Abstract
Renin, a major regulatory component of the renin-angiotensin system, plays a pivotal role in regulating blood pressure and electrolyte homeostasis and is predominantly expressed in the kidney. Several cAMP-responsive elements have been identified within renin gene promoters. Here, we study how 2 such elements, renin proximal promoter element-2 (RP-2) and overlapping cAMP and negative regulatory elements (CNRE), affect the transcriptional regulation of renin. We generated Tg mice (TgM) bearing BACs containing either WT or mutant RP-2 or CNRE, integrated at single chromosomal loci. Analysis of the TgM revealed that RP-2 was essential to basal promoter activity in the kidney, while renin mRNA levels did not significantly change in any tissues tested in the CNRE mutant TgM. To evaluate the physiological significance of these mutations, we used the BAC Tg to rescue hypotensive Renin-null mutant mice. As predicted, no renin expression was observed in the kidneys of RP-2 mutant/Renin-null compound mice, whereas renin expression in CNRE mutant compound mice was indistinguishable from that in control mice. Consistent with this, RP-2 mutant animals were hypotensive, while CNRE mutants had normal blood pressure. Thus, transcriptional regulation of renin expression via RP-2 but not CNRE is critical for blood pressure regulation by this gene.
Introduction
Renin is an aspartyl protease synthesized mainly in the juxtaglomerular (JG) cells of the kidney. It catalyzes cleavage of a unique substrate, angiotensinogen, a plasma protein synthesized predominantly in the liver; cleavage yields the decapeptide angiotensin I (AI). AI is further processed by angiotensin-converting enzyme to produce the octapeptide angiotensin II (AII), which mediates vasoconstriction and aldosterone secretion through angiotensin receptors (ATs), leading to increased BP. Renin catalyzes the first, rate-limiting reaction of the renin-angiotensin system and thus plays a central role in regulating overall activity of the renin-angiotensin system. Our lab and others have shown by gene ablation in the mouse that renin is essential to BP homeostasis (1, 2).
A number of physiological stimuli, such as renal sympathetic activity, sodium depletion, and low BP, induce renin transcription and secretion in order to maintain BP homeostasis. Several of these stimuli utilize cAMP as an intracellular signal (reviewed in ref. 3). In JG cells, noradrenalin released from sympathetic nerve endings activates adenylate cyclase. Intracellular cAMP concentration in JG cells can also be modified directly or indirectly by low distal tubular salt load, increased NO and prostaglandins, and/or decreased adenosine release from the macula densa. Pharmacological blockade of AII synthesis or receptor function also increases renin secretion and expression in vivo. In this process, systemic influences such as BP rather than direct AII action on JG cells may be a major cause of renin induction (4).
Most inbred mouse strains can be classified into 2 groups in terms of renin alleles, one carrying a single renin gene, Ren-1C (e.g., C57BL/6), and the other carrying 2 tandemly arrayed genes, Ren-2 and Ren-1D (e.g., DBA/2). The Ren-2 allele arose from gene duplication of Ren-1 (5, 6). Because of variations in their 5′ regulatory sequences, the tissue specificity of expression of the Ren-1 and Ren-2 genes is similar but not identical. As noted above, the most abundant sites of renin synthesis are the kidney JG cells, where both Ren-1 and Ren-2 genes are expressed at approximately equal levels (7). Other extrarenal renin production sites, such as the adrenal gland, testis, ovary, heart, and submandibular gland (SMG), have also been identified, though expression levels in these tissues are lower. Curiously, Ren-1 and Ren-2 expression differ dramatically in the SMG, where Ren-2 expression is more than 100-fold higher than that of Ren-1 (7).
Because of a lack of suitable cell lines, progress in the analysis of renin gene transcription has been slow and has generally utilized renin-expressing cells from outside the kidney or cells that are ordinarily non–renin producing (8–10). In 1990, however, a JG cell–like, renin-secreting cell line, As4.1, was isolated from the kidney of Tg mice (TgM) carrying a Ren-2 promoter–SV40–T antigen construct (11). Using As4.1 and other cell lines, several potential cAMP-responsive sequences have been identified within renin gene promoters (reviewed in ref. 12). Among these, we focused on 2 putative cAMP-responsive elements, CNRE (overlapping cAMP-responsive element [CRE] and negative regulatory element [NRE]), and RP-2/PPE (renin proximal promoter element-2/proximal promoter element).
CNRE was identified within the 5′ flanking region of both mouse Ren-1 and Ren-2 but is not present in homologous human genes (at –603 to –591 relative to the transcription start site of Ren-1C). This element confers cAMP responsiveness through CRE-binding factor (CREB) (13) or LXRα transcription factor (14, 15). It has been proposed that CNRE may also govern differential Ren-1 and Ren-2 expression in the SMG (13, 16–18).
The RP-2/PPE has been identified in the proximal promoter region of the mouse Ren-1C gene (at –75 to –47; refs. 19, 20); analogous elements are conserved in Ren-1D, Ren-2, rat, and human promoters (21). By transiently transfecting Ren-1C promoter constructs into cultured cells, RP-2/PPE was shown to be involved in basal (19, 20) as well as cAMP-inducible transcriptional activity (22). Subsequently, it was proposed that a complex of HOXD10, PBX1b, and PREP1 binds to the RP-2/PPE and activates Ren-1C promoter in As4.1 cells (21).
Due to position effects, qualitative and quantitative traits of Tg expression differ even between independent TgM lines bearing identical DNA constructs. Therefore, in order to compare the levels of Tg expression between WT and mutant (mut) loci, a large number of TgM lines must be analyzed. To overcome this problem, we adopted a combination of 2 technologies: BAC transgenesis and coplacement (23). BAC inserts (~200 kb) carry more genetic information than commonly used plasmids (~20 kb). Furthermore, genes carried on a BAC appear to be more resistant to position effects. Accordingly, for example, a 145–kilo base pair (145-kbp) mouse BAC clone encompassing Ren-1D and Ren-2 alleles exhibited appropriate expression pattern in TgM (24). Tg coplacement, which we describe below, is based on Cre/loxP-mediated recombination. We employed the technique in this study in order to place WT and mut loci into exactly the same position in the genome, thus eliminating possible position effects.
We generated TgM lines harboring a 155-kbp Ren-1C BAC DNA, either with WT or mut Ren-1C promoter sequences, both integrated at the same chromosomal site (23, 25). Analyses of these animals revealed that RP-2/PPE in the Ren-1C gene promoter is essential to its basal transcriptional activity in the kidney. On the other hand, no significant change in Ren-1C mRNA accumulation was observed in any tissues tested in the mut CNRE TgM. Furthermore, the mut CNRE locus responded normally to modulators of renin gene transcription, such as high-salt loading, dehydration, and a potent angiotensin-converting enzyme inhibitor (captopril) that severely reduces BP. These results demonstrate that the CNRE is dispensable or redundant for both basal and activated Ren-1C gene expression in response to typical renin-transcriptional stimuli.
We then generated Ren-1C-BAC-Tg:Ren-1C–null compound mice to evaluate the physiological significance of cis mutations by determining whether they could functionally rescue the low BP phenotype of Ren-1C–null (Ren-1C–/–) mut mice (1). The compound Tgmut CNRE BAC:Ren-1C–/– mice displayed a renin expression profile that was indistinguishable from that of the TgWT CNRE BAC:Ren-1C–/– or heterozygous Ren-1C–null (Ren-1C+/–) mice. In contrast, compound Tgmut RP-2 BAC:Ren-1C–/– mice showed no renin expression in the kidney. Accordingly, the plasma renin activity (PRA), AI concentration, and systolic BP (SBP) of the Tgmut RP-2 BAC:Ren-1C–/– mice were significantly lower than those of WT control mice.
These results demonstrate that BP homeostasis can be regulated through transcriptional mechanisms controlling renin gene expression and that RP-2 but not CNRE plays a crucial role in this mechanism.
Results
A Tg coplacement strategy for direct comparison of gene expression.
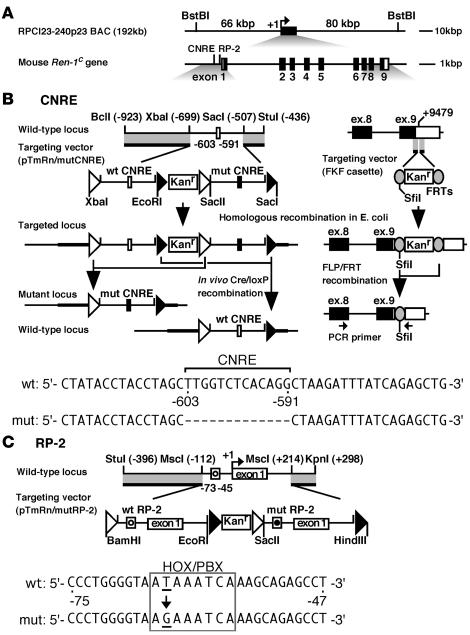
The combined use of BAC Tg and coplacement strategy allowed us to precisely evaluate in vivo activity of 2 candidate cis-regulatory elements in the renin gene, i.e., CNRE and RP-2/PPE (Figure 1, B and C). The CNRE is a composite element, consisting of the CRE and NRE. To determine whether the CNRE might be involved in cAMP responsiveness and/or cell-type specificity, we deleted the entire CNRE sequences from the Ren-1C promoter (Figure 1B). It has been reported that the RP-2/PPE binds to a HOXD10-PBX1b-PREP1 transcription factor complex and that a single nt mutation in the element abrogated factor binding and transcriptional activation in As4.1 cells (21). We therefore introduced this mutation into the RP-2/PPE sequences (Figure 1C).
Figure 1. Tg coplacement strategy.
(A) Mouse Ren-1C BAC. The 155-kbp BstI restriction enzyme fragments with 66 kbp and 80 kbp of 5′ and 3′ flanking sequences, respectively, were used for microinjections. Shown are cis-regulatory elements and 9 exons of the gene. (B, upper left) Map of the Ren-1C promoter region and structure of the targeting vector (pTmRn/mutCNRE), in which WT (open box) and mut CNRE (mut; filled box) sequences as well as homologous sequences to the target locus (thick lines) are included. Restriction enzyme sites with their positions relative to the transcriptional start site (+1) are shown. The loxP2272 and loxP5171 sequences are shown as open and filled triangles, respectively. The targeted locus was generated in E. coli and used to establish TgM lines. Following intercross with Cre-expressing TgM, selective excision in utero of the DNA segment between a pair of loxP5171 or loxP2272 sites generated either WT or mut loci, respectively. (B, upper right) Introduction of the FRT (shaded ovals) and SfiI sites into 3′-UTR of the gene. The targeting and flippase/FRT (FLP/FRT) recombination was carried out in the E. coli strain EL250 (26). The PCR primer set that simultaneously amplifies Tg and endogenous renin transcripts is shown by arrows. (B, bottom) Comparison of WT and mut CNRE promoter sequences. (C, top) Targeting vector (pTmRn/mutRP-2) for modification of the RP-2. Both WT (open circle) and mut (filled circle) RP-2 sequences as well as homologous sequences to the target (thick lines) are included. Tg coplacement was performed as described in B. (C, bottom) Comparison of the WT and mut RP-2 sequences. The HOX/PBX motif is boxed.
The Ren-1C coding sequences (9 kb) are centrally located within the RPCI23-240p23 BAC (Figure 1). To distinguish Tg Ren-1C from endogenous mouse renin sequences in expression and structural analyses, we first introduced flippase recombination target (FRT) and SfiI restriction enzyme sites (133 bp) into the 3′–untranslated region (3′-UTR) of the Ren-1C Tg by employing homologous recombination in E. coli (Figure 1B; see Methods for details) (26). This FRT-marked BAC was then modified independently in the promoter region (Figure 1, B and C). Targeting vectors included the kanamycin resistance (Kanr) gene cassette as a selectable marker in E. coli as well as both WT and mut promoter sequences, which were flanked by homologous sequences to the Ren-1C gene. In the RP-2–targeting vector (Figure 1C), for example, WT and mut promoter sequences were identical except for 1 bp, and both are flanked by a set of loxP sequence variants, loxP2272 and loxP5171 (27). These loxP variants each recombined efficiently with identical sequences but not with one another. Mutations were targeted into the 5′ regulatory region of the Ren-1C by homologous recombination in E. coli (26), and successful mutagenesis was confirmed by Southern blot analysis followed by direct sequencing of the BAC DNA (data not shown). We hereafter refer to these BACs (WT plus mut promoter inserted) loci as “parental.”
Generation of TgM with modified Ren-1C BAC.
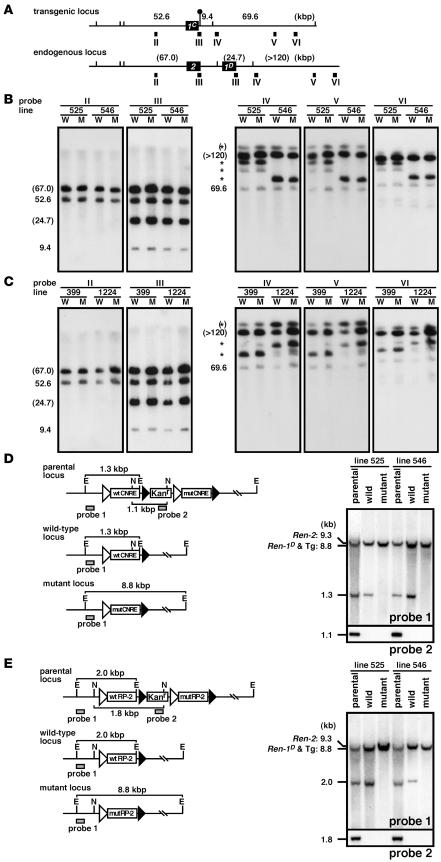
We injected a purified BAC DNA (BstBI fragment, 155 kbp; Figure 1) into fertilized mouse eggs collected from Ren-2 mice (ICR mice). Tail DNA of the offspring was screened by Tg-specific PCR and detailed Southern blot analyses (data not shown). For each construct, 2 TgM lines that carried single copies of the parental BAC DNA were obtained. The integrity of the BACs in these lines was determined by long-range Southern blot analysis (Figure 2, A–C). High molecular weight DNA from the thymi was digested with SfiI, separated on pulsed field gels (PFGs), and independently hybridized to 5 probes spanning the locus (Figure 2A). Because of the introduced SfiI restriction fragment length polymorphism (Figure 2A), DNA fragments from the Tg Ren-1C and endogenous Ren-2 and Ren-1D loci could be clearly discriminated. Most probes detected DNA fragments of the expected sizes, though some detected fragments that resulted from partial digests (Figure 2, B and C). Although we only show representative results on Cre/loxP-manipulated sublines (described below), we also performed the same analysis on each of the parental lines and obtained the expected results (data not shown).
Figure 2. Structural analysis of the Tg lines.
(A) Comparison of the Tg Ren-1C and endogenous Ren-2 and Ren-1D loci. The renin genes and SfiI sites are shown as filled boxes and vertical lines, respectively. The artificially introduced SfiI site is marked by a lollipop. Expected fragment sizes (in kbp) after complete enzyme digestion are shown above the line. The probes used for Southern blot analysis in B and C are indicated by solid rectangles (II–VI). (B and C) Southern blot analyses of the TgM carrying either CNRE (B) or RP-2 (C) modifications. Thymic cells from WT (W) and mut (M) TgM lines were embedded in agarose plugs, digested with SfiI, and separated by PFG electrophoresis. DNA blots were hybridized separately to probes (II–VI) shown in A. The sizes of the expected bands are indicated (in kbp) on the left of each panel. Those expected from the endogenous Ren2 and Ren-1D loci are in parentheses, and partially digested DNA is marked by asterisks. (D and E) Fine structural analyses of the CNRE (D) and RP-2 (E) promoter regions. (Left panels) In vivo Cre/loxP recombination removes the Kanr gene plus either mut or WT promoter sequences from the parental locus to generate either the WT or the mut locus, respectively. E, EcoRI; N, NcoI. (Right panels) Tail DNA from parental, WT, and mut TgM was digested with EcoRI (top) or NcoI (bottom), separated on agarose gels, transferred to a nylon membrane, and hybridized with probes 1 and 2 (shaded rectangles in the left panels), respectively.
As shown in Figure 1, Cre recombinase randomly recognizes either a pair of loxP2272 or loxP5171 and removes the intervening WT or mut sequences as well as the Kanr cassette to generate mut or WT Ren-1C loci, respectively. To initiate this reaction in vivo, we crossed parental Ren-1C TgM lines with TgM ubiquitously expressing Cre recombinase and obtained pups carrying both Tgs (Cre-F0). Individual mice that had undergone recombination, as determined by Southern blot analysis (data not shown), were crossed with non-Tg animals, and the genotypes of the offspring (Cre-F1) were discriminated by Southern blot analysis as shown in Figure 2, D and E. In the CNRE TgM line (Figure 2D), for example, a 1.1-kbp NcoI fragment (detected by probe 2 from the Kanr gene) in the parental locus disappeared in both WT and the mut loci after recombination. In addition, a 1.3-kbp EcoRI fragment (detected by probe 1) in the parental locus remained the same size in the WT and changed to 8.8 kbp in the mut locus. Analysis of these diagnostic fragments in the tail DNA from parental and sibling TgM in each line revealed that recombination took place properly (Figure 2, D and E). In addition, promoter regions of each TgM line were cloned by PCR and verified by sequencing analysis (data not shown).
In summary, we established 2 sets of WT and mut TgM lines for each of the CNRE (lines 525 and 546) and RP-2 (lines 399 and 1224) constructs, each of which carried at least 132 kbp of the BAC DNA (Figure 2, A–C).
Basal level Tg expression in the kidney.
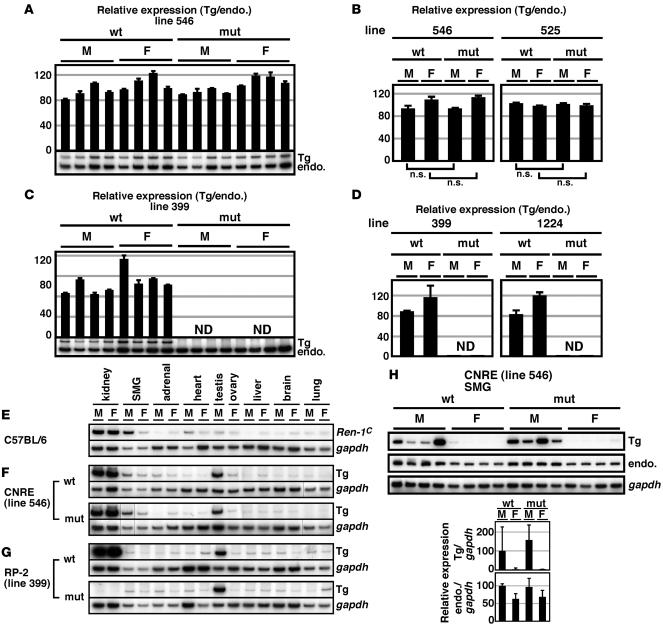
Total RNA was prepared from kidneys of BAC TgM, and expression of the Tg Ren-1C as well as endogenous Ren-2 and Ren-1D genes was analyzed by semiquantitative RT-PCR. A PCR primer set, common to all these genes, was designed to span the FRT sequences in the 3′-UTR of the Tg (Figure 1B) so that Tg as well as the endogenous gene could be simultaneously amplified; their products can be distinguished on a gel because of their size difference (133 bp; Figure 3, A and C). To eliminate individual differences originating from possible environmental effects, the signal from the Tg (single copy) was normalized to that of the endogenous Ren-2 and Ren-1D genes (4 copies). Summarized results are graphically represented in Figure 3, B and D. No significant difference in renal Tg expression was seen between the WT and mut CNRE TgM (Figure 3B). In contrast, in the mut RP-2 TgM, Ren-1C mRNA accumulation was barely detectable (Figure 3D). Essentially the same results were obtained in 2 sets of lines for each construct (Figure 3, B and D). These results demonstrated that RP-2/PPE, but not CNRE, is indispensable for basal transcriptional activity of the Ren-1C gene in the kidney.
Figure 3. Renin expression in TgM.
(A–D) Kidneys were isolated from 8-week-old Ren-1C BAC TgM (4 males [M] and 4 females [F] for each genotype [WT and mut] in lines 546, 525, 399, and 1224), and the RNA was analyzed. (A and C) Each value represents the ratio of Tg Ren-1C (Tg) expression to that of endogenous Ren-2 and Ren-1D (endo.), which served as an internal control. Relative Tg/endo. values for each individual are shown after normalization to the average value of the WT group, which was arbitrarily set at 100. Each sample was analyzed at least 3 times, and the means ± SD are shown only for lines 546 and 399. ND, not detected. (B and D) The means ± SD of each group in A and C are summarized. Results are shown also for lines 525 and 1224. P values were determined by 2-tailed Student’s t test. (E–G) RNA was isolated from various tissues of non-Tg (E), CNRE Tg (F), and RP-2 Tg (G) animals (8-week-old). Endogenous Ren-1C, Tg Ren-1C (Tg), and Gapdh gene expression was analyzed. Lines in F indicate that lanes were run on the same gel but were noncontiguous. (H) The SMG was isolated from Ren-1C TgM (8-week-old), and Tg Ren-1C (Tg), endogenous Ren-2 and Ren-1D (endo.), and Gapdh gene expression was analyzed. Below, the relative renin/GAPDH value for each individual mouse was calculated for both Tg and endogenous (endo.) renin genes and normalized to the average value of the WT male group, arbitrarily set at 100. Values are mean ± SD.
Extrarenal Tg expression.
Renin gene expression has been reported in extrarenal tissues, such as SMG, adrenal, heart, testis, and ovary (Figure 3E) (3, 12). We extracted total RNA from various extrarenal tissues of the TgM and analyzed gene expression using RT-PCR primer sets specific for the endogenous Ren-1C (Figure 3E), Tg Ren-1C (Figure 3, F–H), and Gapdh (Figure 3, E–H) genes. Although accurate quantitative comparison was difficult because of the low expression level in these samples, no difference in extrarenal Tg expression was seen between the WT and mut CNRE TgM (Figure 3F). To analyze renin gene expression in the SMG more precisely, we collected RNA from 4 male and 4 female TgM carrying either WT or mut CNRE sequences (Figure 3H). Because the level of Ren-2 mRNA in the SMG was roughly 100-fold higher than that of Ren-1D and because our PCR primer set for amplifying the endogenous renin transcripts was common to both Ren-2 and Ren-1D sequences, endogenous renin in Figure 3H probably reflects mostly Ren-2 expression. As expected from the fact that renin gene expression in the SMG is regulated by testosterone (28), the level of Tg Ren-1C as well as endogenous Ren-2 gene expression was higher in males than in females (Figure 3H). Although Tg Ren-1C gene expression was variable, no significant difference was seen between the WT and mut CNRE TgM. These results demonstrate that CNRE has no significant role in basal renin promoter activity in extrarenal tissues, including SMG. In sharp contrast to its indispensable role in renal gene expression, no significant difference in extrarenal Ren-1C gene expression was observed between the WT and mut RP-2 TgM (Figure 3G). This result demonstrates that RP-2/PPE is a JG cell–specific activator element; furthermore, single nt substitution within the RP-2/PPE does not completely disrupt the Ren-1C promoter activity in extrarenal tissues.
Renal Tg expression in response to physiological stimuli.
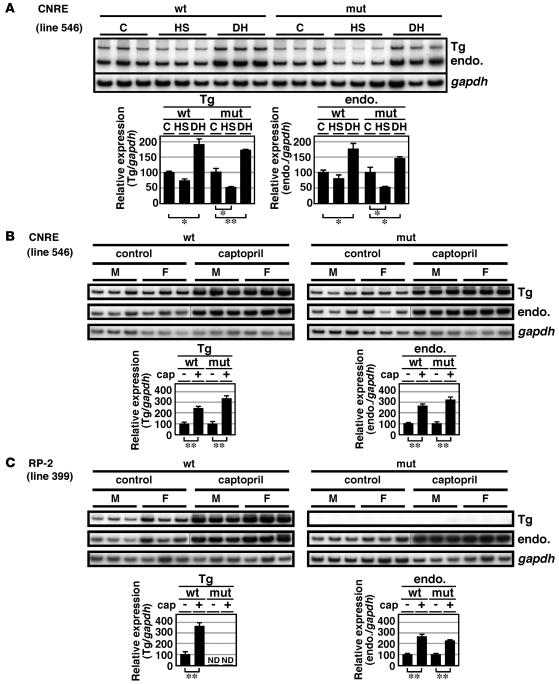
We tested to determine whether Tg CNRE loci were responsive to the physiological stimuli that are known to modify renin gene expression. Semiquantitative RT-PCR analysis of the RNA from kidneys of 8-week-old male TgM revealed that high-salt diet reduced and dehydration induced endogenous Ren-2 and Ren-1D gene expression by 20%–50% and 50%, respectively (Figure 4A). Ren-1C gene expression from both mut and WT CNRE Tg loci responded normally to both stimuli (Figure 4A), demonstrating that the CNRE is dispensable for response to these stimuli.
Figure 4. Effects of physiologic stimuli on renin gene expression in WT and TgM.
(A) Renin mRNA expression in the WT CNRE and mut CNRE TgM treated with high-sodium diet (HS) or dehydration (DH). Two groups of male TgM (8-week-old) were fed high-sodium (8%) or normal-sodium (0.6%; C for control) diets for 5 days. In another group, access to drinking water was restricted for 1 day (DH). RNA was isolated from the kidney and analyzed by semiquantitative RT-PCR using 2 sets of primer pairs, one coamplifying Tg Ren-1C (Tg) and endogenous Ren-2 and Ren-1D (endo.) genes and another specific for the Gapdh gene (top). Relative amount of renin mRNA after normalization to that of Gapdh was determined by 3 independent RT-PCR for 3 individuals in each group (bottom). Expression value of untreated control animals in each group was arbitrarily set at 100. (B and C) Renin mRNA expression in the TgM treated with or without captopril. Six pairs each of WT and mut TgM (8-week-old) were used in the study. One group was treated for 7 days with captopril dissolved in drinking water (0.5 mg/ml). Kidneys were isolated, and RNA was analyzed as described in A. Lines in B and C indicate that lanes were run on the same gel but were noncontiguous. *P < 0.05; **P < 0.01.
We then tested to determine whether Tg Ren-1C expression in the kidney could be increased by captopril administration. Semiquantitative RT-PCR analysis revealed that the level of endogenous Ren-2 and Ren-1D mRNA accumulation increased by 2- to 4-fold upon treatment of 8-week-old mice with captopril for 1 week (Figure 4, B and C). In both WT and mut CNRE TgM, captopril treatment of animals increased Tg Ren-1C mRNA to the same extent as endogenous genes, demonstrating that the CNRE is dispensable in the induction of renin gene expression by captopril (Figure 4B). In the mut RP-2 TgM, Tg Ren-1C mRNA accumulation was barely detectable, even after the animals were treated with captopril, whereas it increased by 3- to 4-fold in the WT RP-2 TgM (Figure 4C). Since RP-2/PPE is required for basal Ren-1C gene transcription in the kidney, we cannot determine from this result whether this element is involved in captopril induction of renin gene expression.
We analyzed CNRE mut lines 525 and 546 and RP-2 mut lines 399 and 1224 and obtained essentially the same results in both lines, although data are shown only for lines 546 and 399 (Figure 4, A–C). We decided to use lines 546 and 399 for the rescue experiment.
Generation of Ren-1C-BAC-Tg:Ren-1C–null compound mice.
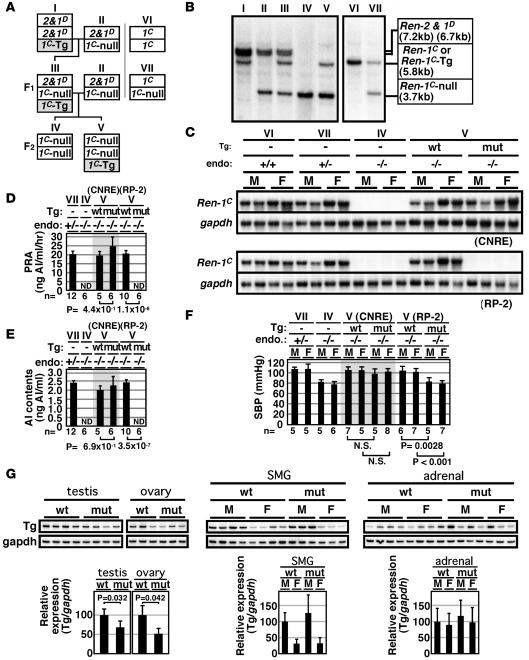
To evaluate the physiological significance of the Ren-1C cis elements, we set up breeding to introduce 1 copy of the BAC Tg (WT or mut) into the Ren-1C–null (Ren-1C–/–) genetic background, which was originally generated in the C57BL/6 inbred mouse strain (1). To facilitate genotyping, we first backcrossed the Ren-1C–/– mouse with an ICR mouse (5 generations; Figure 5A), then cross-mated it with an ICR mouse carrying the Ren-1C BAC Tg (TgRen-1C BAC). The F1 progeny (III), which are heterozygous for Ren-1C–null and Ren-2 and Ren-1D alleles and hemizygous for the BAC Tg (TgRen-1C BAC:Ren-1C+/–), was crossed with a mouse heterozygous for Ren-1C–null and Ren-2 and Ren-1D alleles (Ren-1C+/–). The F2 progeny were screened to identify a mouse homozygous for the Ren-1C–null alleles, either with (TgRen-1C BAC:Ren-1C–/–) or without (Ren-1C–/–) the Ren-1C BAC Tg. In the crosses, the loss of the endogenous Ren-2 and Ren-1D fragments (7.2 and 6.7 kbp, respectively) and the gain of the Ren-1C BAC Tg fragment (5.8 kbp) were confirmed by allele-specific Southern blot analysis of tail-tip DNA (Figure 5B).
Figure 5. BAC Tg–mediated rescue of the Ren-1C–null mouse.
(A) Breeding strategy for introducing a single copy of WT or mut Ren-1C BAC Tg into the Ren-1C–null genetic background. 2&1D, endogenous Ren-2 and Ren-1D loci; 1C-Tg, Ren-1C BAC Tg; 1C-null, targeted Ren-1C locus; 1C; endogenous Ren-1C locus. (B) Southern blot analysis for discriminating the genotype. Tail-tip DNA was digested with BamHI and hybridized with a Ren-1C genomic DNA probe (PstI-XbaI fragment around exon 5), which hybridized to diagnostic bands from the Ren-2 (7.2 kb) and Ren-1D (6.7 kb), Ren-1C or Tg (5.8 kb), and Ren-1C–null (3.7 kb) loci. (C) Northern blot analysis of renin gene expression in the compound mice. Total RNA from the kidney of 2-month-old animals was hybridized with a mouse Ren-1C cDNA probe (KpnI-NcoI fragment, 820 bp) and mouse Gapdh cDNA probe (453 bp, nt 565 to 1,017; MUSGAPDH, GenBank accession no. M32599). endo, endogenous Ren-1C genotype. (D–F) PRA (8-week-old), AI contents (8-week-old), and SBP (6-week-old) of the compound mice. Data are means ± SD; n is shown below each bar. (G) Renin mRNA levels in the testis, ovary, SMG, and adrenal of WT and mut RP-2 compound mice (8-week-old). Graph shows means ± SD; n = 3–4 per group. Expression values in the WT male animals were arbitrarily set at 1.0. Roman numerals refer to the genotypes shown in A.
Expression of the Ren-1C BAC Tgs in the absence of endogenous renin genes.
We analyzed renin gene expression in the kidney of the compound mouse by Northern blot analysis (Figure 5C). Endogenous Ren-1C gene expression was not detectable in the Ren-1C–/– mice, while Ren-1C+/– or WT control mice (Ren-1C+/+) expressed high levels that were indistinguishable from each other (Figure 5C) (29). In the TgWT CNRE BAC:Ren-1C–/– or the TgWT RP-2 BAC:Ren-1C–/– (Figure 5C) mice, abundant Ren-1C gene expression was observed at levels similar to those in the animals heterozygous (Ren-1C+/–) or homozygous (Ren-1C+/+) for the intact Ren-1C allele. Importantly, in situ hybridization analysis of the WT BAC-Tg:Ren-1C–null compound mouse revealed that Tg Ren-1C expression was restricted to JG cells of the kidney (data not shown). The level of Tg Ren-1C gene expression in the kidney of the Tgmut CNRE BAC:Ren-1C–/– mouse was comparable to that seen in the TgWT CNRE BAC:Ren-1C–/– mouse (Figure 5C), which is consistent with the expression profile of the CNRE BAC TgM in the presence of endogenous allele (Figures 3 and 4). In the Tgmut RP-2 BAC:Ren-1C–/– mouse (Figure 5C), Tg failed to rescue renin gene expression (Figure 5C). This is consistent with the observation that the mut RP-2 locus did not respond to captopril-induced hypotension (Figure 4C).
Rescue of PRA, AI contents in the plasma, and BP.
We then measured PRA (Figure 5D) and serum concentration of AI (Figure 5E) in the compound mice by radioimmunoassays. As expected from the renal renin gene expression (Figure 5C), these parameters were rescued in TgWT CNRE BAC:Ren-1C–/–, TgWT RP-2 BAC:Ren-1C–/–, and Tgmut CNRE BAC:Ren-1C–/– mice (Figure 5, D and E). These parameters were statistically indistinguishable from those in the Ren-1C+/– mouse. Notably, both PRA and AI contents were below detectable level in the Tgmut RP-2 BAC:Ren-1C–/– mouse (Figure 5, D and E), just as in the Ren-1C–/– mouse.
We next measured the SBP of the animals by the tail-cuff method (Figure 5F). The SBP of the TgWT CNRE BAC:Ren-1C–/– and TgWT RP-2 BAC:Ren-1C–/– compound mice (WT) was comparable to that of the Ren-1C+/– as well as the WT (Ren-1C+/+, data not shown) animals. While SBP of the Tgmut CNRE BAC:Ren-1C–/– mice was normal, the Tgmut RP-2 BAC:Ren-1C–/– mice exhibited a hypotensive phenotype (Figure 5F) with BPs similar to those of the Ren-1C–/– mice and 20–25 mmHg lower than those of the Ren-1C+/– mice (1).
The BP phenotype observed in the compound animals was correlated with the pattern of Tg Ren-1C gene expression in the kidney (Figure 5C). We also compared Tg Ren-1C gene expression in extrarenal tissues (testis, ovary, SMG, and adrenal) of the TgWT RP-2 BAC:Ren-1C–/– and Tgmut RP-2 BAC:Ren-1C–/– compound mice (Figure 5G). Small but significant differences were observed in the testis and ovary, where expression was 30% and 50% lower, respectively, in the mut animals. No significant difference in Tg Ren-1C gene expression was observed in the SMG or adrenal glands. Thus, lack of PRA and low BP in the Tgmut RP-2 BAC:Ren-1C–/– mice is most likely attributable to almost complete loss of renin gene expression in the kidney.
Discussion
Studies using cultured cells have identified a large number of candidate cis-regulatory elements in the renin proximal promoter region. Among these, elements that may govern cell-type specificity and inducibility by cAMP have been most intensively studied, since these are the most obvious features of renin gene transcription. In the current work, we assessed the in vivo significance of 2 such elements, CNRE and RP-2/PPE, in the BAC TgM. We also characterized the phenotype resulting from BAC rescue of the Ren-1C–null mouse.
It has been proposed that CNRE may be involved in cell-type–restricted transcription of renin genes (13, 16–18). The model is based on the competition between CREB and the NRE-binding factor for the CNRE, in which CREB binds with higher affinity. In the SMG, this model proposes that CREB is sequestered by an inhibitory protein. Therefore, only NRE-binding factor can bind to the CNRE and suppress renin gene transcription (30). The fact that Ren-2 but not Ren-1 carries a 160-bp insertion adjacent to the NRE, which would disrupt the suppressive function of NRE on Ren-2, explains why expression of Ren-2 is much more abundant than that of Ren-1 in the SMG. It is therefore predicted that deletion of the CNRE from the Ren-1C promoter would affect only NRE function and lead to upregulation of the mut Ren-1C promoter in the SMG, as seen for the Ren-2. In the kidney, CREB preferentially activates the CNRE of both Ren-1 and Ren-2 genes, and the expression level of these genes is almost equivalent. Thus, it is predicted that deletion of the CNRE from the Ren-1C would lead to downregulation of the mut Ren-1C promoter in the kidney. In our experimental design, however, no significant difference in the activity of the WT and mut CNRE Ren-1C promoters was observed, indicating that CNRE either has no function or its activity is redundant. The promoter sequences of mut CNRE Ren-1C and Ren-2 are not exactly the same (31); the lack of obvious phenotype in the Ren-1C BAC TgM carrying mut CNRE might be attributed to this difference. In apparent conflict with the proposed model, however, it has been reported that the Ren-2 allele of the Mus hortulanus strain produces abundant Ren-2 mRNA in the SMG, even though its promoter has no insertion next to the NRE (31). Therefore, the mechanism by which locus-specific differences of renin gene expression are achieved in the SMG and kidney remains an open question.
Tamura et al. reported that LXRα, a transcription factor of the nuclear receptor family, could bind to the CNRE and confer cAMP responsiveness to the mouse renin gene promoter in AS4.1 cells (14, 15). In basal as well as induced conditions under several physiological stimuli, we did not observe significant differences in the WT and mut CNRE Ren-1C promoter activities. We therefore speculate that several other cAMP-responsive elements in the mouse distal enhancer (mdE at –2,697 to –2,690 of the transcriptional start site of the Ren-1C gene) (32, 33) and/or RP-2/PPE (at –75 to –47) (21, 22) may be compensating for the function of CNRE in the mut allele. An alternative explanation for our findings might be that the CNRE is not a cAMP-responsive element in vivo.
The RP-2/PPE was originally identified in the mouse Ren-1C promoter by using a cell line not derived from JG cells (19) and was shown to be involved in cAMP regulation of the mouse Ren-1C promoter (22). Subsequently, activity of this element in the regulation of mouse renin gene transcription was further demonstrated by reporter assays in As4.1 cells (20). These assays showed that mdE was capable of efficiently activating the Ren-1C promoter only when RP-2/PPE was intact (32). Pan et al. analyzed RP-2/PPE binding proteins in As4.1 nuclear extract and identified a HOXD10-PBX1b-PREP1 complex (21), although the relevance of this finding in JG cells of the kidney has not yet been confirmed. In this study, we introduced a single nt substitution into the RP-2/PPE sequences that disrupts factors binding in gel-shift assay and diminishes transcriptional activity in reporter assays (21). Although we clearly showed that RP-2/PPE sequences are critical for mouse Ren-1C promoter activity in vivo, it remains to be seen whether loss of HOXD10-PBX1b-PREP1 complex binding to the site is in fact responsible for the observed phenotype. ChIP assays on the WT and mut RP-2 promoters in the compound mice may be helpful in clarifying this issue; these experiments, however, are technically difficult because the kidney consists of many cell types and the renin-producing JG cell population is quite small (~1%) (7). Conventional gene ablation experiments showed that lack of either Pbx1 or Prep1 resulted in embryonic lethality (34, 35), and the BP phenotype of the Hoxd-10–null mouse has not yet been investigated (36). Conditional gene ablation experiments in the JG cells of the kidney may shed light on the functions of these and other transcription factors in the regulation of renin gene transcription and BP homeostasis, although possible complications originating from genetic redundancy must be taken into consideration.
Although our study did not reveal an important function of the CNRE, one may yet be uncovered by challenging TgM physiological stimuli different from those examined here. The RP-2/PPE sequence is conserved in human and rat promoter regions, and it is thus plausible that this element also plays a role in these species (37). The results presented here demonstrate that a single point mutation within the RP-2/PPE disrupts renin gene transcription. Therefore, subtle mutations in regulatory regions of human renin genes could potentially lead to BP dysregulation.
Methods
BAC clone and mutagenesis.
A BAC clone carrying the mouse Ren-1C gene (RPCI23-240p23; GenBank accession number AC068906) was screened (high-density replica filter of female C57BL/6J mouse DNA) and purchased from BAC/PAC Resources. In this clone (Figure 1), the transcriptional start site of the Ren-1C gene corresponded to nt position 89005; all the nt positions in this report were expressed relative to this site (+1). We introduced the 240p23 BAC DNA into the E. coli strain EL250 (a gift from N.A. Jenkins, National Cancer Institute, Frederick, Maryland, USA) in order to employ the prophage-recombination system, in which we essentially followed the method outlined by E.-C. Lee et al. (26).
Modification of 3′-UTR region of the Ren-1C gene.
To distinguish Tg Ren-1C from endogenous mouse renin sequences by RT-PCR or Southern blot analyses, we introduced a SfiI recognition site as well as FRT sequences into the 3′-UTR of the Ren-1C Tg by homologous recombination in E. coli. The targeting DNA fragment was PCR amplified from pIGCN21, which carries a Kanr gene flanked by FRT sequences (a gift from N.A. Jenkins), using the following primer set: 5′-ATTGGATTCGCCTTGGCCCGCTAAGGCCCTCTGCCACCCAGGCCAGCTGGGCCATCCACCGGATCTAGATAAC-3′ (SfiI site is underlined) and 5′-TGGCCCCCAGGACTGCCAGCTTGGCTTGGCCTAGGGTTACTTCCAGAAGTAGTGAGGAGG-3′. Sequences homologous to the Ren-1C gene (at nt positions 9440–9479 and 9480–9519) and template DNA are in plain text and italics, respectively, in each oligonucleotide sequence. EL250 carrying 240p23 BAC DNA was transformed with the targeting DNA, and the correct recombination event was confirmed by Southern blot and sequencing analyses. The Kanr DNA cassette was removed by flippase/FRT recombination in E. coli, leaving a 133-bp insertion with the SfiI recognition sequence at the 3′-UTR region of the Ren-1C gene (Figure 1B).
Introduction of mutations into the 5′ regulatory region of the Ren-1C gene.
To facilitate cloning procedures, we replaced the XhoI-SacI portion of the pBluescript II KS (+) with double-stranded oligonucleotides containing XhoI-StuI-MscI-BclI restriction enzyme sites; hereafter, we refer to this modified plasmid as pBluescript II KAS (+). A floxed Kanr cassette was PCR amplified and digested with EcoRI and SacII, as described previously (23).
WT CNRE DNA fragment (Figure 1B) was PCR amplified from 240p23 BAC DNA, using the following primer set, and digested with XbaI and EcoRI: 5′-GCTCTAGAATAACTTCGTATAGGATACTTTATACGAAGTTATGTCATTGGGCTCAGCCACCCT-3′ (XbaI) and 5′-GGAATTCTCTCGCCCACCTCCTTTATTTC-3′ (EcoRI). To make the mut CNRE DNA fragment, the following 2 primer sets were used for first-round PCR: (set 1) 5′-GTCCCCGCGGGTCATTGGGCTCAGCCACCCT-3′ (SacII; primer A) and 5′-TCTGATAAATCTTAGGCTAGGTAGGTATAGG-3′; (set 2) 5′-CTATACCTACCTAGCCTAAGATTTATCAGAGCTG-3′ and 5′-GGCGAGCTCTGGTGAGATAACTTCGTATAATGTGTACTATACGAAGTTATTCTCGCCCACCTCCTTTATTTC-3′ (SacI; primer B). Restriction enzyme sites are underlined and loxP sequences are italicized in each oligonucleotide sequence. The first-round PCR products were gel purified and subjected to second-round PCR with primers A and B, followed by digestion with SacII and SacI. 5′ regulatory sequences from the mouse Ren-1C gene (BclI-StuI, from –923 to –436) were cloned into BclI/StuI sites of pBluescript II KAS (+). The WT CNRE, Kanr, and mut CNRE DNA fragments were linked in that order (from 5′ to 3′) and used for substituting the corresponding portion (XbaI-SacI, from –699 to –507) of the above plasmid to derive pTmRn/mutCNRE (Figure 1B). A BclI-StuI fragment from the pTmRn/mutCNRE was used for BAC mutagenesis.
The WT RP-2 DNA fragment (Figure 1C) was PCR amplified with the following primer set and digested with BamHI and EcoRI: 5′-CGGGATCCATAACTTCGTATAGGATACTTTATACGAAGTTATGAAAACAGGCTGCCTTTCATGG-3′ (BamHI) and 5′-GGAATTCAAAGGACTGGATGATCCAAC-3′ (EcoRI). Mut RP-2 DNA fragment was prepared by first-round PCR with 2 primer sets shown below, followed by second-round PCR with the primers C and D and digestion with SacII and HindIII: (set 1) 5′-GTCCCCGCGGGAAAACAGGCTGCCTTTCATGG-3′ (SacII; primer C) and 5′-GCTCTGCTTTGATTTCTTACCCCAGGGCCT-3′; (set 2) 5′-CAGGCCCTGGGGTAAGAAATCAAAGCAGAG-3′ and 5′-GGCCCAAGCTTATAACTTCGTATAATGTGTACTATACGAAGTTATAAAGGACTGGATGATCCAACG-3′ (HindIII; primer D). Two segments from the mouse Ren-1C gene, 5′ homology (StuI-MscI, from –396 to –112) and 3′ homology (MscI-KpnI, from +214 to +298), were separately PCR amplified from 240p23 BAC DNA using the following primer sets, then digested with StuI/BamHI and HindIII/KpnI, respectively: (5′ homology) 5′-CAGGGGAAGGCCTATTCCA-3′ and 5′-CGGGATCCTGGCCAAGGACCCAGCTCCAT-3′ (BamHI); (3′ homology) 5′-GGCCCAAGCTTTGGCCAGCCAGTCAGGGATG-3′ (HindIII) and 5′-GGAGTTGTGGTACCCCTTAAT-3′. The 5′ homology, WT RP-2, Kanr, mut RP-2, and 3′ homology DNA fragments were linked in that order (from 5′ to 3′) and cloned into StuI/KpnI sites of pBluescript II KAS (+) to derive pTmRn/mutRP-2 (Figure 1C). A StuI-KpnI fragment from the pTmRn/mutRP-2 was used for BAC mutagenesis.
TgM.
Circular BAC DNA was purified from 500 ml culture of E. coli using NucleoBond BAC 100 kit (BD Biosciences — Clontech). To release insert DNA from the vector, BAC DNA was completely digested with BstBI restriction enzyme (sites are located at nt –66,295 and +89,190 relative to a transcription start site of renin gene; Figure 1A) in solution. Following fractionation of DNA fragments on PFG, low melting-point agarose blocks containing BAC inserts were excised from the gel and equilibrated at 4°C for 2 hours with injection buffer (10 mM Tris-Cl [pH 7.5], 0.1 mM EDTA, 100 mM NaCl, 3 μM spermine, 7 μM spermidine), including 1 buffer change. After gels were melted at 68°C for 30 minutes and incubated with β-agarase (New England BioLabs) at 42°C for 3 hours, the DNA solution was concentrated by centrifugation through a Millipore MC-filter (Ultrafree-MC 100,000 NMWL filter unit; Amicon) at 1,000 g. After allowing the DNA to resuspend on the filters for 2 days at 4°C, the solution was recovered and an aliquot of it was loaded onto a PFG to verify the integrity of the recovered DNA (38).
Purified DNA fragments were microinjected into the pronuclei of fertilized eggs from ICR mice (Charles River Laboratories). Since the ICR (closed colony strain) mouse available in Japan is heterogeneous at the renin locus, we only used a genotypically screened Ren-2 pool for our experiments. Tg founders were screened by Tg-specific PCR and Southern blot analyses. For genotyping the Cre TgM, in which Cre recombinase expression is under the control of CMV promoter (39), the following PCR primers were used: 5′-GGCTGCCACGACCAAGTGAC-3′ and 5′-CGTGAGATATCTTTAACCCTGATC-3′.
Animal experiments were carried out in a humane manner under approval from the Institutional Animal Experiment Committee of the University of Tsukuba. Experiments were in accordance with the Regulation of Animal Experiments of the University of Tsukuba and the Fundamental Guidelines for Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Structural analysis of the Tg.
For long-range structural analysis of the Tg, agarose-embedded thymus DNA was treated with SfiI, fractionated by PFG electrophoresis, and capillary transferred onto nylon membrane (PerkinElmer). Blots were hybridized with [α-32P]-labeled DNA probes (Figure 2A), which were PCR amplified, subcloned, and verified by DNA sequencing. nt positions of each probe in the Ren-1C allele were as follows: II (–21,443 to –20,878), III (+9,143 to +9,709), IV (+19,167 to +19,563), V (+60,999 to +61,493), and VI (+75,024 to +75,511). All of these probes cross-hybridize with endogenous Ren-2 and Ren-1D sequences (Figure 2A).
Cre/loxP-mediated in vivo recombination was confirmed by Southern blot analysis of tail-tip DNA, which was digested with EcoRI and hybridized with renin-specific probe 1 (–1,737 to –1,224). Removal of the Kanr gene cassette was confirmed by NcoI digestion of tail DNA followed by hybridization with probe 2 (from Kanr gene).
Semiquantitative RT-PCR.
Total RNA (2.7 μg) was converted to cDNA using random hexamers and reverse transcriptase (ReverTra Ace; TOYOBO). One-twentieth of the cDNA was analyzed by semiquantitative RT-PCR in the presence of [α-32P]dCTP (94°C for 5 minutes; 25 [renin] or 18 [GAPDH] cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute; and 72°C for 5 minutes). Products were separated on 8% polyacrylamide gels and analyzed by phosphorimaging.
For the analysis of renal basal level expression, signals for Tg Ren-1C mRNA were normalized to those of the endogenous Ren-2 and Ren-1D genes. Primer sequences common to all 3 genes are as follows: 5′-CAGTACGGACTACGTGCTACA-3′ (+8,736 to +8,756) and 5′-AGCTTGGCTTGGCCTAGGGT-3′ (+9,483 to +9,502). Since the primers were designed to span the 3′ FRT of the Tg (Figure 1B), the PCR products from Ren-2 and Ren-1D transcripts (223 bp) and Tg Ren-1C transcripts (356 bp) are separable on the gel.
For analyzing extrarenal expression as well as expression in induction experiments, the endogenous and Tg Ren-1C, endogenous Ren-2 and Ren-1D, and Gapdh genes were separately analyzed by the following primer sets specific for each gene: Tg Ren-1C, 5′-TAACTTCTCCATGGTAGCCTC-3′ (this primer was set in the 3′-FRT region) and 5′-CAAAGCCAGACAAAATGGCCC-3′ (+9,514 to +9,534), product size 104 bp; endogenous Ren-1C, Ren-2, and Ren-1D, 5′-GCCCTCTGCCACCCAGTAA-3′ (+9,465 to +9,483) and 5′-CAAAGCCAGACAAAATGGCCC-3′ (+9,514 to +9,534), product size 70 bp; Gapdh, 5′-TCACTGGCATGGCCTTCC-3′ and 5′-CAGGCGGCACGTCAGATC-3′, product size 65 bp (nt 723 to 787, MUSGAPDH; GenBank accession no. M32599). Signals for Ren-1C, Ren-2, and Ren-1D transcripts were normalized to that of Gapdh.
Radioimmunoassay.
Blood samples were withdrawn from anesthetized mice (8 weeks old) and collected into ice-cold tubes containing EDTA, which were immediately centrifuged to isolate plasma. PRA was estimated by measuring the rate of AI formation. The concentration of AI was determined by radioimmunoassays (40).
BP.
SBP was measured by a programmable sphygmomanometer (BP-98A; Softron) using the tail-cuff method as described previously (41). Statistical analysis for comparison of BP was performed using 2-tailed Student’s t test. A P value less than 0.05 was considered significant.
Acknowledgments
We thank James Douglas Engel (University of Michigan, Ann Arbor, Michigan, USA) and Jorg Bungert (University of Florida, Gainesville, Florida, USA) for critical reading of the manuscript. This work was supported by grants from the Astellas Foundation for Research on Metabolic Disorders (to K. Tanimoto), the Japan Heart Foundation (to K. Tanimoto), the Uehara Memorial Foundation (to K. Tanimoto), the Tokyo Biochemical Research Foundation (to K. Tanimoto), the Takeda Science Foundation (to K. Tanimoto), the Grant-in-Aid for Exploratory Research from the Japan Society for the Promotion of Science (to K. Tanimoto), and the Special Coordination Fund for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K. Tanimoto).
Footnotes
Nonstandard abbreviations used: AI, angiotensin I; CNRE, overlapping cAMP and negative regulatory elements; CRE, cAMP-responsive element; CREB, CRE-binding factor; FRT, flippase recombination target; JG, juxtaglomerular; Kanr, kanamycin resistance; kbp, kilo base pair(s); mut, mutant; NRE, negative regulatory element; PFG, pulsed field gel; PPE, proximal promoter element; PRA, plasma renin activity; RP-2, renin proximal promoter element-2; SBP, systolic BP; SMG, submandibular gland; TgM, Tg mouse (mice); UTR, untranslated region.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:1006–1016 (2008). doi:10.1172/JCI33824
References
- 1.Yanai K., et al. Renin-dependent cardiovascular functions and renin-independent blood-brain barrier functions revealed by renin-deficient mice. J. Biol. Chem. 2000;275:5–8. doi: 10.1074/jbc.275.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Clark A.F., et al. Renin-1 is essential for normal renal juxtaglomerular cell granulation and macula densa morphology. J. Biol. Chem. 1997;272:18185–18190. doi: 10.1074/jbc.272.29.18185. [DOI] [PubMed] [Google Scholar]
- 3.Bader M., Ganten D. Regulation of renin: new evidence from cultured cells and genetically modified mice. J. Mol. Med. 2000;78:130–139. doi: 10.1007/s001090000089. [DOI] [PubMed] [Google Scholar]
- 4.Matsusaka T., et al. Chimeric mice carrying ‘regional’ targeted deletion of the angiotensin type 1A receptor gene. Evidence against the role for local angiotensin in the in vivo feedback regulation of renin synthesis in juxtaglomerular cells. J. Clin. Invest. 1996;98:1867–1877. doi: 10.1172/JCI118988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abel K.J., Gross K.W. Close physical linkage of the murine Ren-1 and Ren-2 loci. Nucleic Acids Res. 1988;16:2111–2126. doi: 10.1093/nar/16.5.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abel K.J., Gross K.W. Physical characterization of genetic rearrangements at the mouse renin loci. Genetics. 1990;124:937–947. doi: 10.1093/genetics/124.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Field L.J., Gross K.W. Ren-1 and Ren-2 loci are expressed in mouse kidney. Proc. Natl. Acad. Sci. U. S. A. 1985;82:6196–6200. doi: 10.1073/pnas.82.18.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura K., Tanimoto K., Murakami K., Fukamizu A. Activation of mouse renin promoter by cAMP and c-Jun in a kidney-derived cell line. Biochim. Biophys. Acta. 1993;1172:306–310. doi: 10.1016/0167-4781(93)90218-3. [DOI] [PubMed] [Google Scholar]
- 9.Germain S., Konoshita T., Philippe J., Corvol P., Pinet F. Transcriptional induction of the human renin gene by cyclic AMP requires cyclic AMP response element-binding protein (CREB) and a factor binding a pituitary-specific trans-acting factor (Pit-1) motif. Biochem. J. 1996;316:107–113. doi: 10.1042/bj3160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ying L., Morris B.J., Sigmund C.D. Transactivation of the human renin promoter by the cyclic AMP/protein kinase A pathway is mediated by both cAMP-responsive element binding protein-1 (CREB)-dependent and CREB-independent mechanisms in Calu-6 cells. J. Biol. Chem. 1997;272:2412–2420. doi: 10.1074/jbc.272.4.2412. [DOI] [PubMed] [Google Scholar]
- 11.Sigmund C.D., et al. Isolation and characterization of renin-expressing cell lines from transgenic mice containing a renin-promoter viral oncogene fusion construct. J. Biol. Chem. 1990;265:19916–19922. [PubMed] [Google Scholar]
- 12.Pan L., Gross K.W. Transcriptional regulation of renin: an update. Hypertension. 2005;45:3–8. doi: 10.1161/01.HYP.0000149717.55920.45. [DOI] [PubMed] [Google Scholar]
- 13.Horiuchi M., Pratt R.E., Nakamura N., Dzau V.J. Distinct nuclear proteins competing for an overlapping sequence of cyclic adenosine monophosphate and negative regulatory elements regulate tissue-specific mouse renin gene expression. J. Clin. Invest. 1993;92:1805–1811. doi: 10.1172/JCI116770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K., et al. LXRalpha functions as a cAMP-responsive transcriptional regulator of gene expression. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8513–8518. doi: 10.1073/pnas.100519097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morello F., et al. Liver X receptors alpha and beta regulate renin expression in vivo. J. Clin. Invest. 2005;115:1913–1922. doi: 10.1172/JCI24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura N., Burt D.W., Paul M., Dzau V.J. Negative control elements and cAMP responsive sequences in the tissue-specific expression of mouse renin genes. Proc. Natl. Acad. Sci. U. S. A. 1989;86:56–59. doi: 10.1073/pnas.86.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett G., et al. Identification of a negative regulatory element involved in tissue-specific expression of mouse renin genes. Proc. Natl. Acad. Sci. U. S. A. 1992;89:885–889. doi: 10.1073/pnas.89.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada T., et al. In vivo identification of a negative regulatory element in the mouse renin gene using direct gene transfer. J. Clin. Invest. 1995;96:1230–1237. doi: 10.1172/JCI118156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K., Tanimoto K., Murakami K., Fukamizu A. A combination of upstream and proximal elements is required for efficient expression of the mouse renin promoter in cultured cells. Nucleic Acids Res. 1992;20:3617–3623. doi: 10.1093/nar/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrovic N., et al. Role of proximal promoter elements in regulation of renin gene transcription. J. Biol. Chem. 1996;271:22499–22505. doi: 10.1074/jbc.271.37.22499. [DOI] [PubMed] [Google Scholar]
- 21.Pan L., et al. An Abd-B class HOX.PBX recognition sequence is required for expression from the mouse Ren-1c gene. J. Biol. Chem. 2001;276:32489–32494. doi: 10.1074/jbc.M011541200. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K., et al. Mechanism of cAMP regulation of renin gene transcription by proximal promoter. J. Clin. Invest. 1994;94:1959–1967. doi: 10.1172/JCI117547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu T., et al. Identification of cis-regulatory sequences in the human angiotensinogen gene by transgene coplacement and site-specific recombination. Mol. Cell. Biol. 2005;25:2938–2945. doi: 10.1128/MCB.25.8.2938-2945.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullins L.J., et al. Granulation rescue and developmental marking of juxtaglomerular cells using “piggy-BAC” recombination of the mouse ren locus. J. Biol. Chem. 2000;275:40378–40384. doi: 10.1074/jbc.M007315200. [DOI] [PubMed] [Google Scholar]
- 25.Siegal M.L., Hartl D.L. Transgene Coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E.C., et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 27.Lee G., Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 28.Catanzaro D.F., Mesterovic N., Morris B.J. Studies of the regulation of mouse renin genes by measurement of renin messenger ribonucleic acid. Endocrinology. 1985;117:872–878. doi: 10.1210/endo-117-3-872. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi N., et al. Ren1c homozygous null mice are hypotensive and polyuric, but heterozygotes are indistinguishable from wild-type. J. Am. Soc. Nephrol. 2005;16:125–132. doi: 10.1681/ASN.2004060490. [DOI] [PubMed] [Google Scholar]
- 30.Horiuchi M., Nakamura N., Tang S.S., Barrett G., Dzau V.J. Molecular mechanism of tissue-specific regulation of mouse renin gene expression by cAMP. Identification of an inhibitory protein that binds nuclear transcriptional factor. J. Biol. Chem. 1991;266:16247–16254. [PubMed] [Google Scholar]
- 31.Abonia J.P., et al. Evaluating a model of an NRE mediated tissue-specific expression of murine renin genes. Hypertension. 2001;37:105–109. doi: 10.1161/01.hyp.37.1.105. [DOI] [PubMed] [Google Scholar]
- 32.Pan L., et al. Critical roles of a cyclic AMP responsive element and an E-box in regulation of mouse renin gene expression. J. Biol. Chem. 2001;276:45530–45538. doi: 10.1074/jbc.M103010200. [DOI] [PubMed] [Google Scholar]
- 33.Klar J., Sandner P., Muller M.W., Kurtz A. Cyclic AMP stimulates renin gene transcription in juxtaglomerular cells. Pflugers Arch. 2002;444:335–344. doi: 10.1007/s00424-002-0818-9. [DOI] [PubMed] [Google Scholar]
- 34.Selleri L., et al. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- 35.Ferretti E., et al. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol. Cell. Biol. 2006;26:5650–5662. doi: 10.1128/MCB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carpenter E.M., Goddard J.M., Davis A.P., Nguyen T.P., Capecchi M.R. Targeted disruption of Hoxd-10 affects mouse hindlimb development. Development. 1997;124:4505–4514. doi: 10.1242/dev.124.22.4505. [DOI] [PubMed] [Google Scholar]
- 37.Pan L., Glenn S.T., Jones C.A., Gross K.W. Activation of the rat renin promoter by HOXD10.PBX1b.PREP1, Ets-1, and the intracellular domain of notch. J. Biol. Chem. 2005;280:20860–20866. doi: 10.1074/jbc.M414618200. [DOI] [PubMed] [Google Scholar]
- 38.Tanimoto K., Liu Q., Grosveld F., Bungert J., Engel J.D. Context-dependent EKLF responsiveness defines the developmental specificity of the human epsilon-globin gene in erythroid cells of YAC transgenic mice. Genes Dev. 2000;14:2778–2794. doi: 10.1101/gad.822500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu R.N., Ito M., Saunders T.L., Camper S.A., Jameson J.L. Role of Ahch in gonadal development and gametogenesis. Nat. Genet. 1998;20:353–357. doi: 10.1038/3822. [DOI] [PubMed] [Google Scholar]
- 40.Fukamizu A., et al. Chimeric renin-angiotensin system demonstrates sustained increase in blood pressure of transgenic mice carrying both human renin and human angiotensinogen genes. J. Biol. Chem. 1993;268:11617–11621. [PubMed] [Google Scholar]
- 41.Tanimoto K., et al. Angiotensinogen-deficient mice with hypotension. J. Biol. Chem. 1994;269:31334–31337. [PubMed] [Google Scholar]