Abstract
Monocyte differentiation involves the participation of lineage-restricted transcription factors, although the mechanisms by which this process occurs are incompletely defined. Within the hematopoietic system, members of the Kruppel-like family of factors (KLFs) play essential roles in erythrocyte and T lymphocyte development. Here we show that KLF4/GKLF is expressed in a monocyte-restricted and stage-specific pattern during myelopoiesis and functions to promote monocyte differentiation. Overexpression of KLF4 in HL-60 cells confers the characteristics of mature monocytes. Conversely, KLF4 knockdown blocked phorbol ester-induced monocyte differentiation. Forced expression of KLF4 in primary common myeloid progenitors (CMPs) or hematopoietic stem cells (HSCs) induced exclusive monocyte differentiation in clonogenic assays, whereas KLF4 deficiency inhibited monocyte but increased granulocyte differentiation. Mechanistic studies demonstrate that KLF4 is a target gene of PU.1. Consistently, KLF4 can rescue PU.1−/− fetal liver cells along the monocytic lineage and can activate the monocytic-specific CD14 promoter. Thus, KLF4 is a critical regulator in the transcriptional network controlling monocyte differentiation.
Keywords: hematopoietic progenitors, Kruppels, leukemia, monocyte differentiation, PU.1
Introduction
Hematopoietic stem cells (HSCs) may generate committed progenitor cells that lose the capacity to self-renew and ultimately differentiate along a specific lineage to form mature blood cells. Control of hematopoiesis is a complex process requiring the coordinated expression of stage-specific transcription factors that allow for subsequent induction of lineage-restricted genes and cell surface receptors (Tenen et al, 1997; Friedman, 2002). Cytokines and growth factors may then increase lineage-committed progenitor cell populations depending, in part, on the expression levels of lineage determination transcription factors. In the myeloid pathway, mature monocytes and granulocytes arise from bipotential granulocyte/macrophage progenitors (GMPs) that, in turn, arise from multipotential common myeloid progenitors (CMPs); CMPs may also give rise to bipotential megakaryocyte/erythrocyte progenitors (MEPs) (Akashi et al, 2000; Traver et al, 2001). Identification of transcription factors that participate in controlling progenitor cell fate decisions in the myeloid pathway has been of considerable interest with therapeutic implications for a variety of conditions including leukemia, anemia, and chronic inflammatory diseases, among others.
Gene knockout experiments in mice have identified several transcription factors as critical regulators of different aspects of myeloid development (Tenen et al, 1997; Friedman, 2002). For example, disruption of the Ets transcription factor PU.1 in mice resulted in multiple hematopoietic defects, including a reduction of not only mature macrophages and granulocytes, but also B and T lymphocytes and NK cells (Scott et al, 1994; McKercher et al, 1996; Colucci et al, 2001). Indeed, the presence of PU.1 is required for the development of CMPs and common lymphoid progenitors (CLPs) from HSCs (Iwasaki et al, 2005). These findings place PU.1 upstream in the transcriptional hierarchy of specifying progenitor cell fate and raise the possibility that additional factors may participate at specific stages to restrict lineage commitment along single myeloid pathways. For example, GATA-1, a transcription factor essential in specifying progenitors along erythrocyte, megakaryocyte, mast cell, and eosinophil lineages (at the expense of monocyte/granulocyte lineages), interacts with PU.1 to inhibit its function and vice versa (Rekhtman et al, 1999; Zhang et al, 1999; Nerlov et al, 2000). Examples of this type of mutual antagonism exist throughout the hematopoietic system (Orkin, 2000). The balance between monocyte and granulocyte differentiation may be regulated, in part, by mutual antagonism between PU.1 and the CCAAT enhancer-binding protein-α (C/EBP-α). C/EBP-α-deficient mice exhibit a complete block in neutrophil differentiation with normal monocyte maturation (Zhang et al, 1997). Recent mechanistic studies have revealed that C/EBP-α physically interacts with PU.1 and may contribute to the specification of myeloid progenitors to the granulocyte lineage depending on the expression ratio of these two factors (Reddy et al, 2002; Dahl et al, 2003) (Radomska et al, 1998; Reddy et al, 2002). Interestingly, mice carrying a graded reduction of PU.1 expression to 20% of normal levels develop acute myeloid leukemia (Rosenbauer et al, 2004). Thus, stage-specific expression of various transcription factors helps dictate not only cell fate decisions in the myeloid lineage, but also the development of malignant transformation (Tenen, 2003; Rosenbauer et al, 2004).
The Kruppel-like family of transcription factors (KLFs) are critical regulators of cellular development, growth, and differentiation. (Bieker, 1996; Feinberg et al, 2004). Two examples highlight the critical role of this family of genes in hematopoietic development. KLF1, or EKLF (erythroid Kruppel-like factor), is expressed primarily in red blood cells. Gene targeting experiments revealed that KLF1 is necessary for γ- to β-globin switching during erythrocyte development (Nuez et al, 1995; Perkins et al, 1995). KLF2, or LKLF (lung Kruppel-like factor), is highly expressed in T cells, and targeted disruption of KLF2 verified an essential role for this factor in programming and maintaining naïve T-cell quiescence (Kuo et al, 1997). Because of the importance of KLF1 and KLF2 in different hematopoietic lineages, we hypothesized that a related Kruppel-like zinc-finger (ZF) protein may regulate the differentiation of precursor cells along the monocyte/macrophage cell pathway. Using a low-stringency homology screening strategy, we identified KLF4 as being highly expressed in monocytes. KLF4 was initially identified in the epithelial lining of the gut and skin (Garrett-Sinha et al, 1996; Shields et al, 1996) and gene targeting experiments have verified a critical role for this factor in these tissues (Segre et al, 1999; Katz et al, 2002) as well as in embryonic cells (Takahashi and Yamanaka, 2006) and cornea (Swamynathan et al, 2006). Recently, we demonstrated that KLF4 can regulate iNOS expression and TGF-β signaling in activated macrophages (Feinberg et al, 2005) and Noti et al (2005) identified that KLF4 can repress the CD11d promoter in leukemic cell lines. However, the functional role of KLF4 in myeloid cell development has not been defined.
Herein, we found that among myeloid cells, KLF4 is expressed principally in monocytes and is induced in a stage-specific manner during myelopoiesis. Overexpression of KLF4 in promyelocytic HL-60 cells or in primary CMPs or HSCs from bone marrow restricts these cells along the monocyte/macrophage pathway at the expense of other myeloid lineages and confers the morphologic, genetic, and functional characteristics of a mature monocyte. We further show that KLF4 is a downstream target gene of PU.1 and is capable of binding to the monocyte-specific CD14 promoter. Taken together, these data support an important role for KLF4 as a key transcriptional regulator of monocyte differentiation.
Results
Identification of KLF4
The full-length cDNA of KLF4 was identified through low-stringency homology screening of a rat monocyte/macrophage cDNA library using the ZF domain of KLF1/EKLF as a probe. Analysis of the 1422-base pair open reading frame revealed a 474-amino-acid protein with three Cys2/His2 zinc fingers at the C-terminus and a proline-rich N-terminus (data not shown). A GenBank search of three out of nine clones isolated revealed that our factor is identical to the rat homolog of GKLF/EZF, also known as KLF4 (Garrett-Sinha et al, 1996; Shields et al, 1996; Higaki et al, 2002).
Expression of KLF4 in human hematopoietic cell lines
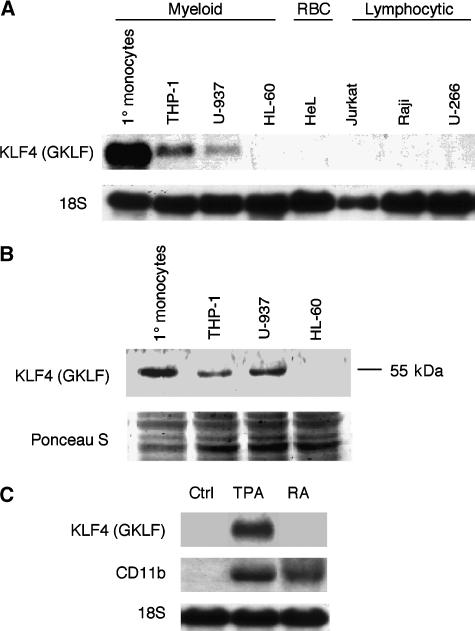
To understand the pattern of KLF4 expression in human hematopoietic cells, we analyzed total RNA isolated from primary human peripheral blood monocytes and seven human hematopoietic cell lines by Northern blotting (Figure 1A). A single, intense 3.5-kb band corresponding to KLF4 was detected in RNA from human peripheral blood monocytes, the monocyte-like THP-1 cell line, and the histiocytic U-937 cell line. In contrast, KLF4 mRNA was undetectable in RNA derived from the HeL (erythrocyte), Jurkat (T lymphocyte), Raji (B lymphocyte, immature), and U-266 (B lymphocyte, mature) cells. To assess the expression of KLF4 protein in myeloid cells, we harvested total cell extracts from human peripheral blood monocytes and THP-1, U-937, and HL-60 cells and performed Western blot analysis with a polyclonal KLF4 antiserum. KLF4 protein was detected in human peripheral blood monocytes and THP-1 and U-937 cells but not in promyelocytic HL-60 cells (Figure 1B). Thus, among hematopoietic cells, KLF4 mRNA and protein are expressed in monocytes and monocyte-like cell lines.
Figure 1.
Expression of KLF4 in human monocytes, hematopoietic cell lines, and during monocyte and granulocyte differentiation. (A) Northern blot analysis of KLF4 demonstrates a monocyte-enriched expression pattern. The cell types tested were human peripheral blood monocytes (1° Monocytes), THP-1 (monocytic leukemia), U-937 (histiocytic leukemia), HeL (erythrocyte), Jurkat (T cell), Raji (immature B cell), and U-266 (mature B cell). (B) Western blot analysis of KLF4 protein expression in human monocytes and human myeloid cell lines. (C) Northern blot analysis of HL-60 cells shows expression of KLF4 in TPA-differentiated monocytes, but not in RA-differentiated granulocytes, whereas CD11b is expressed in both cell types.
As shown in Figure 1A and B, KLF4 is expressed in several cell lines committed to the monocytic lineage (THP-1 and U-937 cells). In contrast, KLF4 is not expressed in the uncommitted bipotential cell line HL-60. These cells can differentiate along the monocytic or the granulocytic pathway when treated with 12-O-tetradecanoyl phorbol-13 acetate (TPA) or retinoic acid (RA), respectively. Therefore, we reasoned that HL-60 cells would provide a useful in vitro system for studying the role of KLF4 in monocytic differentiation. First, we determined whether TPA and RA treatment led to KLF4 expression in HL-60 cells. KLF4 mRNA was undetectable in HL-60 control cells (vehicle alone) and in cells treated with RA, but was markedly induced in cells treated with TPA (Figure 1C). In contrast, CD11b mRNA (a marker of differentiation that does not distinguish monocytes from granulocytes) was induced by TPA as well as RA. Taken together, these observations demonstrate that KLF4 expression is restricted to monocytes but not granulocytes (Figure 1C).
KLF4 induces a monocytic phenotype in HL-60 cells
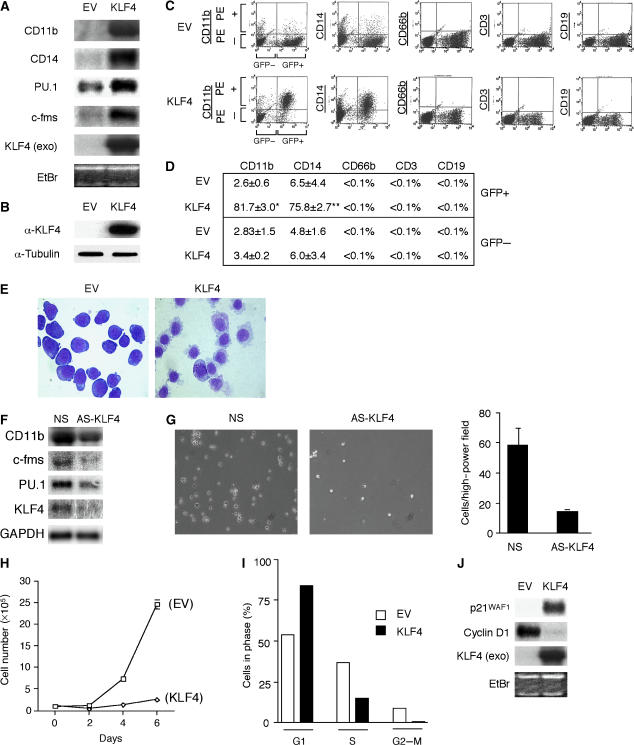
To determine whether KLF4 participates directly in monocytic differentiation, we retrovirally infected HL-60 cells with either full-length KLF4 or an empty virus control (EV) and analyzed the cells for various myeloid markers 4 days later. Exogenous expression of KLF4 was verified by Northern and Western analyses (Figure 2A and B). In comparison with EV-infected cells, we noticed a marked induction of the myeloid markers CD11b and CD14 (Figure 2A). Consistent with a monocytic phenotype, KLF4 expressing cells also expressed higher levels of c-fms (Figure 2A). To quantitate the induction of various hematopoeitic cell surface markers, we performed fluorescence-activated cell sorter (FACS) analyses on both EV- and KLF4-infected HL-60 cells. In comparison with EV-infected cells, KLF4 induced monocytic markers for CD11b (81.7 vs 2.6%) and CD14 (75.8 vs 6.5%), while having no effect on the granulocytic marker CD66b or the lymphocytic markers CD3 and CD19 (Figure 2C). As expected, there were no significant differences in the expression of cell surface markers between the GFP-negative cell populations (Figure 2D). Thus, KLF4 induces monocytic but not granulocytic or lymphocytic markers in HL-60 cells.
Figure 2.
Retroviral overexpression of KLF4 in HL-60 cells promotes features of mature monocytes. HL-60 cells were retrovirally infected with either an empty virus (EV) control or KLF4 construct as described in Materials and methods. (A) Northern blot analysis shows that KLF4 overexpression was capable of inducing a number of myeloid differentiation markers such as CD11b, CD14, PU.1, and c-fms. KLF4 (exo) is exogenous KLF4 mRNA expression. (B) Western blot analysis of exogenous KLF4 protein. (C) FACS analysis was performed on EV or KLF4 transduced cells and revealed high induction for myeloid differentiation markers CD11b (81.7 vs 2.6%; P<0.000002) and CD14 (75.8 vs 6.5%; P<0.00003) in response to KFL4 overexpression. There were no differences using antibodies to CD66b (granulocytes), CD3 (T lymphocytes), or CD19 (B lymphocytes). (D) Percent positivity for each marker in EV or KLF4-overexpressing cells from three independent experiments. (E) Cytospin preparations from EV or KLF4-overexpressing HL-60 cells were stained by Wright–Giemsa staining and viewed at × 100. (F) KLF4 knockdown inhibits HL-60 TPA-induced monocyte differentiation. HL-60 cells were incubated with morpholino oligonucleotide specific to KLF4 or nonspecific (NS) control and then allowed to differentiate in the presence of TPA (100 ng/ml) for 48 h. (G) Marked reduction (∼5-fold, right) of adherent and differentiated HL-60 cells after KLF4 knockdown. Light microscopy (left, × 100) of HL-60 cells after NS or AS-KLF4 incubation as described in panel F. (H) The growth rate of EV or KLF4-infected cells counted over 6 days. (I) Cells overexpressing EV or KLF4 were analyzed for DNA contents. (J) Northern blot analysis shows that KLF4 induces p21WAF1 and inhibits cyclin D1. EtBr, ethidium bromide.
Both monocytes and granulocytes bear characteristic morphologic features. To determine if KLF4 overexpression affects cellular morphology, we performed cytospin analyses. As shown in Figure 2E, HL-60 cells infected with KLF4 exhibited marked morphologic changes. In comparison with EV-infected cells, the KLF4-overexpressing cells are larger, with increased cytoplasmic size and smaller more condensed indented nuclei. In addition, KLF4-overexpressing cells bear ruffled edges, are less basophilic, and contain cytoplasmic vacuoles. These characteristics are consistent with a monocytic phenotype in these cells (Collins, 1987). To verify whether KLF4 directly participates in regulating PU.1 and monocytic gene expression, we used morpholino antisense oligonucleotides to knockdown KLF4 expression during HL-60 cell TPA-induced monocytic differentiation. As shown in Figure 2F and G, in comparison with the nonspecific control morpholino, KLF4 knockdown resulted in a significant reduction in PU.1 and monocytic differentiation markers, as well as impairment of cell adhesion and cytoplasmic spreading. Furthermore, KLF4 overexpression induced HL-60 cell G1-growth arrest, p21WAF1, and inhibited cyclin D1 (Figure 2H–J). Finally, we demonstrate that overexpression of KLF4 allows precursor HL-60 cells to become functionally mature monocytes capable of adhering to a stimulated endothelial monolayer and undergoing phagocytosis (Supplementary Figure 1). Taken together, these data indicate that KLF4 is a critical regulator of HL-60 monocytic differentiation.
KLF4 transactivates the monocytic CD14 promoter
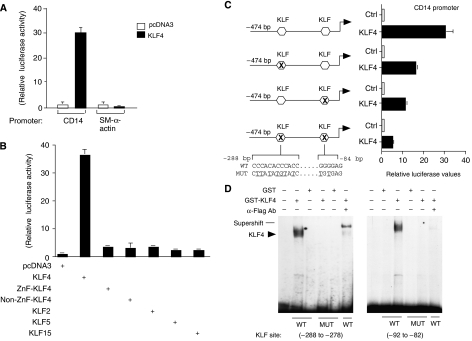
To define the mechanism(s) underlying the ability of KLF4 to induce expression of monocytic markers, we performed transient transfection studies using the proximal −474-bp CD14 in HeLa cells. We observed an ∼30-fold induction of the CD14 promoter by KLF4 (Figure 3A). In contrast, KLF4 repressed the smooth muscle cell-specific promoter SM-α-actin (Figure 3A). To assess whether other KLF family members are capable of activating a myeloid promoter, we cotransfected KLF4, KLF2/LKLF, KLF5/IKLF, or KLF15 with the monocytic-specific CD14 promoter. As demonstrated in Figure 3B, KLF4 induced the CD14 promoter by 35-fold, whereas KLF2, KLF5, or KLF15 transactivated the CD14 promoter no more than the ZF domain of KLF4 alone. Finally, the induction by KLF4 required both the ZF DNA-binding domain of KLF4 as well as the non-ZF domain (aa 1–388), as each construct alone had little effect on the CD14 promoter (Figure 3B). Collectively, these data suggest that in comparison with several other KLF family members, KLF4 is able to promote the monocytic differentiation marker CD14 and this requires intact KLF4.
Figure 3.
KLF4 transactivates the monocytic CD14 promoter and binds to DNA through KLF sites. Transient transfection experiments were performed with either 0.5 μg of pcDNA3 or KLF4, along with the respective promoter-luciferase reporter constructs in HeLa cells. Relative luciferase values are reported after correcting for β-galactosidase. (A) KLF4 induces the CD14 promoter, whereas it represses the non-myeloid smooth muscle (SM) α-actin promoter. (B) Transient transfection studies were performed comparing KLF4, KLF4 DNA-binding domain only (ZnF-KLF4), and several other KLF family members (KLF2, KLF5, and KLF15). Only full-length KLF4 and not other KLFs can transactivate the monocytic CD14 promoter. (C) Loss of the proximal and distal KLF sites results in marked reduction of KLF4 induction of CD14 promoter. (D) Electrophoresis mobility shift assays (EMSAs) were performed using GST or GST-KLF4-Flag-purified protein on the proximal and distal KLF DNA-binding sites. A specific band (arrow) for KLF4 demonstrates binding only to a radiolabeled oligonucleotide probe containing either the wild-type KLF proximal (−92 to −82) or distal (−288 to −278) site, but not to a mutant site, and may be supershifted in the presence of an α-Flag antibody.
KLF4 induces the CD14 promoter through DNA binding
To better define how KLF4 may induce monocytic-specific markers, we analyzed the −474-bp CD14 promoter, as it is active almost exclusively in monocytes/macrophages. Analysis of this promoter region revealed two potential KLF4-binding sites (open boxes, Figure 3C). One of these sites (−288 to −278) contains three partially overlapping KLF-binding sites. A 5′ deletion downstream to this site (−277-bp CD14 promoter) revealed a ∼48% decrease in KLF4 transactivation (data not shown). Site-directed mutation of the proximal (−88 to −83) KLF site within the −474-bp CD14 promoter also resulted in a ∼61% reduction of activity by KLF4 (Figure 3C). However, when the two KLF sites were mutated within the −474-bp CD14 promoter, we found that there was an ∼84% reduction in activity by KLF4 (Figure 3C). These data suggest that KLF4 can transactivate the CD14 promoter by binding to KLF sites.
Members of the Kruppel-like family bind to specific DNA elements (5′-CNCCC-3′) to exert their function. To assess the ability of KLF4 to bind DNA within the −474-bp CD14 promoter, we performed electrophoretic mobility shift assays using Flag-tagged GST-KLF4 and a radiolabeled oligonucleotide probe containing the KLF sites (−288 to −278) and (−92 to −82) of the −474-bp CD14 promoter. As shown in Figure 3D, in comparison to GST alone, incubation of GST-KLF4 resulted in a dominant DNA–protein complex (arrow) that bound to each of these sites. These complexes are specific as they cannot bind to mutated radiolabeled probes and can be supershifted with an α-Flag antibody. Thus, KLF4 is able to bind to KLF sites within the −474-bp CD14 promoter.
Expression of KLF4 mRNA in primary bone marrow-derived myeloid progenitor cells
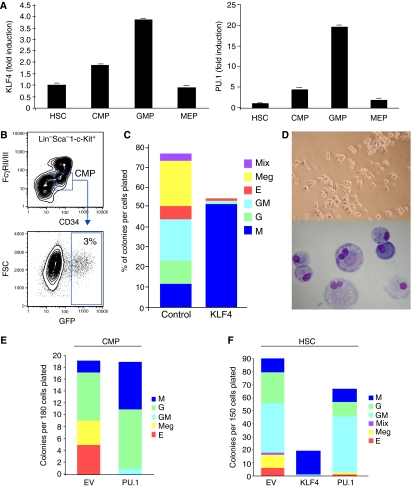
To assess KLF4 expression in primary progenitor cells, we isolated several populations of myeloid progenitors or HSCs by multicolor FACS system, as described previously (Akashi et al, 2000; Miyamoto et al, 2002). As shown in Figure 4A (left), real-time PCR analyses revealed that KLF4 mRNA expression progressively increased from the HSC to the GMP stage, whereas it decreased in MEPs. Expression of the Ets transcription factor PU.1 also increased in a similar manner as KLF4 from the HSC to the GMP stage, with weaker expression in MEPs (Figure 4A, right). These findings raise the possibility that KLF4 may participate in regulating myeloid differentiation.
Figure 4.
Enforced KLF4 instructs CMPs to preferentially induce monocyte differentiation. (A) Stage-specific expression of endogenous KLF4 during myeloid differentiation. qPCR analysis for KLF4 (left) or PU.1 (right) was performed on mouse bone marrow-derived myeloid progenitors (HSC, hematopoietic stem cell; CMP, common myeloid progenitor; GMP, granulocyte macrophage progenitors; MEP, megakaryocyte erythrocyte progenitors). (B–F) Transduction of CMPs or HSCs. Bone marrow-derived CMPs or HSCs were isolated, transduced with control (EV), KLF4, or PU.1 retrovirus as indicated, sorted for GFP positivity, and assessed for differentiation in methylcellulose colony assays. Approximately 3–5% of cells were GFP positive (B). (C) Effect of KLF4 overexpression in CMPs on various hematopoietic lineages identified based on morphology. Control retrovirus-infected cells demonstrated a spectrum of all the various myeloid lineages, whereas KLF4-overexpressing cells exhibited predominant monocytic differentiation. (D) Morphology of KLF4-overexpressing CMPs. Light microscopy (top) and Wright–Giemsa staining (bottom) show that KLF4-transduced cells exhibit morphologic characteristics of monocytes. (E) PU.1 overexpression in CMPs promotes both monocytic and granulocytic differentiation. (F) KLF4 overexpression in HSCs promotes monocytic differentiation, whereas PU.1 promotes both monocytic and granulocytic differentiation. Data are representative of three independent experiments and the same results were obtained.
KLF4 overexpression restricts CMPs or HSCs to the monocytic lineage
Because KLF4 expression is induced in a stage-specific manner during primary myeloid differentiation, we hypothesized that overexpression of KLF4 in CMPs may promote monocytic differentiation at the expense of other lineages. To address this, we retrovirally infected CMPs with either EV (Ctrl) or KLF4, isolated the GFP positive cells (Figure 4B), and allowed them to grow in methylcellulose medium in a cocktail of cytokines capable of differentiating the CMPs along each of the lineages after 5 days of culture. Remarkably, overexpression of KLF4 in CMPs resulted in the preferential commitment and differentiation of nearly all cells to the monocytes/macrophage lineage in comparison with EV (Ctrl) cells (Figure 4C). Furthermore, this was accompanied by a reduction of cells from other lineages (granulocytes, erythrocytes, or megakaryocytes). Light microscopy of KLF4-overexpressing cells demonstrated many adherent cells, and cytospun preparations verified that these cells had enlarged vacuolated cytoplasms with condensed, often indented, nuclei—morphological characteristics typical of mature monocytes and macrophages in these assays (Figure 4D) (Akashi et al, 2000). A similar pattern of monocyte-restricted differentiation occurred when KLF4 was overexpressed in HSCs (Figure 4F). In contrast, PU.1 overexpression in either CMPs or HSCs promoted both monocyte and granulocyte differentiation (Figure 4E and F).
KLF4 rescues monocyte differentiation in PU.1-null cells
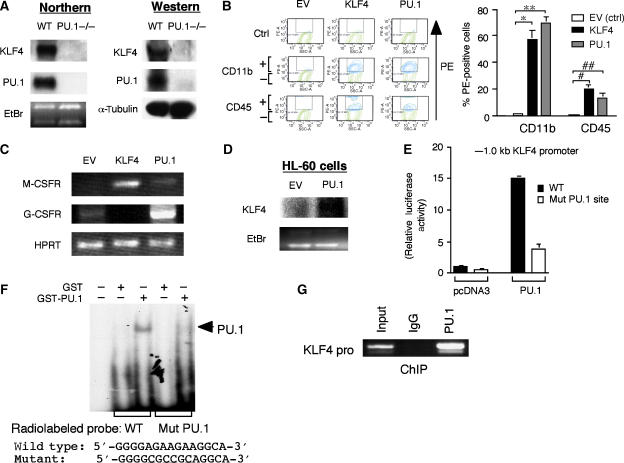
To assess if KLF4 is capable of monocyte differentiation independent of PU.1, we used fetal liver cells from neonatal PU.1-null mice. These cells give rise to a reduced number of hematopoietic progenitors and mature myeloid cells; however, in the presence of IL-3, they can form neutrophils but not monocytes/macrophages (Anderson et al, 1999). In PU.1-null fetal liver cells, expression of KLF4 mRNA and protein is absent in comparison with wild-type bone marrow-derived macrophages (Figure 5A). To assess whether KLF4 requires PU.1 to induce monocyte differentiation, we retrovirally overexpressed EV (Ctrl) or KLF4 in PU.1-null fetal liver cells and grew them in methylcellulose for 7 days with GM-CSF. These cells were also transduced with PU.1 as a positive control, as previous reports have shown that rescue with PU.1 allows the formation of mature monocytes/macrophages and neutrophils, as well as the cell surface markers CD11b and CD45 (Anderson et al, 1999, 2001). As demonstrated in Figure 5B, KLF4 was capable of inducing these differentiation markers in the PU.1-deficient cells to a level similar as that achieved by PU.1 alone. Consistently, KLF4-overexpressing cells possessed morphological features of monocytes in comparison with EV control cells (Supplementary Figure 2). Indeed, PCR analyses demonstrated that KLF4 markedly induced expression only of M-CSFR, whereas PU.1 induced both M-CSFR and G-CSFR (Figure 5C). Taken together, these results suggest that KLF4 can rescue the differentiation defect of PU.1-null myeloid cells along the monocyte/macrophage pathway.
Figure 5.
KLF4 rescues monocyte differentiation in PU.1-null cells and is a PU.1 target gene. (A) Northern and Western blot analyses demonstrate absence of KLF4 expression in PU.1−/− fetal liver cells (Anderson et al, 2001, 1999) in comparison to wild-type (WT) macrophages. (B) Retroviral overexpression of KLF4 in PU.1−/− fetal liver cells induces myeloid differentiation markers CD11b and CD45 to levels achieved by PU.1 itself. (C) Semi-quantitative RT–PCR analysis shows that KLF4-overexpressing cells express M-CSFR and not G-CSFR. In contrast, PU.1-overexpressing cells induce both M-CSFR and G-CSFR. HPRT is shown as a loading control. (D) Northern blot analysis of PU.1 overexpression in HL-60 cells induces KLF4 mRNA expression. EtBr, ethidium bromide. (E) PU.1 induces the −1.0 kb KLF4 promoter ∼15-fold, whereas mutation of a putative-PU.1 DNA-binding site markedly decreases PU.1 transactivation. (F–G) PU.1 can bind to the PU.1 site in the KLF4 promoter, as verified by (F) electrophoretic mobility shift assay (EMSA) and by ChIP studies (G).
KLF4 is a target gene of PU.1
The fact that KLF4 expression is absent in PU.1-null cells raised the possibility that KLF4 may be a downstream target of PU.1 in the transcriptional hierarchy of specifying macrophage cell fate. To assess whether PU.1 overexpression can induce KLF4, we retrovirally infected HL-60 cells with empty-virus GFP-control (EV) or PU.1-GFP for 5 days. As shown in Figure 5D, in comparison to EV, PU.1-overexpressing cells induced KLF4 mRNA expression. Conversely, cells deficient in PU.1 have markedly decreased expression of KLF4 (Figure 5A; Supplementary Figure 3). PU.1 is a member of the ets family of transcription factors that bind to consensus GGAA-like motifs (Rosenbauer et al, 2005). Examination of the KLF4 promoter identified a potential PU.1 site at −118 bp. To assess whether PU.1 can induce the KLF4 promoter, cells were cotransfected with pcDNA3 or PU.1, in the presence of the wild-type KLF4 promoter or the KLF4 promoter bearing a mutant PU.1 site (KLF4 pro-Mut-PU.1). As demonstrated in Figure 5E, PU.1 induced the KLF4 promoter ∼15-fold. However, in the presence of a KLF4 pro-Mut-PU.1, this induction was markedly attenuated to ∼3.8-fold. To determine whether this site is capable of binding PU.1, we performed gel shift studies using a GST-PU.1 fusion protein. As shown in Figure 5F, GST-PU.1, but not GST alone, bound to this site and was abolished in the presence of a radiolabeled mutant PU.1 site. Chromatin immunoprecipitation (ChIP) studies also verified that PU.1 can bind to the KLF4 promoter (Figure 5G). To gain further insight, we validated these observations using quantitative real-time PCR (qPCR). As shown in Supplementary Figure 4, only primer sets inclusive of the PU.1 site demonstrated binding to the KLF4 promoter; in addition, no PU.1 binding was identified using an IgG control antibody or an irrelevant antibody to c-Jun. Taken together, these data indicate that KLF4 is a PU.1 target gene that helps specify monocyte commitment and differentiation.
KLF4 deficiency alters myeloid differentiation
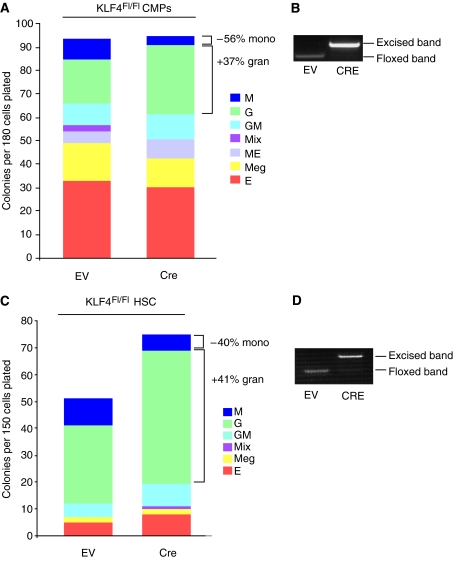
Because overexpression of KLF4 exclusively promoted monocyte/macrophage differentiation in CMPs and HSCs, we hypothesized that KLF4 deficiency may reduce monocytes and possibly shift CMP differentiation toward granulocytes. To assess this, we isolated CMPs derived from wild-type or Klf4loxP mice (Katz et al, 2005) and retrovirally infected them with retrovirus containing Cre recombinase or EV (Ctrl). GFP+ cells were isolated by FACS and subsequently grown in culture for up to 7 days in the presence of a cocktail of cytokines capable of differentiating the cells along all myeloid pathways (Akashi et al, 2000). CMPs derived from wild-type mice showed no differences in myeloid differentiation toward monocytes or granulocytes (data not shown). In contrast, CMPs-overexpressing Cre recombinase from Klf4loxP mice showed a marked reduction of ∼56% in the formation of mature monocytes, whereas granulocyte formation was increased by ∼36% (Figure 6A and B). Similarly, Cre-excision of KLF4 in HSCs from Klf4loxP resulted in a ∼40% reduction in the number of monocytes and an ∼41% increase in granulocyte formation (Figure 6C and D). Collectively, these findings indicate that varying levels of KLF4 may dynamically regulate the balance between monocytes and granulocytes.
Figure 6.
KLF4 deficiency in CMPs or HSCs decreases monocyte and increases granulocyte differentiation. (A–D) Clonogenic analyses of CMPs or HSCs from Klf4loxP mice. CMPs were transduced with retrovirus carrying Cre recombinase or EV (Ctrl). GFP-positive CMPs or HSCs were isolated 36 h after the retroviral infection and subjected to methylcellulose assays for 7 days. Cre-mediated excision of KLF4 (B, D) corresponded with a reduction in monocytes and increase in granulocytes (A, C). Data are representative of three independent experiments, and the same results were obtained.
Discussion
We have shown here that KLF4 is a novel regulator of myeloid differentiation. In support, KLF4 is expressed in a monocyte-restricted and stage-specific pattern during myelopoiesis, programs myeloid progenitor cell fate towards the monocyte lineage, and acts as a PU.1 target gene. Moreover, KLF4 deficiency is associated with impaired monocyte and enhanced granulocyte differentiation in bone marrow clonogenic assays. Mechanistically, KLF4 may mediate these effects by inducing monocytic-specific promoter activity. These findings represent a new paradigm involving KLF4 in the regulatory network of monocyte development.
KLFs in hematopoiesis
Kruppel-like factors have been shown to play critical roles in various aspects of hematopoietic cell differentiation (Bieker, 1996; Feinberg et al, 2004). For example, the founding member, KLF1, is highly expressed in erythrocyte lineages and gene deletion experiments demonstrated an essential role for KLF1 in β-globin synthesis (Nuez et al, 1995; Perkins et al, 1995). KLF2 is induced during the differentiation of immature double positive T cells (CD4+CD8+) to single positive T cells (CD4+ or CD8+), which circulate in the bloodstream. Indeed, targeted disruption of KLF2 verified an essential role for this factor in programming the quiescent phenotype of single positive T cells (Kuo et al, 1997). Recently, KLF2 was also shown to regulate T-cell egress from the thymus and peripheral trafficking (Carlson et al, 2006). KLF3/BKLF was originally identified after screening a mouse erythroleukemia cDNA library, and has a broad tissue expression pattern (Crossley et al, 1996). Interestingly, KLF3 expression was markedly and specifically reduced in erythroid cells from KLF1−/− mice. In contrast to KLF1−/− mice, preliminary data suggest that KLF3−/− mice have a less severe defect in hematopoiesis and display a myeloproliferative disorder (Perkins et al, 1997). Collectively, these observations suggest that KLFs can have profound, often cell type-specific, phenotypic effects within the hematopoietic system. The studies presented in this paper extend a participatory role of KLF proteins in monocyte development.
KLF4 as a PU.1 target gene: role of transcription factor gradients for specifying myeloid differentiation
The presence of PU.1 is required for the formation of both CMPs and CLPs, raising the possibility that gradients of specific transcription factors may help direct lineage commitment at various stages (Rosenbauer et al, 2005). For example, GATA-1 antagonizes PU.1 to drive megakaryocyte/erythrocyte maturation (Rekhtman et al, 1999), whereas C/EBP-α antagonizes PU.1 to promote granulocytic maturation (Dahl et al, 2003). Consistently, loss of C/EBP-α in mice blocks granulocyte maturation and is important for the transition of the CMP to GMP stage; however, after the GMP stage, loss of C/EBP-α has no effect on granulocyte or monocyte formation, indicating that instructive cues or ‘progenitor priming' for monocyte versus granulocyte development likely occur before GMP differentiation (Zhang et al, 2004). Because PU.1-null mice have defects in multiple cell lineages (Scott et al, 1994; McKercher et al, 1996; Colucci et al, 2001), it raises the possibility that downstream factors may be necessary for sustaining progenitors along the monocytic pathway. Several lines of evidence support KLF4 as an important target gene of PU.1 in this process: (1) KLF4 expression was induced in an analogous manner as PU.1 in the CMP-to-GMP transition (Figure 4A); (2) KLF4 modulated the balance between monocytes and granulocytes with exclusive monocyte differentiation in CMPs, HSCs, and the bipotential HL-60 cell line (Figures 2 and 4); (3) KLF4 expression is absent in PU.1−/− fetal liver hematopoietic progenitors and overexpression of KLF4 endowed PU.1−/− progenitors to express M-CSFR, whereas rescue of these cells with PU.1 induced both M-CSFR and G-CSFR (Figure 5A–C); (4) overexpression of PU.1 induced KLF4 expression in HL-60 cells (Figure 5D); (5) mutation of a PU.1 site in the proximal KLF4 promoter significantly inhibited the ability of PU.1 to transactivate the promoter (Figure 5E); and (6) gel shift and ChIP studies both verify that PU.1 can bind to the proximal KLF4 promoter (Figure 5F and G; Supplementary Figure 4). Taken together, based on these observations, we propose a transcriptional hierarchy whereby PU.1 induces KLF4, which can, in turn, promote monocyte and antagonize granulocyte development. Indeed, accumulating evidence supports a role for the order of expression of transcription factors in controlling hematopoietic lineage commitment (Iwasaki et al, 2006).
Role of KLF4 in hematopoiesis and implications for leukemogenesis
During hematopoiesis, KLF4 is expressed in a stage-specific manner— with highest expression in GMPs and lowest expression in HSCs and MEPs (Figure 4A). Disruption of KLF4 at either the HSC or CMP stage reduced the number of monocytes with a shift toward increased granulocytes, whereas overexpression of KLF4 induced only monocyte differentiation at both of these stages (Figures 4 and 6). Because the relative levels and order of transcription factor expression can have profound effects in cell lineage fate, dysregulation of KLF4 may be important in leukemogenesis. Mice with a genetic deletion of the PU.1 URE (a.k.a PU.1-knockdown mice), develop acute myeloid leukemia as a result of an ∼80% reduction in PU.1 expression and absence of M-CSFR and GM-CSFR expression (Rosenbauer et al, 2004). Recent analyses of HSCs from preleukemic PU.1-knockdown mice demonstrate a reduction of a number of transcriptional regulators including KLF4 (approximately −2.73-fold) (Steidl et al, 2006), raising the possibility that altered ratios of PU.1/KLF4 may influence leukemic transformation. Indeed, JunB expression was also reduced (approximately −1.71) in PU.1-knockdown HSCs and overexpression of JunB rescued the myelomonocytic block and leukemogenic properties in these cells (Steidl et al, 2006). Although our data suggest that KLF4 can promote monocyte differentiation independent of PU.1 as shown in Figure 5, we cannot rule out PU.1-dependent effects such as the possibility of KLF4 augmenting PU.1 expression in a positive feedback loop. Future studies will be necessary to explore the potential relationship of KLF4 and JunB as PU.1-target genes and potential regulators of the PU.1 URE in normal myeloid differentiation and leukemogenesis.
Materials and methods
Cell culture and reagents
Human peripheral blood monocytes were isolated from healthy donors (Blood Bank, Children's Hospital, Boston) by the Ficoll–Hypaque centrifugation technique, as described previously (Feinberg et al, 2000). HL-60, U-937, THP-1, Jurkat, Raji, and U-266 cells were obtained from the American Type Culture Collection (ATCC) and cultured as recommended. HL-60 cells were treated with TPA (20 nM; Sigma) to induce monocytic differentiation or with all-trans RA (1 μM; Sigma) to induce granulocytic differentiation. Primary antibodies recognizing Flag M2, Myc, IgG1, and α-tubulin were purchased from Sigma; a rabbit polyclonal KLF4 antibody was obtained from Active Motif. Immunohistochemical analyses were performed on paraffin-fixed tissues (spleen or tumor) for the presence of GFP staining. PU.1−/− fetal liver cells were generously provided by B Torbett (Scripps) and were cultured as described (Anderson et al, 1999, 2001). Adenoviral constructs were generated by the Harvard Gene Therapy Initiative (Boston). Mice carrying a KLF4 loxP allele (Klf4loxP) were kindly provided by K Kaestner (U Penn) (Katz et al, 2005).
Retroviral transduction
For retroviral studies, the indicated cDNA was cloned into the retroviral vector GFP-RV (gift from K Murphy) and retrovirus generated as described (Feinberg et al, 2005). For infection of HL-60 cells, retroviral supernatant and culture medium (10% FCS/DMEM+4 μg/ml polybrene) were mixed at a 1:1 ratio and added to 2 × 106 HL-60 EcoR cells (gift from G Nolan, Stanford, CA, USA). Within 72 h ∼20–40% infectivity was noted by assessment for GFP. Cells underwent FACS sorting for GFP positivity, resulting in a nearly 100% pure GFP population before use in experiments.
Northern and Western blot analyses
Total RNA was isolated from cultured cells using Trizol methods as previously described(Feinberg et al, 2005). Cellular protein extraction and Western blot analyses were performed as described (Feinberg et al, 2005).
Flow cytometric analysis and cell sorting
For FACS analysis, HL-60 cells were incubated with phycoerythrin (PE)-conjugated monoclonal antibodies specific for CD11b, CD14, CD3, or CD19 as described by the manufacturer's protocol (BD Pharmingen). A monoclonal antibody for CD66b was used, followed by incubation with a PE-conjugated goat anti-mouse IgM secondary antibody (BD Pharmingen). FACS analysis was performed on a FACSCalibur flow cytometer and analyzed with CellQuest (Becton Dickinson, Franklin Lakes, NJ, USA). For analysis and sorting of mouse bone marrow progenitor cells, total RNA was purified from ∼5000 double sorted cells from each population and analyzed by qPCR, as reported (Akashi et al, 2000). qPCR analysis was repeated for at least two separately prepared sets of samples.
In vitro differentiation assay
CMPs or HSCs were purified from C57BL/6 or Klf4loxP mice bone marrow by using multicolor FACS, as previously described (Akashi et al, 2000). CMPs were transduced with the KLF4-retrovirus vector carrying eGFP reporter in the presence of Slf (20 ng/ml) and IL-11 (10 ng/ml) (R&D Systems, Minneapolis, MN, USA), on a fibronectin-coated dish (RetroNectin dish, Takara, Japan). After 30 h of transduction, the eGFP-positive CMPs or HSCs were purified by FACS, and clonal cultures were performed on methylcellulose medium (Methocult H4100; Stem Cell Technologies, Vancouver, BC, Canada) supplemented with Slf, IL-3 (10 ng/ml), IL-11, GM-CSF (10 ng/ml), Epo (2 U/ml), and Tpo (10 ng/ml) (R&D Systems). After 6 days of culture, the colonies were picked up individually and evaluated by their morphologies. CMPs or HSCs transduced with the empty vector were used as a control.
Supplementary Material
Supplementary Materials and methods
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Acknowledgments
This work was supported by funding from the National Institutes of Health HL67755 (MWF), HL 080174 (MWF) and HL03747 (MKJ), HL69477(MKJ), HL-72952 (MKJ), HL-75427 (MKJ), HL-76754 (MKJ), American Heart Association Awards 0355691T (MWF), and American Cancer Society Research Scholar Grant RSG0719501-LIB (MWF). This work was also supported by a Carl J and Ruth Shapiro Scholar Award (MWF), and a Lerner Junior Faculty Scholar Award (MWF, SG).
References
- Akashi K, Traver D, Miyamoto T, Weissman IL (2000) A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193–197 [DOI] [PubMed] [Google Scholar]
- Anderson KL, Nelson SL, Perkin HB, Smith KA, Klemsz MJ, Torbett BE (2001) PU.1 is a lineage-specific regulator of tyrosine phosphatase CD45. J Biol Chem 276: 7637–7642 [DOI] [PubMed] [Google Scholar]
- Anderson KL, Smith KA, Perkin H, Hermanson G, Anderson CG, Jolly DJ, Maki RA, Torbett BE (1999) PU.1 and the granulocyte- and macrophage colony-stimulating factor receptors play distinct roles in late-stage myeloid cell differentiation. Blood 94: 2310–2318 [PubMed] [Google Scholar]
- Bieker JJ (1996) Isolation, genomic structure, and expression of human erythroid Kruppel-like factor (EKLF). DNA Cell Biol 15: 347–352 [DOI] [PubMed] [Google Scholar]
- Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC (2006) Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442: 299–302 [DOI] [PubMed] [Google Scholar]
- Collins SJ (1987) The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood 70: 1233–1244 [PubMed] [Google Scholar]
- Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, Di Santo JP (2001) Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood 97: 2625–2632 [DOI] [PubMed] [Google Scholar]
- Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin SH (1996) Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol Cell Biol 16: 1695–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC (2003) Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol 4: 1029–1036 [DOI] [PubMed] [Google Scholar]
- Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK (2005) Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem 280: 38247–38258 [DOI] [PubMed] [Google Scholar]
- Feinberg MW, Jain MK, Werner F, Sibinga NE, Wiesel P, Wang H, Topper JN, Perrella MA, Lee ME (2000) Transforming growth factor-beta 1 inhibits cytokine-mediated induction of human metalloelastase in macrophages. J Biol Chem 275: 25766–25773 [DOI] [PubMed] [Google Scholar]
- Feinberg MW, Lin Z, Fisch S, Jain MK (2004) An emerging role for Kruppel-like factors in vascular biology. Trends Cardiovasc Med 14: 241–246 [DOI] [PubMed] [Google Scholar]
- Friedman A (2002) Transcriptional regulation of granulocyte and monocyte development. Oncogene 21: 3377–3390 [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B (1996) A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem 271: 31384–31390 [DOI] [PubMed] [Google Scholar]
- Higaki Y, Schullery D, Kawata Y, Shnyreva M, Abrass C, Bomsztyk K (2002) Synergistic activation of the rat laminin gamma1 chain promoter by the gut-enriched Kruppel-like factor (GKLF/KLF4) and Sp1. Nucleic Acids Res 30: 2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K (2006) The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev 20: 3010–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, Fenyus ML, Elf S, Chan S, Kastner P, Huettner CS, Murray R, Tenen DG, Akashi K (2005) Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106: 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, Kaestner KH (2005) Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology 128: 935–945 [DOI] [PubMed] [Google Scholar]
- Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH (2002) The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129: 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Veselits ML, Leiden JM (1997) LKLF: a transcriptional regulator of single-positive T cell quiescence and survival [see comments]. Science 277: 1986–1990 [DOI] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA (1996) Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 15: 5647–5658 [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K (2002) Myeloid or lymphoid promiscuity as a critical step in hematopeietic commitment. Dev Cell 3: 137–147 [DOI] [PubMed] [Google Scholar]
- Nerlov C, Querfurth E, Kulessa H, Graf T (2000) GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95: 2543–2551 [PubMed] [Google Scholar]
- Noti JD, Johnson AK, Dillon JD (2005) The leukocyte integrin gene CD11d is repressed by gut-enriched Kruppel-like factor 4 in myeloid cells. J Biol Chem 280: 3449–3457 [DOI] [PubMed] [Google Scholar]
- Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F (1995) Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375: 316–318 [DOI] [PubMed] [Google Scholar]
- Orkin SH (2000) Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet 1: 57–64 [DOI] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH (1995) Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375: 318–322 [DOI] [PubMed] [Google Scholar]
- Perkins AC, Yang H, Crossley PM, Fujiwara Y, Orkin SH (1997) Deficiency of the CACC-element binding protein, BKLF, leads to a progressive myeloproliferative disease and impaired expression of SHP-1. Blood 90: 575a [Google Scholar]
- Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG (1998) CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol 18: 4301–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VA, Iwama A, Iotzova G, Schulz M, Elsasser A, Vangala RK, Tenen DG, Hiddemann W, Behre G (2002) Granulocyte inducer C/EBPalpha inactivates the myeloid master regulator PU.1 possible role in lineage commitment decisions. Blood 100: 483–490 [DOI] [PubMed] [Google Scholar]
- Rekhtman N, Radparvar F, Evans T, Skoultchi AI (1999) Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev 13: 1398–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Koschmieder S, Steidl U, Tenen DG (2005) Effect of transcription-factor concentrations on leukemic stem cells. Blood 106: 1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG (2004) Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet 36: 624–630 [DOI] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H (1994) Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265: 1573–1577 [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E (1999) Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 22: 356–360 [DOI] [PubMed] [Google Scholar]
- Shields JM, Christy RJ, Yang VW (1996) Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem 271: 20009–20017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl U, Rosenbauer F, Verhaak RG, Gu X, Ebralidze A, Otu HH, Klippel S, Steidl C, Bruns I, Costa DB, Wagner K, Aivado M, Kobbe G, Valk PJ, Passegue E, Libermann TA, Delwel R, Tenen DG (2006) Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet 38: 1269–1277 [DOI] [PubMed] [Google Scholar]
- Swamynathan SK, Katz JP, Kaestner KH, Ashery-Padan R, Crawford MA, Piatigorsky J (2006) Conditional deletion of mouse Klf4 gene results in corneal epithelial fragility, stromal edema and loss of conjunctival goblet cells. Mol Cell Biol 27: 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tenen DG (2003) Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer 3: 89–101 [DOI] [PubMed] [Google Scholar]
- Tenen DG, Hromas R, Licht JD, Zhang DE (1997) Transcription factors, normal myeloid development, and leukemia. Blood 90: 489–519 [PubMed] [Google Scholar]
- Traver D, Miyamoto T, Christensen J, Iwasaki-Arai J, Akashi K, Weissman IL (2001) Fetal liver myelopoiesis occurs through distinct, prospectively isolatable progenitor subsets. Blood 98: 627–635 [DOI] [PubMed] [Google Scholar]
- Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG (1997) Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci USA 94: 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z (1999) Negative crosstalk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA 96: 8705–8710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Himes JA, Akashi K, Tenen DG (2004) Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity 21: 853–863 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and methods
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3