being understood. We have identified a novel function for the GA protein, GM130, in the regulation of centrosome morphology, position and function during interphase. RNA interference–mediated depletion of GM130 from five human cell lines revealed abnormal interphase centrosomes that were mispositioned and defective with respect to microtubule organization and cell migration. When GM130-depleted cells entered mitosis, they formed multipolar spindles, arrested in metaphase, and died. We also detected aberrant centrosomes during interphase and multipolar spindles during mitosis in ldlG cells, which do not contain detectable GM130. Although GA proteins have been described to regulate mitotic centrosomes and spindle formation, this is the first report of a role for a GA protein in the regulation of centrosomes during interphase.
INTRODUCTION
The centrosome, the major microtubule-organizing center (MTOC) of the cell, can be formed by two pathways involving template-based and de novo formation of centrioles (La Terra et al., 2005; Kleylein-Sohn et al., 2007; Peel et al., 2007; Rodrigues-Martins et al., 2007). Template-based centrosome duplication during S-phase involves the formation of a procentriole next to a pre-existing centriole, followed by elongation and maturation through the sequential recruitment of centrosomal proteins (Kleylein-Sohn et al., 2007). Duplicated centrosomes, each containing a pair of centrioles, separate upon entry into mitosis to form the poles of the mitotic spindle. In the absence of a centriole, de novo centriole formation can occur through a template-independent pathway in which Centrin2-positive foci recruit other centrosomal marker proteins, followed by maturation into a normal centriole during the next cell cycle (Khodjakov et al., 2002; Uetake et al., 2007). These pathways of centrosome formation have to be tightly controlled because dysregulation may result in supernumerary centrosomes, which lead to multipolar mitoses, chromosome loss, and aneuploidy (Nigg, 2006).
There is evidence that the organization of the centrosome can be regulated by Golgi apparatus (GA)-associated proteins. We have shown that the GA protein, GRASP65, is necessary for bipolar spindle formation and cell cycle progression as depletion of this protein resulted in multiple aberrant spindles, metaphase arrest, and cell death (Sutterlin et al., 2005). Tankyrase-1 and Rint-1, which are associated with the GA and the early secretory pathway, respectively, are also required for mitotic spindle formation (Chang et al., 2005; Lin et al., 2007).
To understand how GRASP65 regulates spindle formation, we investigated whether the GRASP65-interacting protein, GM130, is also involved in the formation of a bipolar spindle. GM130 is a large coiled-coil protein of the cis-GA that has recently been shown to regulate the stability and localization of GRASP65 (Puthenveedu et al., 2006). GM130 has also been proposed to regulate mitotic GA fragmentation (Nakamura et al., 1997; Lowe et al., 1998), endoplasmic reticulum-to-GA transport (Seemann et al., 2000), GA ribbon formation (Puthenveedu et al., 2006; Marra et al., 2007), and cell migration (Preisinger et al., 2005). Although these studies have focused on the roles of GM130 in membrane trafficking and GA biogenesis, they have not examined an involvement of this protein in spindle formation and cell cycle progression.
In this report, we show that GM130 regulates the association of GRASP65 with the GA and is necessary for the formation of a bipolar mitotic spindle. Surprisingly, however, GM130 does not appear to exert its effect on spindle formation via GRASP65. Instead, we found that GM130 regulates centrosome morphology and function before entry into mitosis. Thus, the effect of GM130 on spindle formation is indirect and a consequence of aberrant centrosomes. These findings provide evidence of a second, GRASP65-independent pathway by which a GA-associated protein is able to regulate the centrosome.
MATERIALS AND METHODS
Antibodies
Antibodies were from the following sources: GRASP65, GRASP55, and Giantin (Dr. Vivek Malhotra, University of California, San Diego), GM130 (Dr. Adam Linstedt, Carnegie Mellon University, Pittsburgh, PA), Golgin 97 (Dr. Ed Chan, University of Florida, Gainesville), Centrin2 hCten2.4 and 20H5 (Dr. Jeffrey Salisbury, Mayo Clinic), Kendrin (Dr. Mikiko Takahashi, University of Koke, Japan), GM130 (BD Biosciences, San Jose, CA), α-tubulin, acetylated-tubulin, GM130 (Sigma, St. Louis, MO), GRASP65 (Santa Cruz Biotechnologies, Santa Cruz, CA), phospho-Histone H3 (Upstate Biotechnology, Lake Placid, NY), α-tubulin, γ-tubulin, Ninein (AbCam, Cambridge, MA), bromodeoxyuridine (BrdU; Calbiochem, LA Jolla, CA), EB1 (BD Transduction Laboratories, Lexington, KY), BrdU (MP Biomedicals, Solon, OH), fluorochrome and HRP-conjugated secondary antibodies (Molecular Probes, Eugene, OR/Invitrogen, Carlsbad, CA).
Molecular Biology
Knockdown plasmids were constructed in pSUPER as described (Brummelkamp et al., 2002). Targeting sequences within the human GM130 cDNA were GM130-1 (nucleotides 985-1005) and GM130-2 (nucleotides 1646–1665; Puthenveedu et al., 2006) or a scrambled control sequence with similar nucleotide composition (Sutterlin et al., 2005; Puthenveedu et al., 2006). The H1 promoter-short hairpin cassette was amplified by PCR and cloned two more times into pSuper, generating plasmids with three sequential H1 promotor-short hairpin cassettes. Human GM130 cDNA was provided by Dr. Antonino Colanzi (Consozorio Mario Negri Sud, Italy). Rescue experiments were carried out by coexpressing the triple GM130 knockdown or control cassette and green fluorescent protein (GFP), GFP-tagged GM130, and GFP-tagged ΔN690 from the same plasmid.
Cell Culture
HeLa, U2-OS, SaOS-2 (all from ATCC, Manassas, VA), HeLa GalNAc-T2 (Dr. Adam Linstedt), and A549 (Dr. Ingrid Ruf, University of California, Irvine) cells were grown in Advanced DMEM (Invitrogen) supplemented with 2 or 4% fetal FBS and 2 mM GlutaMAX-I (GIBCO, Rockville, MD). Cells at 50% confluency grown in six-well dishes were transfected using Fugene 6 (Roche, Indianapolis, IN). The low plating confluency at the time of transfection was important to achieve complete knockdown. Small interfering RNA (siRNA; Qiagen, Chatsworth, CA) was transfected using Oligofectamine (Invitrogen) as described (Sutterlin et al., 2005). Twenty-four hours later, cells were split onto a coverslips and processed for immunofluorescence.
The mitotic index and spindle organization were determined as described (Sutterlin et al., 2005). In brief, cells were harvested using warm EDTA-PBS and replated onto poly-l-lysine–coated coverslips, fixed, and analyzed by immunofluorescence with antibodies to phospho-Histone H3 and α-tubulin, followed by the quantification of the observed phenotypes.
Wild-type and mutant IdlG Chinese hamster ovary (CHO) cells (Dr. Monty Krieger, Massachusetts Institute of Technology, Cambridge, MA) were grown in Ham's F-12 medium (Gibco BRL), supplemented with 5% FBS and 2 mM l-glutamine, and plated onto coverslips for immunofluorescence analysis. Although wild-type cells were grown at 37°C, ldlG mutant cells were grown at the permissive temperature of 34°C.
Immunofluorescence Microscopy
Cells were fixed for 8 min with warm 4% formaldehyde or for 3 min with ice-cold methanol at −20°C and blocked in 10% blocking solution (0.1% Triton X-100, 10% FBS). Primary and secondary antibodies were diluted into 2.5% blocking buffer (0.1% Triton X-100, 2.5% FBS). Cells were imaged with a Zeiss Axiovert 200M microscope (Thornwood, NY) and analyzed with linear adjustments with the Zeiss Axiovision software.
Western Blot Analysis
Cells were lysed in 150 mM NaCl, 20 mM Tris-HCl, pH 7.4, 1% NP-40, 2 mM pepstatin, 150 mM aprotinin, and 1 mM leupeptin on ice for 10 min, followed by clearing the lysates by centrifugation. Twenty micrograms of total cell lysate per lane were separated by SDS-PAGE and subjected to Western blot analysis using enhanced chemiluminescence (Amersham Pharmacia, Piscataway, NY) for signal detection.
BrdU Incorporation
The BrdU incorporation assay was carried out as described (Sutterlin et al., 2002). In brief, U2-OS cells, transfected with control and the GM130 knockdown plasmid 48 h after transfection, were pulse-labeled with 10 μM BrdU (Calbiochem) for 30 min, followed by washing the cells three times with each, PBS and medium. Twenty-four hours after the incubation with BrdU, cells were fixed in 4% formaldehyde, denatured with 4 N HCl, quenched in 0.1 M sodium borate, and stained with an antibody to BrdU.
Wound Healing Assay
siRNA-transfected A549 were plated as a confluent monolayer onto 35-mm MatTek (Ashland, MA) glass-bottom dishes. Scratch wounds were introduced, followed by washing the cells with medium to remove debris and mitotic cells (Geiser et al., 2004). Wound healing was imaged with a 20× long-working phase objective (Zeiss). After the last time point, cells were fixed and processed for immunofluorescence.
Fluorescence-activated Cell Sorting Analysis
For fluorescence-activated cell sorting (FACS) analysis, transfected U2-OS cells were incubated for 30 min in complete media containing 10 μg/ml Hoechst 33342 (Molecular Probes/Invitrogen). Cells were then harvested with Tryp LE (Gibco BRL), and resuspended in PBS containing 10 μg/ml propidium iodide (Molecular Probes/Invitrogen). Cells were collected using a Becton Dickinson LSRII (Franklin Lakes, NJ) and analyzed using FlowJo (Tree Star, Ashland, OR).
RESULTS
GM130 Regulates the Association of GRASP65 with the GA
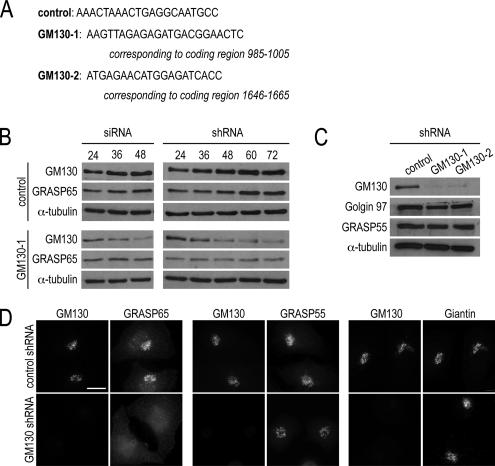
To analyze the relationship between the GA proteins, GRASP65 and GM130, we took an overall approach of depleting GM130 protein levels and assaying the effect on GRASP65 function. We used two complementary RNA interference (RNAi)-based methods to deplete GM130, targeting sequences within the GM130 cDNA that previously led to efficient protein depletion (Figure 1A; Puthenveedu et al., 2006). In the first approach, we transfected GM130-1 siRNA (Figure 1A) into cells and depleted endogenous GM130 by 90% at 48 h, as assayed by Western blots (Figure 1B). In a second approach, we used a knockdown plasmid containing three sequential short hairpins of the GM130-1 sequence (shRNA). GM130 levels were also reduced by 90%, although this level of depletion required 72 h (Figure 1B), which may reflect the additional time needed to generate double-stranded RNA from the plasmid. In these experiments, there were ∼5–10% untransfected cells, which may account for the low levels of residual GM130. For both approaches, a control consisting of a scrambled sequence had no effect on GM130 levels (Figure 1, A–C).
Figure 1.
GM130 regulates the GA association of GRASP65, not its stability. (A) RNAi targeting sequences (GM130-1 and GM130-2) and their positions within the human GM130 cDNA are shown. (B) HeLa cells, transfected with the control or knockdown siRNA (GM130-1 siRNA) or short hairpin-expressing plasmids (GM130-1 shRNA), were harvested at the indicated time points after transfection. Western blot analysis was used to reveal the levels of GM130, GRASP65, and α-tubulin as a loading control. (C) Lysates of control and GM130-depleted cells were examined by Western blot analysis with antibodies to GM130, Golgin 97, GRASP55, and α-tubulin as a loading control. (D) Control or GM130-depleted cells were analyzed by immunofluorescence with antibodies to GRASP65, GRASP55, and Giantin 72 h after knockdown transfections. Scale bar, 10 μm.
Having successfully depleted GM130, we examined the stability of GRASP65 and other GA marker proteins. Depletion of GM130 by siRNA or shRNA did not affect the levels of GRASP65 (Figure 1B). Also, levels of GRASP55, a GRASP65-related protein, and Golgin 97, a Golgin family member, were not altered (Figure 1C). Similar results were obtained with an shRNA construct targeting a different sequence in GM130 (GM130-2, Figure 1A).
We next examined if GM130 depletion could affect the localization of GRASP65 as monitored by immunofluorescence. In these and the following studies, we only present the results obtained with the GM130-1 shRNA although similar observations were made with the GM130-2 shRNA and GM130 knockdown by siRNA. In contrast to the lack of an effect on protein levels, GM130 depletion disrupted the association of GRASP65 with the GA, resulting in a dispersed, cytosolic appearance of GRASP65 (Figure 1D). The effect was specific for GRASP65, because the GA association of GRASP55 and the transmembrane protein, Giantin, were unaltered. Thus, our data indicates that GM130 is necessary for the GA localization, but not the stability of GRASP65. The results have been confirmed by depleting GM130 protein levels with two different RNAi-based methods and by targeting two different GM130 sequences.
GM130 Is Required for Bipolar Spindle Formation
As GRASP65 is required for bipolar spindle formation and cell cycle progression, we examined whether depletion of GM130 could cause a similar phenotype of spindle defects and mitotic arrest. We first compared the growth behavior of GM130-depleted cells over a period of 4 days. Although control cells doubled at an exponential rate, GM130-depleted cells did not proliferate and underwent cell death between days 3 and 4 (data not shown). Before this endpoint, however, we observed an increased mitotic index as measured with the mitotic marker, phospho-Histone H3 (Juan et al., 1998), indicating that cells were arrested in mitosis as discussed in more detail below (Figure 3B).
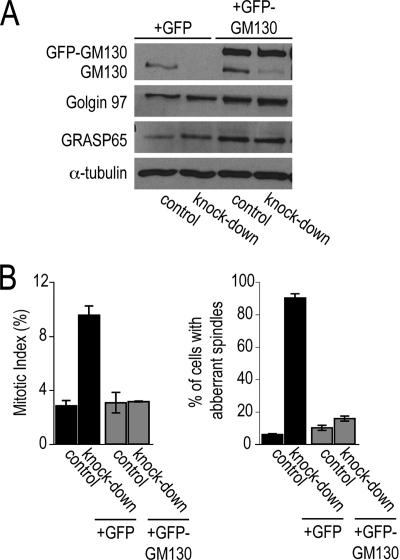
Figure 3.
Defects in spindle formation are corrected by expressing full-length GM130. (A) HeLa cells were transfected with the control or GM130 knockdown plasmid, which also expressed cytosolic GFP (+GFP) or GFP-tagged RNAi-resistant GM130 (+GFP-GM130). Lysates were prepared and analyzed by Western blotting with antibodies to GM130, Golgin 97, GRASP65, and α-tubulin. (B) In parallel, the cells described in Figure 3A were replated onto coverslips, fixed and processed for immunofluorescence with antibodies to phospho-Histone H3 and α-tubulin to reveal the mitotic index and spindle organization, respectively. The percentage of cells in mitosis and with aberrant mitotic spindles is shown (n = 3).
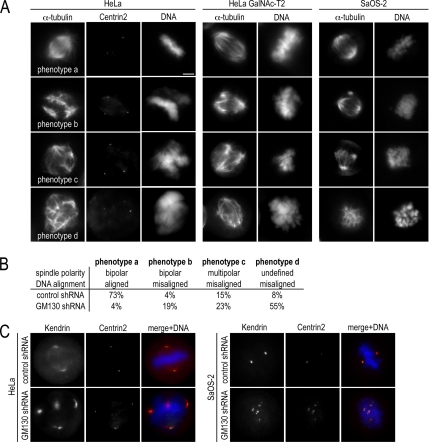
We next examined mitotic spindle organization in GM130-depleted cells by staining microtubules and spindle poles with antibodies to α-tubulin and to the centriole protein Centrin2, respectively, and by visualizing DNA with the dye Hoechst 33342. Based on the organization of microtubules, spindle poles and the DNA, we grouped mitotic, GM130-depleted cells into four phenotypes, which we defined as normal (phenotype a: bipolar spindle with aligned DNA), mild (phenotype b: bipolar spindle that does not align DNA in the metaphase plate), moderate (phenotype c: multipolar spindle that does not align DNA in the metaphase plate), and severe (phenotype d: undefined spindle poles and misaligned DNA; Figure 2A). Control cells predominantly formed bipolar spindles, which were nucleated at two Centrin2-positive spindle poles and which aligned DNA in the metaphase plate (phenotype a; Figure 2B). In contrast, only 4% of mitotic, GM130-depleted cells had normal bipolar spindles (phenotype a). The great majority contained disorganized spindles with 55% of the cells, showing the most severe defects (phenotype d) with disorganized microtubules, undefined poles, and unaligned DNA (Figure 2B). These results were confirmed by depleting GM130 with shRNA targeting a different sequence (GM130-2, Figure 1A) and by using siRNA (data not shown).
Figure 2.
GM130 is required for bipolar spindle formation. (A) HeLa, HeLa GalNAc-T2, and SaOS-2 cells were transfected with control and knockdown plasmids, fixed after 72 h, and stained with antibodies to Centrin2, α-tubulin to monitor the microtubule organization and with the dye Hoechst 33342 to visualize DNA. Spindle organization and DNA alignment were used as criteria to group the mitotic phenotypes into four categories. A representative cell for each phenotype is shown. Scale bar, 5 μm. (B) The distribution of mitotic control and GM130-depleted HeLa among these spindle phenotypes was quantified (n = 3). (C) Spindle poles of mitotic control or GM130-depleted HeLa and SaOS-2 cells were stained with antibodies to Centrin2 and Kendrin. The merged images also reveal the organization of DNA as detected with the dye Hoechst.
To determine if these spindle defects are a general consequence of GM130 depletion, we repeated the spindle analysis in additional cell lines. We did not detect the spindle phenotype in p53-positive cell lines, such as the osteosarcoma cell line, U2-OS, because a loss of GM130 appeared to lead to an overall decrease in the rate of cell proliferation and a cell cycle delay at the G2-M transition (to be discussed in more detail below). However, we were able to examine spindle organization in p53-deficient cell lines, such as a HeLa cell line stably expressing GFP-tagged N-acetylgalactosaminyl-transferase-2 (GalNAc-T2) that has been used in a previous analysis of GM130 (Storrie et al., 1998; Puthenveedu et al., 2006), and SaOS-2, a p53-deficient counterpart of U2-OS (Chandar et al., 1992). The four mitotic knockdown phenotypes were detected in these cell lines at frequencies similar that of HeLa cells (Figure 2A). To further characterize the aberrant spindle poles of GM130-depleted HeLa and SaOS-2 cells, we costained these cells with antibodies to Centrin2 and the pericentriolar markers, Kendrin (Takahashi et al., 1999; Flory and Davis, 2003) and γ-tubulin and found that each spindle pole contained centriole and pericentriolar marker proteins (Figure 2C, data for γ-tubulin not shown). This phenotype is different from the spindle pole composition of mitotic GRASP65-depleted cells, in which Centrin2 is only present in two of the multiple spindle poles, whereas γ-tubulin is detected at each of them (Sutterlin et al., 2005).
To verify that the observed mitotic phenotypes were due to the depletion of GM130 and not to nonspecific off-target effects, we carried out functional rescue experiments. We generated a construct that contained the triple knockdown cassettes together with GFP-tagged GM130 or GFP for control experiments. GFP-GM130 was designed with five silent point mutations within the RNAi-targeting sequence so that it would resist RNAi-mediated degradation. Transfection of this rescue plasmid into HeLa cells led to the replacement of endogenous GM130 by GFP-GM130 expressed at about four- to fivefold higher levels (Figure 3A). GFP-GM130, but not GFP alone, restored cell cycle progression and normal bipolar spindle formation (Figure 3B). Taken together, our results demonstrate that depletion of GM130 blocks cell cycle progression and causes aberrant spindle formation in p53-negative cells, implicating GM130 in a role in bipolar spindle formation during mitosis.
The Role of GM130 in Spindle Formation Appears to be Independent of GRASP65
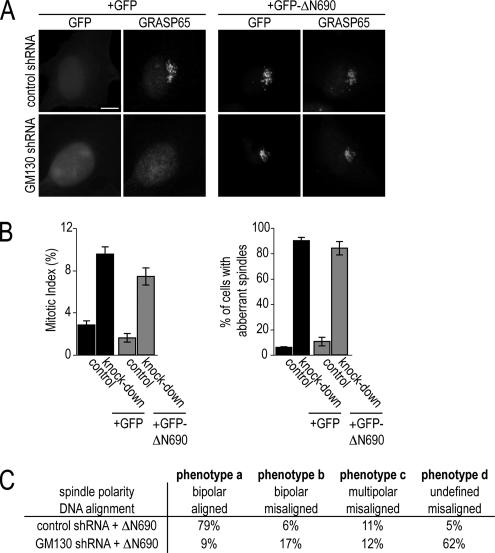
Because both GRASP65 (Sutterlin et al., 2005) and GM130 (Figure 2A) are required for bipolar spindle formation, we next determined whether these two GA proteins function together in the same regulatory pathway. We restored GRASP65 localization to the GA in GM130-depleted cells by expressing the C-terminal domain of GM130 (GFP-ΔN690), which mediates the GA association of GM130 and which is also the interaction domain for GRASP65 (Nakamura et al., 1997; Barr et al., 1998; Figure 4A). Relocalized, GA-bound GRASP65 was functional, as it was able to correct the microtubule acetylation defect of GM130 knockdown cells (see Figure 8C). Intriguingly, correcting GRASP65 localization did not change the GM130 depletion phenotype: cells still arrested in mitosis and formed aberrant spindles that fell into the same four categories detected in GM130-depleted cells with cytosolic GRASP65 (Figure 4C). This result suggests that the function of GM130 in spindle formation is independent of GRASP65.
Figure 4.
The role of GM130 in spindle formation is independent of GRASP65. (A) Control and GM130-depleted (knockdown) HeLa cells expressing GFP alone (+GFP) or the C-terminus of GM130 (+GFP-ΔN690) were fixed 72 after transfection and examined by fluorescence microscopy for the localization of GFP and GRASP65. Scale bar, 5 μm. (B) Control and GM130-depleted cells expressing GFP or the GM130 C-terminus, GFP-ΔN690, were stained with antibodies to phospho-Histone H3 and α-tubulin to reveal the mitotic index and the organization of the mitotic spindle, respectively. The percentage of cells in mitosis and with aberrant spindles was determined from three independent experiments. (C). Microtubule organization and DNA alignment were used as criteria to categorize spindle organization into the same four categories defined in Figure 2B.
Figure 8.
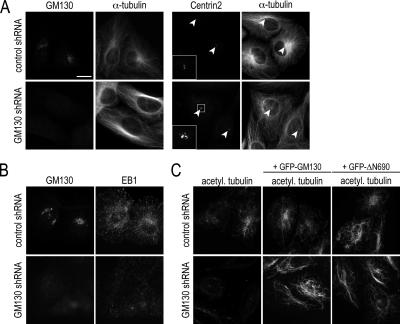
The depletion of GM130 causes defects in centrosome function. (A) Control and GM130-depleted U2-OS cells were analyzed by immunofluorescence with antibodies to GM130, Centrin2, and α-tubulin to reveal the level of GM130 depletion and centrosome and microtubule organization, respectively. Arrows point to the position of the centrosome. (B) Control and GM130-depleted U2-OS cells were stained with antibodies to GM130 and EB1 to observe the growing ends of microtubules. (C) Control and GM130-depleted U2-OS cells expressing RNAi-resistant GM130 (+GFP-GM130) or the C-terminus of GM130 (+GFP-ΔN690) were analyzed for the presence of acetylated microtubules. Scale bars, 10 μm.
GM130 Regulates Centrosome Morphology
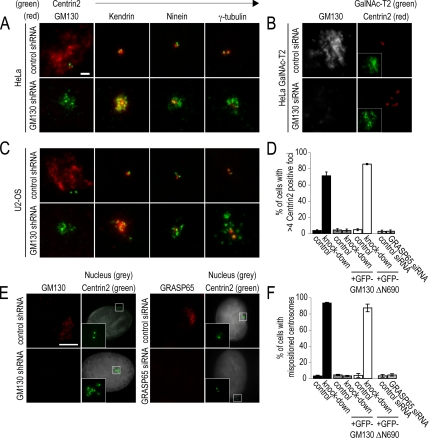
We next investigated whether the aberrant spindle poles present in GM130-depleted cells during mitosis could have resulted from aberrant interphase centrosomes. To address this issue, we examined centrosomes morphology during interphase by immunofluorescence with antibodies to marker proteins of the centrosome and the pericentriolar matrix. Although control cells contained the normal set of two or four centrioles, GM130-depleted cells had supernumerary Centrin2-positive foci (Figure 5A), whereas normal Centrin2 expression levels were maintained (data not shown). Interestingly, these foci did not stain for all markers, as they contained Kendrin, but not the mother centriole-specific protein, Ninein (Ou et al., 2002), or the pericentriolar matrix proteins, γ-tubulin (Figure 5A), GCP2, and GCP-WD (data not shown).
Figure 5.
GM130-depleted cells contain abnormal and mispositioned centrosomes. (A) HeLa cells were transfected with the control (control shRNA) or knockdown plasmid (GM130 shRNA), fixed 72 h later, and processed for immunofluorescence. Centrosome morphology was revealed by staining cells with antibodies to Centrin2 (green) and GM130, Kendrin, Ninein, or γ-tubulin (all in red). (B) GalNAc-T2 cells were stained with antibodies to GM130 (gray) and Centrin2 (red). GFP-GalNAc-T2 (green) is shown as small inset revealing the overall organization of the GA. (C) Interphase control and GM130-depleted U2-OS cells were analyzed by immunofluorescence as described for HeLa cells in A. Scale bar, (A–C) 2 μm. (D) Control and GM130-depleted U2-OS cells expressing RNAi-resistant GM130 (+GFP-GM130) or the C-terminus of GM130 (+GFP-ΔN690) were analyzed for the presence of abnormal centrosomes. We also analyzed and quantified the organization of centrosomes in control and GRASP65-depleted U2-OS cells. The percentage of cells with more than four Centrin2-positive foci is presented (n = 3). (E) Control, GM130-, and GRASP65-depleted U2-OS cells were stained with antibodies to GM130 (red), Centrin2 (green), and the DNA dye Hoechst (gray). The small inset corresponds to the boxed region in each figure. (F) The percentage of cells with mispositioned centrosomes in respect to the nucleus was quantified for each condition. Scale bar, 5 μm. n = 3.
To verify our findings, we examined the morphology of interphase centrosomes in additional GM130-depleted cell lines. We detected aberrant centrosomes in HeLa GalNAc-T2 (Figure 5B), U2-OS (Figure 5C), and SaOS-2 (data not shown) cells that were similar to those seen in HeLa cells. Normal centrosome morphology was restored in U2-OS cells by the expression of GFP-GM130, but not GFP or the GFP-tagged GM130 C-terminus (GFP-ΔN690), which corrects GRASP65 localization. This result suggests that the function of GM130 in centrosome regulation is independent of GRASP65, which is consistent with the detection of normal centrosomes in GRASP65-depleted cells (Figure 5, D and E). Interestingly, the abnormal centrosomes of GM130-depleted cells were mislocalized above the nucleus in more than 90% of GM130-depleted U2-OS cells, whereas control centrosomes localized adjacent to the nucleus (Figure 5E). This mispositioning phenotype was corrected by expression of RNAi-resistant GM130, but not the C-terminus of GM130, GFP-ΔN690 (Figure 5E). We conclude that GM130 is required for the normal organization and positioning of centrosomes during interphase.
Centrosome Defects Precede the Spindle Defects in the Absence of GM130
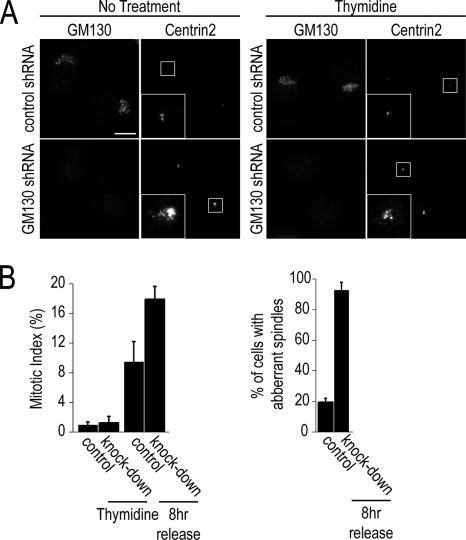
To examine whether the centrosome and the spindle defects of GM130-depleted cells were linked, we carried out cell cycle synchronization studies in HeLa cells, in which both phenotypes are observed. We first treated cells with the nucleotide analog thymidine, which is known to cause cells to arrest in S-phase. Similar to nonsynchronized cells, 70–80% of S-phase–arrested GM130-depleted cells contained multiple Centrin2-positive foci (Figure 6A). We then removed thymidine and allowed control and knockdown cells to progress through the cell cycle and to enter mitosis synchronously. In contrast to control cells, which had normal bipolar spindles, more than 90% of GM130-depleted cells contained aberrant spindles, which resulted in mitotic arrest. Post mitotic cell death, which was frequently seen in such experiments, was prevented by treatment with the caspase inhibitor ZVAD, explaining the higher mitotic index in knockdown versus control cells. Similar results were obtained with GM130-depleted cells in which GRASP65 localization to the GA was corrected (data not shown). These results suggest that aberrant centrosomes are generated independent of cell cycle progression and precede the formation of aberrant spindles. We therefore propose that the primary role of GM130 is the regulation of the interphase centrosome.
Figure 6.
Defects in spindle formation are a consequence of abnormal interphase centrosomes. (A) Control and GM130-depleted HeLa cells were left untreated or incubated with thymidine for 18 h, followed by fixation and processing for immunofluorescence with antibodies to GM130 and Centrin2. The centrosomes of representative cells are shown in insets that correspond to the boxed image area. (B) Cells arrested in S-phase with thymidine or released from a thymidine block for 8 h were fixed and processed for immunofluorescence with antibodies to phospho-Histone H3 and α-tubulin to reveal the mitotic index and spindle organization, respectively. The cells were also incubated with the caspase inhibitor ZVAD to prevent apoptotic cell death. The percentage of cells in mitosis and with aberrant spindles are shown (n = 3).
GM130 Depletion in U2-OS Cells Leads to a Delay at the G2-M Transition
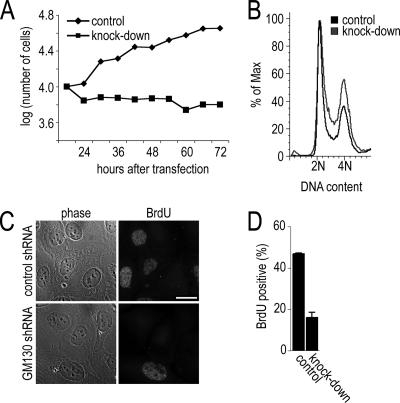
In the course of our experiments, we found that the centrosomal phenotypes in GM130-depleted cells differed depending on whether a cell line was p53-positive or -negative. Although we were able to detect defective mitotic spindles in p53-deficient cell lines, p53-positive cell lines appeared to arrest before mitosis. We first analyzed their growth behavior by monitoring their proliferation rate over a period of 3 d and found that GM130-depleted cells were unable to grow (Figure 7A). To examine whether these cells were arrested at a particular stage in the cell cycle, we determined the DNA profile of Hoechst-labeled control and knockdown cells using flow cytometry. We observed that a nonsynchronized population of GM130-depleted cells contained ∼30% more cells in the G2 peak with 4 N DNA content than control cells (Figure 7B), suggesting that the cells were delayed at the G2-M transition. We also assayed these cells for the incorporation of BrdU in S-phase. Control and GM130-depleted U2-OS cells were pulse-labeled with BrdU and tested for their ability to incorporate BrdU into DNA during replication (Figure 7C). Although 47% of control cells contained BrdU-labeled DNA, only 17% of GM130-depleted cells were able to incorporate BrdU (Figure 7D). Taken together, p53-positive, GM130-depleted cells proliferate at a reduced rate and appear to be delayed at the G2-M transition. These findings are consistent with the reduced incorporation of BrdU and the absence of mitotic cells.
Figure 7.
GM130 depletion leads to an overall reduction in cell growth and cell cycle delay at the G2-M transition. (A) The growth of cells treated with control or GM130-specific siRNA was monitored over a period of 3 days. (B) U2-OS cells were transfected with control or knockdown plasmid, and 72 h later were labeled with Hoechst 33342 for 30 min and subjected to flow cytometry. The profiles of cells are representative of three experiments (>30,000 cells per profile). (C) Control (control shRNA) and knockdown (GM130 shRNA) U2-OS cells were pulse-labeled with BrdU for 30 min, fixed 24 h later, and processed for immunofluorescence with a BrdU-specific antibody. Representative phase-contrast and immunofluorescence images are shown. Scale bars, 10 μm. (D) The percentage of BrdU-positive control and knockdown cells is shown as average values of three independent experiments.
Abnormal Centrosomes of GM130-depleted Cells Are Defective with Respect to Microtubule Organization and Cell Migration
We next investigated whether the effects of GM130 depletion on centrosome morphology were accompanied by functional defects by examining microtubule organization. Control cells contained microtubules that originated predominantly at the centrosome forming a radial cytoskeleton network. In contrast, the majority of microtubules in GM130-depleted cells were organized as thick and elongated bundles next to the nucleus. In addition, relatively few microtubules nucleated at the centrosome below the nucleus (Figure 8A, white arrow). Consistent with this result, the staining of cells with antibodies to the microtubule tip-binding protein EB1 revealed a significant reduction of EB1-positive comets (Figure 8B). GM130 depletion thus appears to affect centrosomally nucleated microtubules, but also the overall assembly of the microtubule array.
We also analyzed the organization of acetylated tubulin and found that GM130-depleted cells were unable to form stable, acetylated microtubules (Figure 8C). This phenotype was, however, corrected by expressing either full-length GM130 or its C-terminus. Although our results demonstrate that the loss of GM130 leads to aberrant centrosomes that are unable to nucleate dynamic microtubules, the role of GM130 in microtubule acetylation appears to be indirect through the regulation of GRASP65 localization.
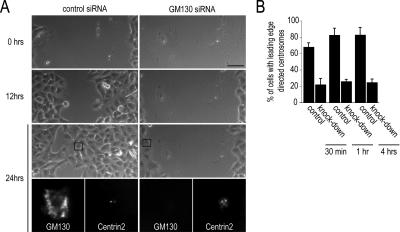
As cell migration has been shown to correlate with centrosome reorientation in many cell types (Yvon et al., 2002), we also tested whether GM130 depletion had an effect on cell migration. We used a wound-healing assay, in which cells migrate into a scratch wound, to evaluate the ability of a cell to reposition its centrosome. Using live imaging, we observed that A549 lung carcinoma cells depleted of GM130 by siRNA were unable to migrate, and the wound in the monolayer remained open (Figure 9A). In contrast, control cells closed the gap in the monolayer within 24 h. Immunostaining with antibodies to GM130 and Centrin2, followed by quantifications, confirmed that the cells that failed to migrate were negative for GM130 and contained aberrant centrosomes that were unable to reorient (Figure 9B). These results, which were confirmed through scratch wound assays in U2-OS cells (data not shown), suggest that GM130 is required for cell migration through its regulatory role on the centrosome, but possibly also through effects on microtubule assembly and dynamics.
Figure 9.
GM130-depleted cells cannot migrate or reorient their centrosomes in wound healing assays. (A) Migration of control and GM130-depleted A549 cells into a scratch wound was monitored by live imaging. After the last time point, cells were fixed and stained with antibodies to GM130 to visualize GM130 depletion and to Centrin2 to observe the organization of the centrosome. The cells in the bottom panel correspond to the cells shown in boxes in the phase-contrast image. Scale bar, 50 μm. (B) The percentage of control and knockdown cells with centrosomes that are oriented toward the wound edge were quantified 30 min, 1 h, and 4 h after introduction of the scratch wound.
ldlG Cells Contain Aberrant Interphase Centrosomes and Defective Mitotic Spindles
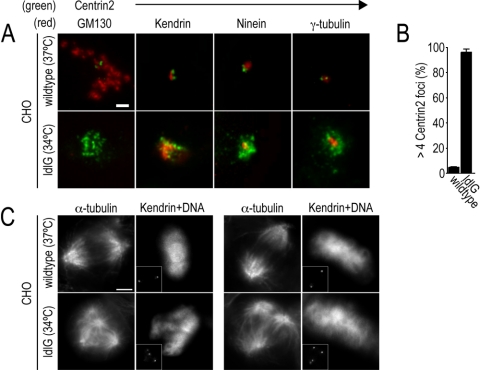
To confirm that the abnormal interphase centrosomes and defective mitotic spindles in GM130-depleted cells are not an artifact of RNAi-mediated protein knockdown, we examined the organization of the centrosome in ldlG cells, a mutant CHO cell line that constitutively lacks detectable GM130 (Vasile et al., 2003). The majority of interphase ldlG cells, but not the parental wild-type CHO cells, contained aberrant Centrin2-positive foci, in which Kendrin, but not Ninein and γ-tubulin was detected (Figure 10, A and B). Furthermore, mitotic ldlG cells contained aberrant spindles that were unable to align chromosomes in the metaphase plate (Figure 10C). In contrast to the other p53-negative cell lines used in this study, ldlG cells with aberrant spindles did not arrest in metaphase and did not undergo cell death. Instead, they were able to proliferate, which may be due to the absence of the spindle checkpoint in CHO cells and the fact that this mutant cell line has been selected for viability (Hobbie et al., 1994; Hut et al., 2003). Overall, these experiments provide compelling evidence for a role of GM130 in the regulation of centrosome organization during interphase and spindle formation during mitosis.
Figure 10.
ldlG cells contain abnormal centrosomes in interphase and defective spindles in mitosis. Wild-type and GM130-deficient (ldlG) CHO cells, grown at 37 or 34°C, respectively, were fixed and processed for immunofluorescence. (A) Representative interphase cells are shown which are stained with antibodies to Centrin2 (green) and to GM130, Kendrin, Ninein, and γ-tubulin (all in red). Scale bar, 2 μm. (B) The percentage of cells with >4 Centrin2-positive foci determined for parental CHO and ldlG cells (n = 3). (C) Wild-type and ldlG cells in mitosis were stained with antibodies to α-tubulin and Kendrin (shown in the small inset) to monitor the organization of the mitotic spindle and spindle poles, respectively. DNA organization was monitored by staining cells with the dye Hoechst 33342. Two representative wild-type and mutant cells are shown. Scale bar, 5 μm.
DISCUSSION
In this study, we have identified a novel and unexpected role for GM130 in the regulation of centrosome morphology, position, and function during interphase. GM130 appears to be necessary for the normal organization of the centrosome as its depletion resulted in aberrant centrosomes during interphase and nonfunctional multipolar spindles during mitosis. We performed a number of confirmatory studies to verify this novel finding because it has not been described in previous GM130 depletion studies (Preisinger et al., 2004; Puthenveedu et al., 2006; Marra et al., 2007). We used two RNAi-mediated approaches, achieving 90% protein knockdown with both siRNA and plasmid-based shRNA. We also replicated our knockdown studies by individually targeting two distinct sequences in the GM130 cDNA. We repeated our knockdown studies with five distinct human cell lines and detected the same centrosome phenotype in each. For example, we observed both aberrant centrosome morphology during interphase and altered spindle formation during mitosis in the HeLa GalNAc-T2 cell line previously used for GM130 depletion studies (Puthenveedu et al., 2006). We showed that the centrosome phenotypes in interphase and mitosis are the specific consequence of GM130 depletion as they could be corrected in functional rescue experiments. Finally, the role of GM130 in centrosome regulation is bolstered by our observation that aberrant interphase centrosome and defective mitotic spindles are seen in ldlG cells, a CHO cell line that does not produce detectable GM130 (Vasile et al., 2003).
A number of possible reasons may explain why this novel role for GM130 in centrosome morphology has not been previously described. Studies of GM130 have focused on its known role in protein transport and GA biogenesis and may not have examined the organization of the centrosome in the absence of GM130 (Nakamura et al., 1997; Lowe et al., 1998; Seemann et al., 2000; Puthenveedu et al., 2006). Furthermore, the ability to detect centrosome phenotypes during interphase and spindle defects during mitosis can be influenced by the choice of cell lines and the use of different experimental parameters. For instance, we found that the spindle phenotype was best detected in p53-deficient cells, because they did not undergo cell cycle arrest in response to changes in centrosome morphology. We also found that the knockdown efficiency was dependent on the cell culture conditions and plating confluency (Materials and Methods).
Although our results suggest that GM130 is necessary for both normal centrosome morphology during interphase and mitotic spindle formation, its primary regulatory role appears to be on the interphase centrosome. We detected alterations in centrosome morphology during interphase in all GM130-depleted cell lines, independent from active cell cycle progression, and in a GM130-deficient cell line. However, we only detected defective multipolar spindles in p53-deficient cell lines, such as HeLa, SaOS-2, and CHO cells, because these cells were able to enter mitosis. In contrast, when we depleted GM130 from p53-positive cell lines, such as U2-OS cells, we observed a delay at the G2-M transition, leading to an overall reduction in the rate of cell proliferation. Our results indicate that loss of GM130 affects cell cycle progression at the G2-M transition in a p53-dependent manner. Further investigations will be necessary to characterize the details of the observed effect on cell cycle progression.
Our findings lead us to propose that GM130 can affect cell cycle–specific centrosome organization and function via two independent pathways: 1) GM130 controls the localization of GRASP65 to the GA via its C-terminal domain. Although we have demonstrated that GA-bound GRASP65 is necessary for the regulation of microtubule acetylation, its significance for mitotic spindle formation is not understood. 2) GM130 also has a role in the regulation of centrosome morphology, position, and function during interphase. This latter function does not appear to depend on GRASP65 for the following reasons. First, in contrast to depletion of GM130, GRASP65 depletion does not affect the organization and function of the centrosome (Figure 5, D–F). Second, relocalizing GRASP65 to the GA in GM130-depleted cells does not correct the aberrant centrosome phenotypes during interphase and mitosis (Figures 4B, 5D, and F). Third, the composition of the spindle poles in GM130-depleted cells and GRASP65-depleted cells is different (Figure 2A; Sutterlin et al., 2005).
In addition to the effects on the centrosome, we found that GM130 depletion resulted in defects in microtubule organization and cell migration. These phenotypes can be explained as the consequence of altered interphase centrosomes. Alternatively, GM130 may control microtubule organization and cell migration independently of its role in regulating centrosome function. For example, the GA has been proposed to behave as a potent MTOC in interphase cells (Chabin-Brion et al., 2001; Efimov et al., 2007), and GM130 from its location on the GA may mediate this effect, possibly via GRASP65. GM130 has also been shown to activate the Ste20-like kinase, YSK1, which has a functional role in cell migration (Preisinger et al., 2004).
Our findings represent the first report of a role for a GA protein in the regulation of the interphase centrosome. We propose that GM130 is necessary for the normal organization of the centrosome as its depletion resulted in aberrant centrosomes during interphase and nonfunctional multipolar spindles during mitosis. There is precedent for a functional link between the GA and the centrosome as we have previously shown that the GA protein, GRASP65, is involved in spindle formation during mitosis (Sutterlin et al., 2005). Several other proteins of the GA or the early secretory pathway, including Tankyrase-1 and Rint-1, have also been shown to be required for the formation of a bipolar spindle (Chang et al., 2005; Lin et al., 2007). A novel functional connection between the GA and the centrosome during interphase is intriguing because it would coincide with the time in the cell cycle when the two organelles are in close physical proximity. Until now, the pericentriolar location of the GA during interphase has been recognized as a feature of mammalian cells that is not found in yeast, insects, or plants, but the significance has not been understood (Colanzi et al., 2003). We propose that from its pericentriolar position, the interphase GA, through associated proteins such as GM130, may regulate the formation and function of centrosomes. It is, however, also conceivable that GA proteins act directly at the centrosome.
ACKNOWLEDGMENTS
We thank Drs. Ming Tan, Tau-Mu Yi, and Naomi Morrissette and members for the Sütterlin lab for comments on the manuscript. We are grateful to Drs. Adam Lindsted, Vivek Malhotra, Monty Krieger, Jeffrey Salisbury, Antonino Colanzi, Mikiko Takahashi, Ed Chan, and Ingrid Ruf for providing critical reagents and to Eigen Peralta for help with the FACS analysis. This work was supported the University of California Cancer Research Coordinating Committee funds to C.S. and by a UCI-HHMI Faculty Fellow Award to C.S. made possible by a grant to UC Irvine and Diane O'Dowd from the Howard Hughes Medical Institute Professors Program.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0847) on November 28, 2007.
REFERENCES
- Barr F. A., Nakamura N., Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T. R., Bernards R., Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K., Marceiller J., Perez F., Settegrana C., Drechou A., Durand G., Pous C. The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandar N., Billig B., McMaster J., Novak J. Inactivation of p53 gene in human and murine osteosarcoma cells. Br. J. Cancer. 1992;65:208–214. doi: 10.1038/bjc.1992.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P., Coughlin M., Mitchison T. J. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- Colanzi A., Suetterlin C., Malhotra V. Cell-cycle-specific Golgi fragmentation: how and why? Curr. Opin. Cell Biol. 2003;15:462–467. doi: 10.1016/s0955-0674(03)00067-x. [DOI] [PubMed] [Google Scholar]
- Efimov A., et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory M. R., Davis T. N. The centrosomal proteins pericentrin and kendrin are encoded by alternatively spliced products of one gene. Genomics. 2003;82:401–405. doi: 10.1016/s0888-7543(03)00119-8. [DOI] [PubMed] [Google Scholar]
- Geiser T., Ishigaki M., van Leer C., Matthay M. A., Broaddus V. C. H(2)O(2) inhibits alveolar epithelial wound repair in vitro by induction of apoptosis. Am. J Physiol. Lung Cell Mol. Physiol. 2004;287:L448–L453. doi: 10.1152/ajplung.00177.2003. [DOI] [PubMed] [Google Scholar]
- Hobbie L., Fisher A. S., Lee S., Flint A., Krieger M. Isolation of three classes of conditional lethal Chinese hamster ovary cell mutants with temperature-dependent defects in low density lipoprotein receptor stability and intracellular membrane transport. J. Biol. Chem. 1994;269:20958–20970. [PubMed] [Google Scholar]
- Hut H. M., Lemstra W., Blaauw E. H., Van Cappellen G. W., Kampinga H. H., Sibon O. C. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol. Biol. Cell. 2003;14:1993–2004. doi: 10.1091/mbc.E02-08-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan G., Traganos F., James W. M., Ray J. M., Roberge M., Sauve D. M., Anderson H., Darzynkiewicz Z. Histone H3 phosphorylation and expression of cyclins A and B1 measured in individual cells during their progression through G2 and mitosis. Cytometry. 1998;32:71–77. doi: 10.1002/(sici)1097-0320(19980601)32:2<71::aid-cyto1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L., Sluder G., Cassels G., Sibon O., Wang C. L. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y. D., Nigg E. A. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- La Terra S., English C. N., Hergert P., McEwen B. F., Sluder G., Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Liu C. C., Gao Q., Zhang X., Wu G., Lee W. H. RINT-1 serves as a tumor suppressor and maintains Golgi dynamics and centrosome integrity for cell survival. Mol. Cell. Biol. 2007;27:4905–4916. doi: 10.1128/MCB.02396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M., Rabouille C., Nakamura N., Watson R., Jackman M., Jamsa E., Rahman D., Pappin D. J., Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- Marra P., Salvatore L., Mironov A., Jr, Di Campli A., Di Tullio G., Trucco A., Beznoussenko G., Mironov A., De Matteis M. A. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol. Biol. Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Lowe M., Levine T. P., Rabouille C., Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nigg E. A. Origins and consequences of centrosome aberrations in human cancers. Int. J. Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- Ou Y. Y., Mack G. J., Zhang M., Rattner J. B. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 2002;115:1825–1835. doi: 10.1242/jcs.115.9.1825. [DOI] [PubMed] [Google Scholar]
- Peel N., Stevens N. R., Basto R., Raff J. W. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C., Korner R., Wind M., Lehmann W. D., Kopajtich R., Barr F. A. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C., Short B., De Corte V., Bruyneel E., Haas A., Kopajtich R., Gettemans J., Barr F. A. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J. Cell Biol. 2004;164:1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D. M., Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- Seemann J., Jokitalo E. J., Warren G. The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol. Biol. Cell. 2000;11:635–645. doi: 10.1091/mbc.11.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B., White J., Rottger S., Stelzer E. H., Suganuma T., Nilsson T. Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C., Hsu P., Mallabiabarrena A., Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Polishchuk R., Pecot M., Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol. Biol. Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Shibata H., Shimakawa M., Miyamoto M., Mukai H., Ono Y. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J. Biol. Chem. 1999;274:17267–17274. doi: 10.1074/jbc.274.24.17267. [DOI] [PubMed] [Google Scholar]
- Uetake Y., Loncarek J., Nordberg J. J., English C. N., La Terra S., Khodjakov A., Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile E., Perez T., Nakamura N., Krieger M. Structural integrity of the Golgi is temperature sensitive in conditional-lethal mutants with no detectable GM130. Traffic. 2003;4:254–272. doi: 10.1034/j.1600-0854.2003.00080.x. [DOI] [PubMed] [Google Scholar]
- Yvon A. M., Walker J. W., Danowski B., Fagerstrom C., Khodjakov A., Wadsworth P. Centrosome reorientation in wound-edge cells is cell type specific. Mol. Biol. Cell. 2002;13:1871–1880. doi: 10.1091/mbc.01-11-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]