Abstract
The initiation of meiotic recombination by the formation of DNA double-strand breaks (DSBs) catalysed by the Spo11 protein is strongly evolutionary conserved. In Saccharomyces cerevisiae, Spo11 requires nine other proteins for meiotic DSB formation, but, unlike Spo11, few of these proteins seem to be conserved across kingdoms. In order to investigate this recombination step in higher eukaryotes, we have isolated a new gene, AtPRD1, whose mutation affects meiosis in Arabidopsis thaliana. In Atprd1 mutants, meiotic recombination rates fall dramatically, early recombination markers (e.g., DMC1 foci) are absent, but meiosis progresses until achiasmatic univalents are formed. Besides, Atprd1 mutants suppress DSB repair defects of a large range of meiotic mutants, showing that AtPRD1 is involved in meiotic recombination and is required for meiotic DSB formation. Furthermore, we showed that AtPRD1 and AtSPO11-1 interact in a yeast two-hybrid assay, suggesting that AtPRD1 could be a partner of AtSPO11-1. Moreover, our study reveals similarity between AtPRD1 and the mammalian protein Mei1, suggesting that AtPRD1 could be a Mei1 functional homologue.
Keywords: Arabidopsis, crossover, DSB, meiosis, recombination
Introduction
In sexually reproducing organisms, meiosis takes place allowing the transition from the sporophytic to the gametophytic state. During the first meiotic division, homologous chromosomes pair and undergo a high level of homologous recombination. Cross-overs (CO) and genetic conversions non-associated with CO (NCO) are the consequences of meiotic recombination between homologous chromosomes, reorganising the allelic combinations among them. CO also establish physical links between homologous chromosomes (chiasmata) necessary for the proper segregation of chromosomes at the first meiotic division (Roeder, 1997).
In budding yeast, meiotic recombination events are initiated by double-strand breaks (DSBs) catalysed by the Spo11 protein (Bergerat et al, 1997; Keeney et al, 1997). The eukaryotic meiosis specific protein Spo11 shares homology with the catalytic subunit (TOP6A) of the archeal type VI topoisomerase from Sulfolobus shibatae (Bergerat et al, 1997), suggesting that Spo11 catalyses the formation of meiotic DSBs via a 5′ phosphotyrosyl linkage.
Meiotic defects resulting from Spo11 disruption have been identified in several species, including fungi, invertebrates, mammals and plants, suggesting that the enzymatic generation of DSBs is evolutionarily conserved (Keeney, 2001). Spo11 is encoded by a single gene in most species and its mutation causes a drastic decrease in meiotic recombination, leading to a sterility phenotype (Dernburg et al, 1998; McKim and Hayashi-Hagihara, 1998; Celerin et al, 2000; Storlazzi et al, 2003). Interestingly, plant genomes (at least Arabidopsis and rice) contain three Spo11 putative homologues (Hartung and Puchta, 2000; Grelon et al, 2001; Hartung and Puchta, 2001). In Arabidopsis, disruption of AtSPO11-1 induces a sterility phenotype, associated with a drastic decrease in meiotic recombination, suggesting that at least this gene encodes a true Spo11 orthologue (Grelon et al, 2001). Recently, Stacey et al (2006) showed that disruption of a second putative ortholog, AtSPO11-2, induces defects similar to those induced by the disruption of AtSPO11-1, suggesting that initiation of meiotic recombination in Arabidopsis requires two Spo11 homologues. By contrast, AtSPO11-3 is not involved in meiosis, but plays a major role during somatic development (Hartung et al, 2002; Sugimoto-Shirasu et al, 2002; Yin et al, 2002), suggesting that plants have retained a topoisomerase VI function in addition to the meiotic specialisation of Spo11 common to higher eukaryotes.
In Saccharomyces cerevisiae, meiotic DSB formation requires nine other proteins in addition to Spo11 (Rad50, Mre11, Xrs2, Rec102, Rec104, Rec114, Ski8, Mer2 and Mei4) (Keeney, 2001). Despite many investigations, very little is known about the molecular function of these proteins, but recent studies tend to group them into distinct subcomplexes, that would interact together to form a large recombination complex in which DSBs are made (Usui et al, 1998; Trujillo and Sung, 2001; Kee and Keeney, 2002; Jiao et al, 2003; Arora et al, 2004; Li et al, 2006; Maleki et al, 2007). Some additional factors as phosphorylation by the cyclin-dependent kinase Cdc28 or chromatin remodelling are also known to modulate DSB formation (Yamada et al, 2004; Henderson et al, 2006; Ogino et al, 2006).
Many investigations have been conducted to identify in other organisms homologues of these DSB-forming proteins. Unfortunately, unlike Spo11, most of these (Rec102, Rec104, Rec114, Mei4 and Mer2) are not conserved across kingdoms, at least at the amino-acid level. Furthermore, even when sequences showing amino-acid similarity to DSB proteins are identified, their role in meiotic DSB formation is generally missing. For example, the Mre11/Rad50/Xrs2 complex was also identified in fission yeast (Rad32/Rad50/Nbs1), in mammals (Mre11/Rad50/Nbs1) and in Arabidopsis (AtMRE11/AtRAD50/AtNBS1), but in fission yeast, as well as in Arabidopsis, different studies revealed that when involved in meiotic recombination, these homologues are required for the repair but not for the induction of Spo11-dependent meiotic DNA breaks (Gallego et al, 2001; Bleuyard et al, 2004; Puizina et al, 2004; Young et al, 2004; Akutsu et al, 2007). Likewise, if Ski8 appears to be conserved, the Arabidopsis ski8 mutant displays no recombination defects, indicating that the meiotic function of SKI8 is not conserved in Arabidopsis (Jolivet et al, 2006).
Given the fact that these reverse genetic approaches have not been helpful to unravel DSB formation mechanisms, new approaches have been used to identify new candidates involved in the initiation of meiotic recombination. In Schizosaccharomyces pombe, five new proteins involved in DSB formation: Mde2, Rec6, Rec10, Rec15 and Rec24, were identified from transcriptomic data associated with a study of mutant phenotypes (Gregan et al, 2005; Martin-Castellanos et al, 2005). These proteins do not show homology with proteins from other organisms, suggesting that they are specific to S. pombe. Very few data are currently available for multicellular species. In Drosophila melanogaster, mei-P22 was isolated in a large screen for P-element insertion (Sekelsky et al, 1999). γH2AX and γH2Av foci (used as DSB markers) were absent in Mei-P22 mutants and X-ray treatment rescued the mutant phenotype, indicating that Mei-P22 is required for DSB formation (Liu et al, 2002; Mehrotra and McKim, 2006). Similarly, the mouse Mei1 protein is a good candidate for being a higher eukaryote DSB-forming protein (Libby et al, 2003; Reinholdt and Schimenti, 2005).
In this study, we present the identification of a new gene, AtPRD1, whose mutation affects meiosis in plants. Our results show that AtPRD1 is involved in meiotic recombination and is required for meiotic DSB formation. Furthermore, we show that AtPRD1 and AtSPO11-1 interact in a yeast two-hybrid assay, suggesting that AtPRD1 could be a partner of AtSPO11-1. Moreover, our study reveals similarities between AtPRD1 and Mei1, suggesting that AtPRD1 could be a Mei1 functional homologue.
Results
Identification and molecular characterisation of Atprd1 mutants
In a screen for T-DNA insertions that generate meiotic mutants (Mercier et al, 2001), a line showing a semi-sterile phenotype was identified. The semi-sterile phenotype segregated 3:1 as a single recessive mutation, and was found to be linked to the T-DNA insertion, allowing us to isolate the tagged meiotic gene (see Materials and methods). The mutant and the corresponding gene were called Atprd1-1 and AtPRD1, respectively (for Arabidopsis thaliana Putative Recombination initiation Defect 1). The T-DNA was found to be inserted in a predicted open reading frame (ORF) of the Arabidopsis genome, At4g14180. We selected from the SALK database (Alonso et al, 2003) a new insertion in At4g14180 (SALK_024703, Atprd1-2; Supplementary data Sup_1A). We observed that this new insertion was also responsible for a meiotic phenotype when homozygous, and that both mutants were allelic (see Materials and methods). Furthermore, the Atprd1-1 mutant was complemented by insertion of a T-DNA construct containing a wild-type genomic DNA fragment, harbouring the AtPRD1 gene and its putative promoter region (see Materials and methods), proving that disruption of At4g14180 was responsible for the isolated meiotic mutation.
As no cDNA was available in the database for this ORF, we sequenced the corresponding cDNA isolated from flower buds (GenBank accession number EF195233). The resulting ORF is 3993 nucleotides long and codes for a protein of 1330 amino acids (aa) containing no putative conserved domains. An alignment of the isolated cDNA with the genomic DNA revealed a total of nine exons and eight introns (Supplementary data Sup_1A). Expression of AtPRD1, analysed by RT–PCR, was mainly found in young buds (Supplementary data Sup_1B). The mutant allele, Atprd1-1, was found to carry an insertion in the second intron and Atprd1-2 in the eighth exon of AtPRD1 (Supplementary data Sup_1A).
Database searches using a BLASTP programme (Blosum 45) revealed significant sequence similarity (38% identity and 57% similarity) to a rice sequence (OsPRD1, NCBI accession number CAE02100). In addition, using tBLASTn programme, AtPRD1 putative homologues were found in Medicago truncatula, Populus trichocarpa and Physcomitrella patens genomes (see Materials and methods). BLASTP searches also revealed sequence similarity with the N-terminal part of a human protein HsMei1 (accession number CAI17882), homologous to the mouse Mei1 protein involved in meiotic recombination (Libby et al, 2003; Reinholdt and Schimenti, 2005). The greatest similarity between AtPRD1 and HsMei1 covers a region of one-third of the protein (22% identity and 39% similarity over aa 50–396 of AtPRD1). Nevertheless, when similarity between the two proteins was scored directly (Blast2 sequence, Matrix Blosum 45) (Tatusova and Madden, 1999), 17% identity and 34% similarity were found over 914 aa. No direct similarity was found between MmMei1 and AtPRD1, because MmMei1 is described as being shorter than the predicted HsMei1, being deleted of the first 379 aa. Nevertheless, genomic MmMei1 could code for a longer protein, according to genescan protein prediction software (Burge and Karlin, 1997), suggesting that MmMei1 is longer than described. We also performed a homology search using HsMei1 as a query and identified several putative vertebrate homologues such as Xenopus tropicalis (JGI sequence: e_gw1.19.261.1 from 40 to 3417 bp).
Multiple alignment of AtPRD1 homologues was carried out using BioEdit software (version 7.0.5, Blosum 62), and revealed an overall alignment of all these proteins. Several stretches of conserved aa between plants and vertebrates can be observed, mainly located in the N-terminal regions of these proteins (Figure 1). Furthermore, all these proteins show a very high leucine content (14.2% for AtPRD1), and those leucines appeared to be scattered throughout the entire sequence of the protein (Figure 1).
Figure 1.
Sequence alignment of AtPRD1 and HsMei1 homologues. OsPRD1 for Oriza sativa; MtPRD1 for Medicago truncatula, PtPRD1 for Populus tricocarpa, PpPRD1 for Physcomitrella patens; HsMei1 for Homo sapiens; XtPRD1 for Xenopus tropicalis, and MmMei1 for Mus musculus homologues. The numbers indicate amino-acid position, identical amino acids are boxed in black, whereas similar amino acids are boxed in grey. The sequence interruption provoked by T-DNA insertions in the mutant alleles is indicated.
The Atprd1 mutants exhibit reduced fertility and meiotic defects
Atprd1-1 and Atprd1-2 mutants did not show any vegetative growth defects, but displayed short silique elongation, suggesting fertility defects (Supplementary data Sup_2). The wild-type plants developed siliques containing an average of 63.5 seeds/silique, whereas Atprd1-1 plants produced only 2.62 (n=78) seeds/silique. The sterility of Atprd1-1 and Atprd1-2 mutants was correlated with abortion of the male and female gametophytes (data not shown). To identify the stages of sporogenesis and/or gametogenesis that were impaired in Atprd1 mutants, we examined developing male gametophytes by DIC microscopy of cleared buds.
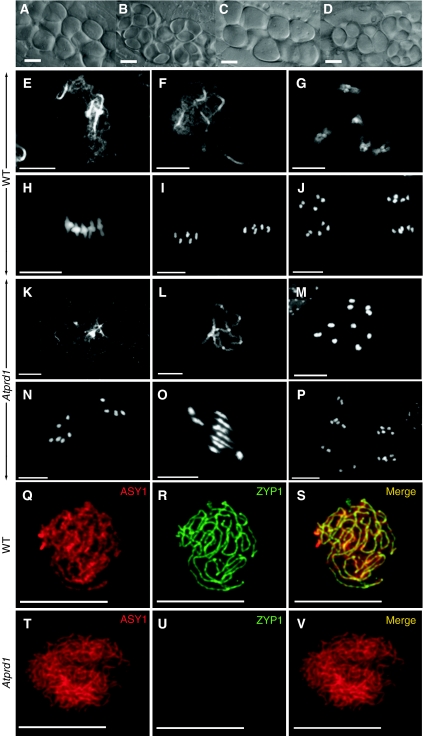
Comparison of the early steps of male sporogenesis did not reveal any difference between wild-type and mutant plants (Figure 2A and C): round pollen mother cells (PMCs, also called meiocytes) were distinguished within the anther locules. In wild-type anthers, these cells underwent two meiotic divisions to produce a characteristic tetrad of microspores enclosed in a callose wall (Figure 2B). Meiotic products were also detected in Atprd1 mutant plants, but lacked the regular tetrahedral structure, and were either asymmetric tetrads or ‘polyads' containing more than four products (Figure 2D), suggesting disturbance of the meiotic program in both Atprd1-1 and Atprd1-2 mutants.
Figure 2.
Male sporogenesis and meiosis of wild-type and the Atprd1-1 plants. (A–D) DIC microscopy of male meiocytes (A, C) or male meiosis products (tetrads of microspores, (B, D)). (A, B): wild type. (C, D) Atprd1-1. Scale bar, 10 μm. (E–J) DAPI staining of wild-type PMCs during meiosis. (E) Zygotene, (F) pachytene, (G) diakinesis, (H) metaphase I, (I) metaphase II, (J) anaphase II. (K–P) DAPI staining of mutant PMCs during meiosis. (K) Pachytene-like stage: homologues remain unsynapsed. (L) Diakinesis stage shows 10 univalents (bright dots correspond to centromeres). (M) Metaphase I with 10 univalents instead of five bivalents as seen in panel H. (N) Anaphase I shows a random segregation of the 10 univalents. (O) Some metaphase/anaphase I transitions show such stretched chromosomes. (P) Anaphase II showing six instead of four batches of chromosomes. (Q–V) Co-immunolocalisation of ASY1 (red) and ZYP1 (green) in wild-type (Q–S) and Atprd1-1 mutant (T–V) PMC. For each cell, each single labelling is shown as well as the overlay of both signals (merge). Scale bar, 10 μm.
We therefore investigated the behaviour of meiotic chromosomes in Atprd1 mutants and in wild-type plants (Figure 2E–P). Wild-type Arabidopsis meiosis is summarised in Figure 2E–J). The 10 Arabidopsis chromosomes appeared as thread-like structures at leptotene (not shown), then underwent synapsis from zygotene (Figure 2E) to pachytene (Figure 2F). After the disappearance of the synaptonemal complex (SC) at diplotene (not shown), the five bivalents condensed through diakinesis (Figure 2G). At metaphase I, the five bivalents were aligned on the metaphasic plate (Figure 2H). During anaphase I, each chromosome separated from its homologue, leading to the formation of dyads corresponding to two pools of five chromosomes (Figure 2I). The second meiotic division then separated the sister chromatids, generating four pools of five chromosomes (Figure 2J), which gave rise to tetrads of microspores (Figure 2B).
In Atprd1 mutants (shown for Atprd1-1), the leptotene stage appeared similar to wild type. The first observable defect was the absence of typical zygotene and pachytene stages. Instead, we observed a pachytene-like stage (Figure 2K), with a clear absence of extended synapsis. In order to further investigate this synapsis defect, we performed co-immunolocalisation of ASY1 and ZYP1. ASY1 is associated with the chromosome axes during prophase I (Armstrong et al, 2002), while ZYP1 is a major component of the central element of the SC (Higgins et al, 2005). We observed a total absence of ZYP1 immunolabelling in Atprd1-1 mutant (Figure 2T–V) in comparison with wild type (Figure 2Q–S), confirming that no synapsis occurs in Atprd1 mutants.
At later stages of meiotic prophase, chromosomes condensed progressively during diakinesis (Figure 2L) until metaphase I, to give rise to 10 univalents instead of five bivalents (Figure 2M). Not a single bivalent was observed in 180 Atprd1-1 or 230 Atprd1-2 metaphase I cells. Consequently the 10 univalents segregated randomly to the two anaphase I poles (Figure 2N). In 13.2% of Atprd1-1 anaphase I (n=129) (12.2% for Atprd1-2; n=147), univalents appeared submitted to a bipolar tension (Figure 2O). But sister chromatid separation at anaphase I occurred rarely, since only 4% of Atprd1-1 (n=129) and Atprd1-2 (n=147) meiocytes at metaphase II showed more than 10 chromosomes (not shown). The second meiotic division then produced unbalanced products (Figure 2P), which generate the previously described polyads (Figure 2D). In conclusions, both Atprd1 mutants show synapsis failure associated with a total absence of chiasmata.
Female meiosis was also investigated after chromosome staining by propidium iodide and subsequent observation by confocal microscopy. The defects observed were similar to those seen in male meiosis. Univalents were also observed at the end of the prophase I, leading to random segregation of chromosomes at anaphase I (Supplementary data Sup_3).
AtPRD1 is required for meiotic recombination
Absence of chiasmata in Atprd1 mutants can be explained either by an absence of recombination or by bivalent instability. In order to test these hypotheses, we measured recombination frequencies in the progeny of Atprd1-1. Therefore, lines heterozygous for the Atprd1-1 mutation (Ws ecotype) were crossed to Columbia (Col-0). Among the mutant F2 plants, we selected by genotyping the ones that were heterozygous for the two chosen linked markers: nga280 and nga111 on chromosome 1 or nga106 and nga76 on chromosome 5 (Bell and Ecker, 1994), in order to measure recombination rates within these two intervals. Mapmaker (Lander et al, 1987) was used to convert data into genetic distances (Table I). The markers were chosen according to the RI map (http://nasc.nott.ac.uk/) as being approximately 30 cM apart, distance confirmed in a previous study (Grelon et al, 2001, Table I). Both distances were dramatically decreased in the Atprd1-1 background: from 36.8 to 0 cM between nga111 and nga280, and from 23.8 to 1 cM between nga76 and nga106 (Table I). These recombination rates are comparable to the residual level of recombination observed in a strong Atspo11-1-2 allele (Table I), suggesting that no recombination was induced in Atprd1 mutant.
Table 1.
Recombination frequencies in Atprd1 and Atspo11-1 mutants
| Genotype | Chr I | Chr V |
|---|---|---|
| Map distancea nga111/nga280 | Map distancea nga76/nga106 | |
| Wild typeb | 36.8 (116) | 23.8 (123) |
| Atprd1-1 | 0 (366) | 1.0 (366) |
| Atspo11-1-2 | 0.7 (251) | 0.8 (206) |
| Map distance calculated in centi-Morgan in the progeny of plants either homozygous for the Atprd1-1 mutation, homozygous for the Atspo11-1-2 mutation or wild type. Total numbers of plants tested for each marker are indicated in parentheses. | ||
| Data from Grelon et al (2001). | ||
DMC1 foci are not formed in Atprd1 mutants
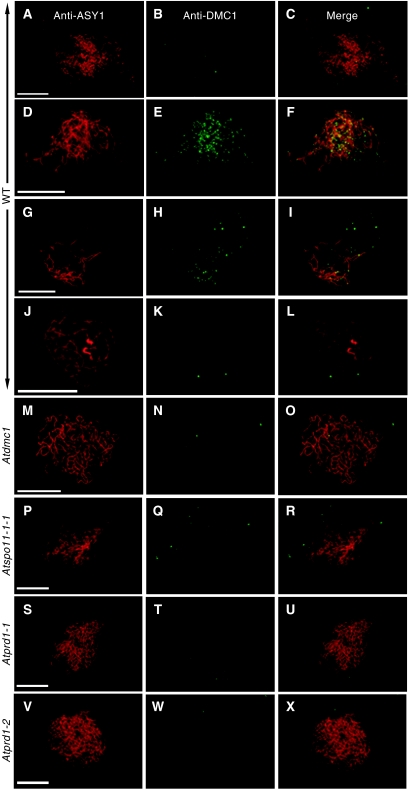
We also analysed the nuclear distribution of the protein DMC1, which is an essential component of the recombination machinery, particularly in meiotic recombination-mediated DSB repair (Masson and West, 2001). To follow the meiotic progression of DMC1 foci during meiosis, co-immunolocalisation was performed with antibodies that recognise the meiotic protein ASY1. Detailed analysis of DMC1 progression in wild-type Arabidopsis meiotic prophase was described in Chelysheva et al (2007). DMC1 foci were not detected during early leptotene, when discontinuous staining of the chromosome axis is observed with the anti-ASY1 antibody (Figure 3A–C). DMC1 signal appeared and tended to a maximum at late leptotene/early zygotene (Figure 3D–F) reaching an average of 240 foci per nucleus (Chelysheva et al, 2007). The number of DMC1 foci decreased during late zygotene (Figure 3G–I) to completely disappear at pachytene (Figure 3J–L). To confirm that the signal was specific to DMC1, a similar dual immunolocalisation was performed in an Atdmc1 mutant (Couteau et al, 1999), where no DMC1 foci was observed at any stage (Figure 3M–O). Similarly, as expected, no DMC1 foci was seen in the Atspo11-1-1 mutant (Figure 3P–R). Dual immnunolocalisation was also performed in Atprd1-1 and Atprd1-2 mutants and we could not detect any DMC1 chromatin-associated foci (Figure 3S–X), revealing a very early defect in the recombination process. A modified version of Figure 3, with enhanced green channel signal, is available as Supplementary data Sup_4, in order to check that no signal above the background (Atdmc1 control) is detected in Atspo11-1 or Atprd1 mutants.
Figure 3.
Immunolocalisation of ASY1 (anti-ASY1) and AtDMC1 (anti-DMC1) in wild type and asynaptic mutants. (A–L) Anti-AtDMC1 labelling (green) in wild type during early leptotene (A–C), late leptotene/early zygotene (D–F), late zygotene (G–I) and pachytene (J–L) stages. No AtDMC1 foci can be seen above background during prophase I of Atdmc1 (M–O), Atspo11-1 (P–R), Atprd1-1 (S–U) and Atprd1-2 (V–X) PMC. For a single antibody, all photographs were taken with the same exposition, but enhanced anti-DMC1 signal in the different mutant backgrounds is provided as Supplementary data. Scale bar, 10 μm.
AtPRD1 is involved in meiotic DSB formation
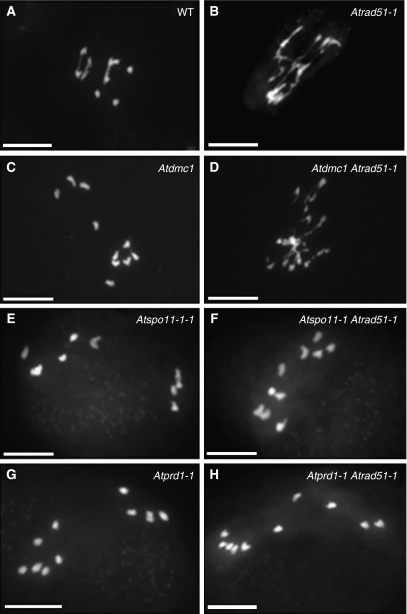
In Arabidopsis two kinds of mutation are known to induce early defects in meiotic recombination: mutations that prevent DSBs such as Atspo11-1 (Grelon et al, 2001; Figure 4E), and mutations in AtDMC1 that prevent normal DSB repair (Figure 4C; Couteau et al, 1999). In the latter case, DSBs are formed as in wild type, but are probably repaired using the sister chromatids as template, and not the homologous chromosomes, explaining the presence of 10 intact univalents at metaphase I (Couteau et al, 1999; Figure 4C).
Figure 4.
DAPI staining of male meiocytes during anaphase I. Wild-type PMC show an equal repartition of chromosomes during anaphase I (A). In the asynaptic mutants Atdmc1 (C), Atspo11-1-1 (E) and Atprd1-1 (G), random segregation of the 10 univalents is observed at the same stage. An Atrad51-1 mutant shows a severe chromosome fragmentation in the first meiotic division (B), which is not abolished in Atdmc1Atrad51 (D). By contrast, fragmentation is abolished in Atspo11-1Atrad51-1 (F) and Atprd1-1Atrad51-1 (H). Scale bar, 10 μm.
In order to discriminate between a role of AtPRD1 in DSB formation per se or in later stages of DSB repair, we used different mutant backgrounds unable of repairing SPO11-induced DSBs such as Atrad51 (Figure 4B) or Atscc3 and Atrec8 (Supplementary data Sup_5). In a Atrad51 mutant, meiotic DSBs are formed but abnormally repaired, yielding aberrant meiotic figures at metaphase I and chromosome fragmentation at anaphase I (Li et al, 2004; Figure 4B). Chromosome fragmentation is abolished in an Atspo11-1 background (Li et al, 2004; Figure 4F), but not in an Atdmc1 background (Figure 4D), demonstrating that this fragmentation is a consequence of DSB repair defects and that DSBs are formed in an Atdmc1 background. Interestingly, chromosomal fragmentation was also abolished in the Atprd1-1Atrad51 double mutant, leading to 10 univalents that segregated randomly at anaphase I (Figure 4H). These results show that Atprd1-1, like Atspo11-1-1, can prevent the meiotic recombination repair defects occurring in Atrad51 and therefore demonstrate that AtPRD1 is involved in DSB formation.
To confirm the role of AtPRD1 in the initiation of meiotic recombination, the Atprd1-1 mutation was introduced into the cohesin Atrec8 and Atscc3-1 mutants that present strong DSB repair defects (Bai et al, 1999; Bhatt et al, 1999; Chelysheva et al, 2005). The introduction of Atprd1 or Atspo11-1 into both cohesin mutants abolished similarly their DSB repair defects (Supplementary data Sup_5).
Lastly, we observed that AtPRD1 was not involved in AtSPO11-1 or AtSPO11-2 transcription. RT–PCR experiments showed no modification of these gene expressions in any of the Atprd1 mutants (Supplementary data Sup_1C).
The N-terminal region of AtPRD1 interacts with AtSPO11-1 in a yeast two-hybrid system
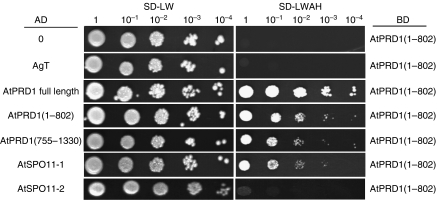
We have shown that AtPRD1 is involved at the same step of meiotic recombination as AtSPO11-1. This prompted us to test the interaction between the two proteins using the Clontech Matchmaker yeast two-hybrid system or derived (see Materials and methods). For this, we cloned the AtPRD1 full length (PRD1FL), AtPRD1 5′ end (encoding aa 1–802), AtPRD1 3′ coding sequence (encoding aa 755–1330) as well as AtSPO11-1 and AtSPO11-2 cDNAs into yeast expression vectors in frame with either the GAL4 DNA-binding domain (pGBKT7 vector) or the GAL4 DNA-activation domain (pGADT7 vector). Two-hybrid assays performed with these different forms of AtPRD1 revealed interactions between AtPRD1(1–802) and AtSPO11-1 when AtPRD1(1–802) is fused to the GAL4 DNA-binding domain (Table II; Figure 5). Moreover, we also detected interactions between AtPRD1(1–802) and AtPRD1 full length, as well as between AtPRD1(1–802) and the two truncated parts of AtPRD1 (Table II; Figure 5), indicating that AtPRD1 can form, at least in yeast, a homodimer. No interaction was observed between AtPRD1(755–1330) and AtSPO11-1, suggesting that the N-terminal part of AtPRD1, but not the C-terminal part of AtPRD1, is essential for interaction with AtSPO11-1. Lastly, we could not detect any interaction between AtPRD1 (1–802) and AtSPO11-2 (Figure 5).
Table 2.
Interaction between AtPRD1 and AtSPO11-1 in yeast two-hybrid assay
| pGBKT7 | pGADT7 | |||||
|---|---|---|---|---|---|---|
| Empty | AgT | AtPRD1FL | AtPRD1 (1–802) | AtPRD1 (755–1330) | AtSPO11-1 | |
| Empty | − | n.t. | − | − | − | − |
| AtPRD1(1–802) | − | − | + | + | + | + |
| AtPRD1(755–1330) | − | n.t. | − | − | − | − |
| AtSPO11-1 | − | n.t. | − | − | − | − |
| AgT: SV40 large T antigen; n.t.: not tested. | ||||||
| pGBKT7 vector contains Gal4-binding domain and pGADT7 contains Gal4 activation domain. AtPRD1FL corresponds to the entire length of AtPRD1. AtPRD1(1–802) and AtPRD1(755–1330) correspond to the N-terminal and the C-terminal part of AtPRD1, respectively. (+) growth or (−) no growth on synthetic drop-out medium without leucine, tryptophan, adenine and histidine. | ||||||
Figure 5.
AtPRD1 interacts with AtSPO11-1 in a yeast two-hybrid assay. AtPRD1 (1–802) and AtPRD1 (755–1330) correspond to the N-terminal and the C-terminal part of AtPRD1, respectively. AgT encodes for the SV40 large T-antigen. The AH109 strain was co-transformed with the constructs indicated, carrying a binding domain (BD) and an activation domain (AD), and grown on synthetic drop-out (SD) media lacking the aa leucine and tryptophan (SD-LW) or leucine, tryptophan, adenine and histidine (SD-LWAH). Serial dilutions of diploid strains were performed before spotting on media. Yeast containing both vectors grew on SD-LW; positive interactions appear as white spot on SD-LWAH.
Discussion
In S. cerevisiae, at least nine proteins in addition to Spo11 are required to form the DSBs that initiate meiotic recombination (Keeney, 2001). Several studies have revealed physical and functional interactions that connect them to one another and to Spo11, suggesting that these proteins are components of several subcomplexes involved together in DSB formation. In other eukaryotes, there are only few data on the existence of such recombination initiation complexes, except for the wide conservation of Spo11.
AtPRD1 is required for meiotic DSB formation in Arabidopsis
In this study, we identified a new gene AtPRD1 and analysed its function through isolation and characterisation of two mutants. We showed that disruption of AtPRD1 prevents synapsis and bivalent formation in male and female meiosis. This absence of association between homologous chromosomes is not due to a premature release of chiasmata leading to univalent formation, as reported for the dy and dsy mutants of maize (Maguire, 1978; Maguire et al, 1993), because meiotic recombination frequencies are severely reduced in Atprd1 mutants. Furthermore, DMC1 foci are absent during the early stages of meiotic prophase I, indicating that meiotic recombination defects in Atprd1 mutants occur very early in the process, likely before CO formation.
The Atprd1 phenotype is characteristic of the asynaptic meiotic mutants in which chromosome homologues fail to synapse and univalents segregate randomly during the first meiotic division (Ross et al, 1997). Different types of mutations present this kind of phenotype in Arabidopsis such as Atdmc1, Atspo11-1 and Atspo11-2, sds or asy1 (Couteau et al, 1999; Caryl et al, 2000; Grelon et al, 2001; Azumi et al, 2002; Stacey et al, 2006). However, this phenotype similarity does not reflect similarity of function. Atspo11-1 mutation leads to an asynaptic phenotype due to a lack of DSBs. In contrast with AtSPO11-1, AtDMC1 is probably not involved in meiotic DSB formation, since chromosome fragmentation is observed in an Atdmc1Atrad51-1 double mutant (this study) and in Atdmc1 pDMC1::RNAi/RAD51 (Siaud et al, 2004), demonstrating that DSBs are formed in Atdmc1, but are repaired probably onto the sister chromatids, instead of homologous chromosomes (Couteau et al, 1999; Siaud et al, 2004). Unlike Atdmc1, but like Atspo11-1, Atprd1 mutants completely abolish DNA fragmentation occurring in Atrad51-1 mutants or other DSB repair mutants. Similarly, Atprd1 mutations (this study) like Atspo11-1 mutation, but not like Atdmc1 ones (Higgins et al, 2005), prevent any ZYP1 loading onto chromosomes. All these data taken together demonstrate that AtPRD1 is necessary for meiotic DSB formation.
We also showed that the AtPRD1 N-terminal region is able to interact with AtSPO11-1 in a yeast two-hybrid assay. If this interaction occurs in planta, it will mean that these two proteins act together in this crucial step of meiotic recombination that is DSB formation.
In S. cerevisiae, the formation of meiotic DSBs requires nine proteins, of which only three are known to interact with Spo11: Ski8, Rec102 and Rec104 (Kee and Keeney, 2002; Jiao et al, 2003; Arora et al, 2004). Spo11 is required for Ski8 relocalisation in the nucleus, and Ski8 stabilises Spo11 association with meiotic chromosomes, suggesting Ski8 plays a role of scaffold protein by recruiting other DSB proteins to meiotic chromosomes (Tesse et al, 2003; Arora et al, 2004). Rec102 and Rec104 are required for nuclear localisation of Spo11, for the association of Spo11 with meiotic hot spots (Prieler et al, 2005) and for Spo11 self-association at meiosis (Sasanuma et al, 2007). Therefore, the Rec102/Rec104 complex has been suggested to promote alterations in local chromatin structure within loops to stabilise Spo11-containing complexes at hotspots, or to activate hotspot-associated Spo11 complexes to cleave DNA. Another possibility is that it participates in loop–axis interactions that promote DSB formation or repair in spatial proximity of the axis (Kee and Keeney, 2002; Jiao et al, 2003; Kee et al, 2004). Even though AtPRD1 does not show any sequence similarities with Ski8, Rec102 or Rec104, it might possess one or several functions of these yeast proteins.
We also found that, in the conditions of our experiments, AtPRD1 and AtSPO11-1 can interact but not AtPRD1 and AtSPO11-2. Even if no direct evidence of absence of DSBs in Atspo11-2 is available so far, the phenotype similarity between Atspo11-1 and Atspo11-2 mutants suggests that AtSPO11-2 as AtSPO11-1 is required for DSB formation in Arabidopsis (Stacey et al, 2006). According to crystallographic data of topoVI-A' (a sub-region of the topoVI-A homologue to Spo11) in Methanococcus jannaschii (Nichols et al, 1999), it is admitted that, in yeast, Spo11 forms a stable homodimer containing a groove where the DNA could be bound before cleavage (Keeney and Neale, 2006). Furthermore, it has been recently shown that several Spo11 subunits interact at the time of DSB formation (Sasanuma et al, 2007). Because the two Arabidopsis Spo11 orthologues, AtSPO11-1 and AtSPO11-2, do not show overlapping functions, they might catalyse meiotic DSB formation by forming a heterodimer and not a homodimer. Nevertheless, such interaction remains to be demonstrated and could not be detected in yeast two-hybrid system (not shown). The asymmetry of the interaction of AtPRD1 with AtSPO11-1 and not AtSPO11-2 we observed in yeast two-hybrid experiments (providing it exists in planta), can be explained in several ways. For example, AtPRD1, like AtSPO11-1 and AtSPO11-2, could be necessary at the site of DSB formation on chromatin. By its specific association with a single AtSPO11 subunit, AtPRD1 would therefore set up (or participate in the setting up) of an asymmetry in the DSB complex that could be recognised later, at the step of DSB processing, in order to allow the asymmetrical release of Spo11-bound oligonucleotides that have been identified in S. cerevisiae and mouse (Neale et al, 2005). Another possibility could be that AtPRD1 is regulating the recruitment of AtSPO11-1 and not AtSPO11-2 to its sites of cleavage, but further experimentations will be necessary to choose between these hypotheses.
AtPRD1 is evolutionary conserved from plants to vertebrates
Spo11 function is particularly well conserved among organisms (Keeney, 2001). However, it is not the case for the other proteins, which participate in the meiotic recombination initiation complex. Only four out of the nine proteins described in S. cerevisiae are conserved among other organisms: Rad50, Mre11, Xrs2 and Ski8. However, these proteins have rarely conserved their role in meiotic DSB formation (see Introduction). Given the limits of reverse genetics, other approaches are necessary to identify proteins involved in DSB formation. We have cloned AtPRD1 and found that it codes for a protein of 1330 aa containing no recognisable putative domains. No yeast homologue of AtPRD1 has been found in the databases, suggesting that AtPRD1 is too divergent to be recognizable among yeast proteins, or that it is not conserved in this kingdom, suggesting that plants require different proteins to create the meiotic DSBs. Nevertheless, BLAST searches and multiple alignments revealed a similarity with the human protein HsMei1. Mei1 is conserved among vertebrates such as Xenopus tropicalis or Mus musculus, where it has been well characterised. mei1 mice mutants show synapsis failure and recombination defects. Furthermore, γH2AX staining was markedly decreased at leptonema, indicating that the meiotic DSBs are probably strongly reduced in a mei1 mutant (Libby et al, 2002, 2003). Furthermore, Reinholdt and Schimenti (2005) have established that mei1 mutant spermatocytes show an earlier phenotype than dmc1 ones. All these data support a role for Mei1 in the initiation of meiotic recombination. These similarities between Arabidopsis Atprd1 and mouse mei1 mutants, as well as the similarity of sequences between both proteins, strongly suggest that AtPRD1 and Mei1 are functional homologues. This conservation of a DSB-forming protein between plants and vertebrates, if it is confirmed and extended to others, will give new insights to understand the evolution of the recombination machinery.
Materials and methods
Plant material
The Atprd1-1 mutant (EEO4 line) was obtained from the Versailles collection of Arabidopsis T-DNA transformants (Ws accession) (Bechtold et al, 1993). Mutant screening was performed as described in Mercier et al (2001). The Atprd1-2 mutant, line SALK_ 024703, was obtained from the collection of T-DNA mutants of the Salk Institute Genomic Analysis Laboratory (Col-0 accession) (SIGnAL, http://signal.salk.edu/cgi-bin/tdnaexpress) (Alonso et al, 2003) and provided by NASC (http://nasc.nott.ac.uk/). Atrec8, Atdmc1, Atspo11-1, Atrad51-1 and Atscc3-1 mutants were described in Bhatt et al (1999); Couteau et al (1999); Grelon et al (2001); Li et al (2004); Chelysheva et al (2005), respectively.
Genetic analyses
Isolation of Atprd1-1: The EEO4 line segregated 3:1 for kanamycin resistance (one of the T-DNA markers), suggesting the presence of a single T-DNA insert. After crossing to wild type, linkage between the T-DNA insert and the meiotic phenotype was checked as described in Grelon et al (2001).
Double mutants and allelism test: See Supplementary data.
Recombination rates: These were calculated as described in Grelon et al (2001).
Molecular biology
Isolation of plant T-DNA flanking sequence: The left border (Lb) of the T-DNA insert of Atprd1-1 was obtained from https://genoplante.infobiogen.fr/flagdb/info (FST 305H03). A T-DNA inversed repeat (Lb–Rb–Rb–Lb) insertion occurred between nucleotides 1393 and 1398 of the EF195233 sequence. The insertion site of the T-DNA in Atprd1-2 was obtained from http://signal.salk.edu/cgi-bin/tdnaexpress (FST SALK_024703.29.25).
Complementation of Atprd1-1: A 7.8 BamHI kb from BAC clone T8H22 (http://www.arabidopsis.org/) was subcloned into pBSK (Fermentas). Then a BamHI-NcoI fragment, containing the full-length genomic sequence, was transferred into the binary vector pCAMBIA1381 (http://www.cambia.org/daisy/bios/585.html). Complementation was checked by transforming Atprd1-1 mutants with the previously described clone.
PCR genotyping: See Supplementary data.
cDNA and RT–PCR: See Supplementary data.
Sequence analyses
Protein sequence similarity searches were performed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/), at the Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/Blast), at JGI (http://genome.jgi-psf.org/), and at PHYSCObase (http://moss.nibb.ac.jp/), using BLOSUM45 matrix and default parameters. Multiple alignments were performed with Bioedit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
AtPRD1 homologues: Oriza sativa PRD1 accession number is CAE02100 (OsPRD1), Physcomitrella patens (PpPRD1) from ASYA488561.b1, Homo sapiens (HsMei1) is CAI17882, Mus musculus MmMei1 is XP_487041 and Xenopus tropicalis (XtPRD1) was obtained from JGI (e_gw1.19.261.1). Medicago truncatula (MtPRD1) and Populus trichocarpa (PtPRD1) were derived from sequences AC147484 and LG_II:20125180–20129370 (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), respectively, after genescan processing (http://genes.mit.edu/GENSCAN.html).
Microscopy
Comparison of early stages of microsporogenesis and the development of PMC was carried out as described in Grelon et al (2001). Preparation of prophase stage spreads for immunocytology was performed according to Armstrong et al (2002), with the modifications described in Chelysheva et al (2005). The ASY1 and ZYP1 polyclonal antibodies (Armstrong et al, 2002; Higgins et al, 2005) were used at a working dilution of 1:500. The AtDMC1 antibody was described in Chelysheva et al (2007) and was used at a working dilution of 1:20. All observations were made using a Leica DM RXA2 microscope; photographs were obtained using a CoolSNAP HQ (Roper) camera driven by Open LAB 4.0.4 software; all images were further processed with Open LAB 4.0.4 or Adobe Photoshop 8.0.
Yeast two-hybrid assay
Plasmids expressing either AtSPO11-1, AtSPO11-2 or AtPRD1 were constructed as described in Supplementary data. They were co-transformed into yeast strain AH109 (Clontech) following the protocol MATCHMAKER GAL4 Two-Hybrid System 3 (Clontech). Co-transformants were selected on SD-LW (synthetic drop-out media lacking the aa leucine and tryptophan). Interactions were tested on selective media lacking leucine, tryptophan, adenine and histidine (SD-LWAH), according to the instructions of the manufacturer. Serial 1:10 dilutions were prepared in water and 5 μl of each dilution was used to yield one spot. Plates were incubated at 30°C for 3 days before scoring and taking photographs. SV40 antigen T (AgT) known to interact with p53 protein but not with Lamin C (LAM) were used as positive and negative controls, respectively (Bartel et al, 1993; Li and Fields, 1993). Plasmid inserts in clones growing onto SD-LWAH were checked by PCR. Expression of AtPRD1, AtSPO11-1 and AtSPO11-2 fusion proteins was checked by Western blot.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Information
Acknowledgments
We are grateful to Christine Mézard, Raphaël Mercier, Denise Zickler and Eric Jenczewski for constructive reading of the manuscript, and to Fabien Nogué for his help in analysing Physcomitrella patens genomic sequences. We thank all the Versailles Arabidopsis group members for the screening the T-DNA transformant collection of Versailles. We thank C Franklin for providing the ASY1 and ZYP1 antibodies and M-P Doutriaux for providing the DMC1 antibody.
References
- Akutsu N, Iijima K, Hinata T, Tauchi H (2007) Characterization of the plant homolog of Nijmegen breakage syndrome 1: involvement in DNA repair and recombination. Biochem Biophys Res Commun 353: 394–398 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Caryl AP, Jones GH, Franklin FC (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Arora C, Kee K, Maleki S, Keeney S (2004) Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell 13: 549–559 [DOI] [PubMed] [Google Scholar]
- Azumi Y, Liu D, Zhao D, Li W, Wang G, Hu Y, Ma H (2002) Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J 21: 3081–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Peirson BN, Dong F, Xue C, Makaroff CA (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P, Chien CT, Sternglanz R, Fields S (1993) Elimination of false positives that arise in using the two-hybrid system. Biotechniques 14: 920–924 [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris 316: 1194–1199 [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P (1997) An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386: 414–417 [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19: 463–472 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI (2004) Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma 113: 197–203 [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Caryl AP, Armstrong SJ, Jones GH, Franklin FC (2000) A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109: 62–71 [DOI] [PubMed] [Google Scholar]
- Celerin M, Merino ST, Stone JE, Menzie AM, Zolan ME (2000) Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J 19: 2739–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Gendrot G, Vezon D, Doutriaux MP, Mercier R, Grelon M (2007) Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet 3: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Marquez-Lema A, Bhatt AM, Horlow C, Mercier R, Mezard C, Grelon M (2005) AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 118: 4621–4632 [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398 [DOI] [PubMed] [Google Scholar]
- Gallego ME, Jeanneau M, Granier F, Bouchez D, Bechtold N, White CI (2001) Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J 25: 31–41 [DOI] [PubMed] [Google Scholar]
- Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, Kohli J, Nasmyth K (2005) Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol 15: 1663–1669 [DOI] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J 20: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Angelis KJ, Meister A, Schubert I, Melzer M, Puchta H (2002) An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr Biol 12: 1787–1791 [DOI] [PubMed] [Google Scholar]
- Hartung F, Puchta H (2000) Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res 28: 1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Puchta H (2001) Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene 271: 81–86 [DOI] [PubMed] [Google Scholar]
- Henderson KA, Kee K, Maleki S, Santini PA, Keeney S (2006) Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao K, Salem L, Malone R (2003) Support for a meiotic recombination initiation complex: interactions among Rec102p, Rec104p, and Spo11p. Mol Cell Biol 23: 5928–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet S, Vezon D, Froger N, Mercier R (2006) Non conservation of the meiotic function of the Ski8/Rec103 homolog in Arabidopsis. Genes Cells 11: 615–622 [DOI] [PubMed] [Google Scholar]
- Kee K, Keeney S (2002) Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics 160: 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K, Protacio RU, Arora C, Keeney S (2004) Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J 23: 1815–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Neale MJ (2006) Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans 34: 523–525 [DOI] [PubMed] [Google Scholar]
- Keeney S (2001) Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52: 1–53 [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Li J, Hooker GW, Roeder GS (2006) Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 173: 1969–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Fields S (1993) Identification of mutations in p53 that affect its binding to SV40 large T antigen by using the yeast two-hybrid system. FASEB J 7: 957–963 [DOI] [PubMed] [Google Scholar]
- Libby BJ, De La Fuente R, O'Brien MJ, Wigglesworth K, Cobb J, Inselman A, Eaker S, Handel MA, Eppig JJ, Schimenti JC (2002) The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev Biol 242: 174–187 [DOI] [PubMed] [Google Scholar]
- Libby BJ, Reinholdt LG, Schimenti JC (2003) Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci USA 100: 15706–15711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jang JK, Kato N, McKim KS (2002) mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162: 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire MP, Riess RW, Paredes AM (1993) Evidence from a maize desynaptic mutant points to a probable role of synaptonemal complex central region components in provision for subsequent chiasma maintenance. Genome 36: 797–807 [DOI] [PubMed] [Google Scholar]
- Maguire MP (1978) Evidence for separate genetic control of crossing over and chiasma maintenance in maize. Chromosoma 65: 173–183 [Google Scholar]
- Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S (2007) Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, Smith GR, Moreno S (2005) A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol 15: 2056–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, West SC (2001) The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem Sci 26: 131–136 [DOI] [PubMed] [Google Scholar]
- McKim KS, Hayashi-Hagihara A (1998) mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev 12: 2932–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, McKim KS (2006) Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet 2: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Grelon M, Vezon D, Horlow C, Pelletier G (2001) How to characterize meiotic functions in plants? Biochimie 83: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S (2005) Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols MD, DeAngelis K, Keck JL, Berger JM (1999) Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J 18: 6177–6188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Hirota K, Matsumoto S, Takeda T, Ohta K, Arai K, Masai H (2006) Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc Natl Acad Sci USA 103: 8131–8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieler S, Penkner A, Borde V, Klein F (2005) The control of Spo11's interaction with meiotic recombination hotspots. Genes Dev 19: 255–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J, Siroky J, Mokros P, Schweizer D, Riha K (2004) Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16: 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholdt LG, Schimenti JC (2005) Mei1 is epistatic to Dmc1 during mouse meiosis. Chromosoma 114: 127–134 [DOI] [PubMed] [Google Scholar]
- Roeder GS (1997) Meiotic chromosomes: it takes two to tango. Genes Dev 11: 2600–2621 [DOI] [PubMed] [Google Scholar]
- Ross KJ, Fransz P, Armstrong SJ, Vizir I, Mulligan B, Franklin FC, Jones GH (1997) Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res 5: 551–559 [DOI] [PubMed] [Google Scholar]
- Sasanuma H, Murakami H, Fukuda T, Shibata T, Nicolas A, Ohta K (2007) Meiotic association between Spo11 regulated by Rec102, Rec104 and Rec114. Nucleic Acids Res 35: 1119–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky JJ, McKim KS, Messina L, French RL, Hurley WD, Arbel T, Chin GM, Deneen B, Force SJ, Hari KL, Jang JK, Laurencon AC, Madden LD, Matthies HJ, Milliken DB, Page SL, Ring AD, Wayson SM, Zimmerman CC, Hawley RS (1999) Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics 152: 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaud N, Dray E, Gy I, Gerard E, Takvorian N, Doutriaux MP (2004) Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J 23: 1392–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, Maxwell A, Roberts K, Sugimoto-Shirasu K (2006) Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J 48: 206–216 [DOI] [PubMed] [Google Scholar]
- Storlazzi A, Tesse S, Gargano S, James F, Kleckner N, Zickler D (2003) Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev 17: 2675–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC (2002) DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr Biol 12: 1782–1786 [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL (1999) BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174: 247–250 [DOI] [PubMed] [Google Scholar]
- Tesse S, Storlazzi A, Kleckner N, Gargano S, Zickler D (2003) Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci USA 100: 12865–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Sung P (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem 276: 35458–35464 [DOI] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T (1998) Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95: 705–716 [DOI] [PubMed] [Google Scholar]
- Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K (2004) Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J 23: 1792–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Cheong H, Friedrichsen D, Zhao Y, Hu J, Mora-Garcia S, Chory J (2002) A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc Natl Acad Sci USA 99: 10191–10196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Hyppa RW, Smith GR (2004) Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Information