Abstract
Discoidin domain receptor (DDR) is a cell-surface receptor tyrosine kinase activated by the binding of its discoidin (DS) domain to fibrillar collagen. Here, we have determined the NMR structure of the DS domain in DDR2 (DDR2-DS domain), and identified the binding site to fibrillar collagen by transferred cross-saturation experiments. The DDR2-DS domain structure adopts a distorted jellyroll fold, consisting of eight β-strands. The collagen-binding site is formed at the interloop trench, consisting of charged residues surrounded by hydrophobic residues. The surface profile of the collagen-binding site suggests that the DDR2-DS domain recognizes specific sites on fibrillar collagen. This study provides a molecular basis for the collagen-binding mode of the DDR2-DS domain.
Keywords: collagen, discoidin domain receptor, NMR, protein–protein interaction, transferred cross-saturation
Introduction
Collagen, a major component of extracellular matrices, is characterized by its long triple-helical structure (polyproline II-like helices) formed by three tightly interwoven polypeptide chains, and the triple helix self-assembles into large fibrillar supermolecules under physiological conditions at neutral pH (Kadler et al, 1996; Okuyama et al, 2006). Collagen plays a critical role not only in providing mechanical strength for tissues, but also in mediating a variety of cellular processes, including cell attachment, cell development, and diseases, through interactions with other matrix proteins and cell-surface receptors (Juliano and Haskill, 1993). Five different types of collagen receptors have been identified: integrins, glycoprotein VI, leukocyte-associated IG-like receptor-1, members of the mannose receptor family, and the receptor tyrosine kinases called discoidin domain receptors 1 and 2 (DDR1 and DDR2) (Vogel, 2001; Leitinger and Hohenester, 2007).
DDR2 is expressed in skin, heart, liver, and kidney connective tissue, and is overexpressed in some tumor cells (Labrador et al, 2001; Vogel et al, 2006). The recognition of collagen by DDR2 results in the regulation of cell proliferation and migration (Labrador et al, 2001; Olaso et al, 2002). In addition, DDR2 controls extracellular matrix remodeling by upregulating both the expression and activity of matrix metalloproteinases (Olaso et al, 2001; Wang et al, 2002; Ferri et al, 2004; Xu et al, 2005).
DDR2 consists of a discoidin (DS) domain and a stalk region in its extracellular portion, and an intracellular tyrosine kinase, connected by a transmembrane region. The direct interaction of collagen with the DS domain of DDR2 (DDR2-DS domain) triggers the activation of its intracellular tyrosine kinase, leading to downstream intracellular signaling (Ikeda et al, 2002; Leitinger, 2003; Leitinger et al, 2004; Yang et al, 2005). The DDR2-DS domain binds to collagen types I, II, and III, but does not recognize collagen type IV, in agreement with its receptor activation studies (Leitinger et al, 2004). DDR2 is reportedly responsible for the G0/G1 cell cycle arrest when tumor cells are in contact with the collagen fibril, but not with a monomeric collagen (Wall et al, 2005).
DS domains are found in a number of functionally unrelated proteins including the coagulation factor V and VIII, neuropilin-1, and lectins. It has been demonstrated based on the structural studies that the diverse ligand recognition among DS domain family is mediated by the highly variable loops clustered in the one face of the DS domain(Fuentes-Prior et al, 2002). Recently, the region encompassing the putative loops L1–L4 was replaced by the corresponding loops of other DS domains (Leitinger, 2003). The loop chimeras of L1, L2, and L4 lost the ability to recognize collagen, suggesting that L1, L2, and L4 are responsible for the collagen binding. However, little is known about the collagen recognition mode of the DDR2-DS domain at atomic resolution, due to the lack of the three-dimensional structure of the DDR2-DS domain.
Here, in order to elucidate the collagen recognition mechanism of the DS domain, we have determined the solution structure of the DDR2-DS domain and the interface on the DS domain for the collagen type II fibril by transferred cross-saturation (TCS) methods. TCS is the only technique that can detect the residues contacting the collagen fibril (Nishida et al, 2003). The fibril of collagen type II binds to the interloop trench and the surrounding loop region of the DDR2-DS domain. The property of the collagen interface on the DDR2-DS domain has been revealed to contain the combination of charged and hydrophobic residues, as observed for the sequence specific interaction of the α2 subunit of integrin with collagen (Emsley et al, 2000). The specific recognition of the collagen by DDR2 is discussed, based on the structural basis of the DDR2–collagen interaction.
Results
Collagen binding analyses of the DS domain
The collagen-binding site of DDR2 is reportedly fully contained within the DS domain of DDR2 (DDR2-DS domain, residues 30–185) (Leitinger, 2003; Leitinger et al, 2004). In this study, we expressed DDR2 residues 26–186 as the DS domain in Pichia pastoris and purified the protein to homogeneity. Size-exclusion chromatography analyses indicated that the purified DDR2-DS domain is monomeric in solution (data not shown).
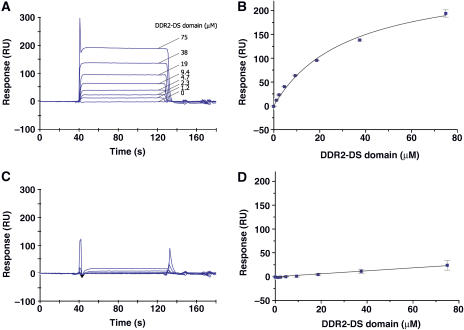
Surface plasmon resonance (SPR) experiments were carried out to examine the collagen binding properties of the DDR2-DS domain. The SPR data for the interaction with collagen type II were fitted using the one-site binding model, resulting in an excellent fit with a dissociation constant of 2.5 × 10−5 M (Figure 1A and B). In contrast, no binding for collagen type IV was detected (Figure 1C and D). Therefore, the binding of the DDR2-DS domain to collagen type II observed under the conditions of the SPR experiments is highly specific.
Figure 1.
Collagen binding analyses of the DDR2-DS domain by SPR experiments. Overlay plots of the sensorgrams obtained for the interaction between 1.2–75 μM of the DDR2-DS domain and the immobilized collagen type II (A) and type IV (C). RU stands for resonance units. The plots based on the steady-state method in SPR for collagen type II and type IV are shown in (B) and (D), respectively. Error bars are standard deviations of three independent experiments.
Structure description
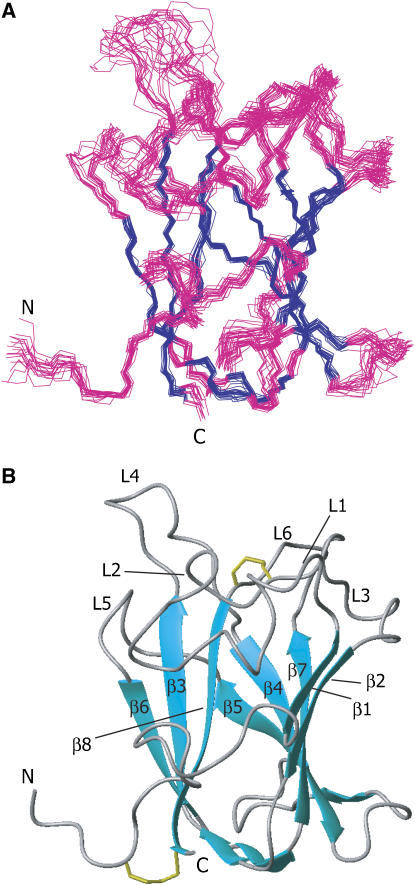
The three-dimensional structure of the DDR2-DS domain was determined, using standard heteronuclear multidimensional NMR techniques (see Materials and methods). The final family of 20 superimposed backbone structures for the DDR2-DS domain is shown in Figure 2A, and the NMR constraints and structure statistics are summarized in Table I.
Figure 2.
NMR structure of the DDR2-DS domain. (A) Twenty backbone models of the DDR2-DS domain (residues 26–186) were superimposed. The β-strand regions are colored blue and the other regions are pink. The overall r.m.s. deviation between the family and the mean coordinate positions is 0.38±0.06 Å for the backbone atoms in the β-stranded regions. (B) Ribbon diagram of the DDR2-DS domain. L1–L6 refer to loops 1–6. The disulfide bonds between Cys30 and Cys185, and between Cys73 and Cys177 are shown as stick models colored yellow. These molecular diagrams were generated with the program MOLMOL (Koradi et al, 1996).
Table 1.
NMR and refinement statistics for protein structures
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 2479 |
| Intra-residue | 617 |
| Inter-residue | |
| Sequential (∣i−j∣=1) | 705 |
| Medium-range (∣i−j∣⩽4) | 258 |
| Long-range (∣i−j∣⩾5) | 899 |
| Hydrogen bonds | 70 |
| Total dihedral angle restraints | |
| ϕ | 110 |
| Structure statistics | |
| Average pairwise r.m.s.d.a (Å) | |
| Backbone atoms | 0.38±0.06 |
| Heavy atoms |
1.2±0.2 |
| NMR, nuclear magnetic resonance; NOE, neuclear overhauser effect; r.m.s.d., root mean-square deviation. | |
| aPairwise r.m.s.d. was calculated among 20 refined structures for the β-sheet region. | |
The overall fold consists of eight major β-strands, β1–β8 (β1, 46–49; β2, 86–90; β3, 99–104; β4, 117–123; β5, 140–142; β6, 151–154; β7, 163–169; β8, 178–185), which are arranged in two antiparallel β-sheets of five (β1–β2–β7–β4–β5) and three (β8–β3–β6) strands packed against each other (Figure 2B). This β-barrel structure with a jellyroll topology is common to the other DS domains: the coagulation factors V and VIII (F5/8) C2 domains and the neuropilin-1 (Npn-1) b1 domain (Macedo-Ribeiro et al, 1999; Pratt et al, 1999; Lee et al, 2003; Vander Kooi et al, 2007), where the r.m.s. deviations of the α-carbon atoms in the β-barrel core region range from 1.2 to 1.4 Å. At the ‘top' of the β-barrel core protrude six juxtaposed loops (L1–L6), among which loops L1, L2, and L4 differ from those of the other DS domains in their length and conformation, as described later. L2 and L6 are linked by a disulfide bridge between Cys73 and Cys177 (Figure 2B). The ‘bottom' of the β-barrel core is closed by three interconnecting straight segments, and the N- and C-termini are connected with a disulfide bond between Cys30 and Cys185 (Figure 2B).
Collagen-binding site revealed by TCS experiments
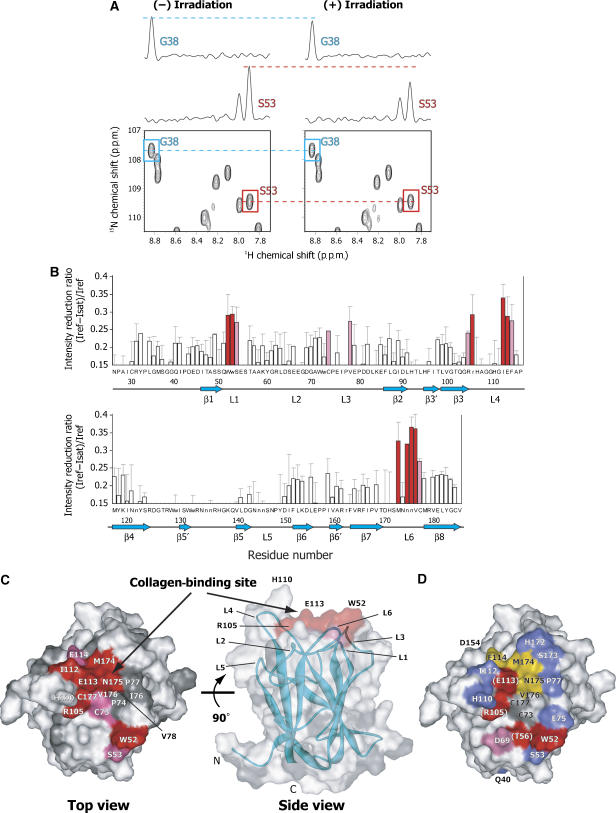
In order to identify the DDR2 residues involved in the direct interaction with the collagen type II fibril, the DDR2-DS domain was studied by TCS experiments (Takahashi et al, 2000; Nakanishi et al, 2002; Nishida et al, 2003). The irradiation with radio frequency pulses of the sample containing the collagen type II fibril resulted in the selective intensity-losses of the 1H–15N HSQC signals originating from the free DDR2-DS domain resonances (Figure 3A). The cross-saturation from collagen to the DDR2-DS domain was quantified as the reduction ratios of the peak intensities (Figure 3B). The residues with high reduction ratios for the backbone amide signals were Trp52, Ser53, Cys73, Val78, Arg105, Ile112, Glu113, Phe114, Met174, Val176, and Cys177. The side-chain signals from Trp52, Arg105, and Asn175 were also affected by the irradiation. Especially significant reductions (reduction ratios >0.29) were observed for Trp52, Arg105, Ile112, Glu113, Met174, Asn175, and Val176. Mapping of these residues on the structure of the DDR2-DS domain shows that they form a contiguous surface at the ‘top' of the DDR2-DS domain in L1, L3, L4, and L6, indicating that this surface is the collagen-binding interface (Figure 3C).
Figure 3.
Collagen-binding residues revealed by TCS experiments and mutagenic scans. (A) A selected portion of the 1H–15N HSQC spectra of uniformly 2H- and 15N-labeled DDR2-DS domain in the presence of collagen type II fibril, which were recorded without (left) and with (right) irradiation. One-dimensional cross-sections are also shown for the signals from Gly38 and Ser53, where the small and significant signal intensity reductions were observed. (B) Plots of the reduction ratios of the signal intensities originating from the HN groups with and without irradiation. Red and pink bars represent the cross-peaks with signal intensity reduction ratios >0.29 and within the 0.29–0.24 range, respectively. The primary sequence of the DDR2-DS domain (residues 26–186) is displayed in the single-letter amino-acid code, and the side-chain HN groups are designated by lower-case characters. Error bars are standard deviations of three independent experiments. (C) Mapping of the affected residues in the TCS experiments on the DDR2-DS domain. The residues with signal intensity reduction ratios >0.29, within the 0.29–0.24 range, and no data are colored red, magenta, and gray, respectively. These molecular diagrams were generated with WebLab Viewer Pro (Molecular Simulations Inc.). In the TCS experiment, the signal intensity reductions of the amide protons correlated well with the distances from the interface (Shimada, 2005). Our in-house program, where TCS effects are simulated based upon protein structures, was used for the determination of the selection criteria. Val78, for which reduction ratio is 0.27, is hidden in these views. (D) Mutagenic scans of the DDR2-DS domain binding to collagen type II. The scanned residues are colored according to the changes in the affinity to collagen at pH 7 by the introduced mutations. The mutants that lost ⩾90% of their affinity relative to the wild type (red) are as follows: W52A, T56A, R105A, and E113Q. The D69A mutant lost 75–90% of its affinity (pink), while the mutants that lost 15–75% (yellow) are F114A, M174A, and N175A, and ⩽15% (blue) are Q40A, S53A, E75A, P77A, H110A, I112A, D154A, H172A, and S173A. The residues that were not mutated are colored gray. The residues in parentheses, T56, R105, and E113, indicate that the mutations of these residues might have caused structural rearrangements of the DDR2-DS domain (Table II).
Mutational analysis
In order to evaluate the contribution of each interacting residue to the collagen binding, point mutations were introduced on and around the binding site identified by TCS, and their effects on the collagen-binding affinity were characterized by SPR. In addition, a 1H–15N HSQC spectrum was observed for each mutant, to determine whether significant conformational changes were caused by the mutations.
Figure 3D and Table II summarize the binding constants for the mutants determined by SPR and their chemical shift differences from those of the wild type. The point mutations of the residues of the collagen-binding surface identified by TCS (W52A, R105A, and E113Q) resulted in significant losses of the affinity of the DDR2-DS domain for collagen type II. It should be noted that the HSQC spectra of the T56A, R105A, and E113Q mutants showed relatively large chemical shift differences from that of the wild type, suggesting that these mutations caused structural rearrangements of the DDR2-DS domain. Therefore, these residues are important for maintaining the three-dimensional structure. For these cases, TCS is a better analytical method than mutagenesis to detect the interacting residues, since TCS does not require any artificial modification that would cause structural rearrangements of the target molecule, and thus can be used to identify cysteine residues, which form a disulfide bond such as Cys73 and Cys177, as contact residues.
Table 2.
Summary of the mutational analysisa
| Ka (× 104 M−1) | Relative affinity (%)b | Location of the mutation | Chemical shift differencec >0.1 p.p.m. | |
|---|---|---|---|---|
| WT | 4.1±0.4 | 100 | — | — |
| Q40A | 4.2±0.6 | 102 | Coil between N-terminus and β1 | 4 |
| W52A | 0.1±0.1 | 3 | L1 | 6 |
| S53A | 6.2±0.4 | 151 | L1 | 9 |
| T56A | 0.1±0.1 | 3 | L1 | 22 |
| D69A | 1.0±0.1 | 23 | L2 | 8 |
| E75A | 4.6±0.4 | 111 | L3 | 2 |
| P77A | 5.9±0.1 | 144 | L3 | 3 |
| R105A | NDd | — | L4 | 42 |
| H110A | 4.2±0.1 | 102 | L4 | 2 |
| I112A | 5.4±0.3 | 132 | L4 | 7 |
| E113Q | 0.4±0.2 | 10 | L4 | 24 |
| F114A | 3.3±0.1 | 80 | L4 | 8 |
| D154A | 4.6±0.7 | 111 | β6 | 5 |
| H172A | 3.9±0.1 | 95 | L6 | 7 |
| S173A | 4.21±0.02 | 103 | L6 | 3 |
| M174A | 3.4±0.7 | 83 | L6 | 16 |
| N175A |
3.5±0.3 |
84 |
L6 |
9 |
| ND, not determined; WT, wild type. | ||||
| aBinding constants (Ka) were determined by SPR experiments. | ||||
| bRelative affinity of each point mutant to the wild-type DDR2-DS domain (WT). The Ka of WT is set to 100%. | ||||
| cNumber of signals with chemical shift difference >0.1 p.p.m. from WT. Chemical shift difference was calculated as follows: δ={(ΔHN)2+(ΔN/5)2}1/2. | ||||
| dThe binding constant for R105A could not be determined because of the weak responses in the SPR sensorgrams. | ||||
Moderate affinity losses were observed for F114A, M174A, and N175A. In addition to the residues identified by TCS, D69A also impaired the binding affinity. The other mutated residues did not show detectable changes in the collagen-binding affinity.
Discussion
Collagen-type-specific binding
An Fc-fused dimer of the DDR2-DS domain reportedly bound to collagen type II, with a dissociation constant (Kd) of 1 × 10−8 M (Leitinger, 2003; Leitinger et al, 2004). The present SPR analyses for the interaction with collagen type II yielded a Kd value of 2.5 × 10−5 M, which is a reasonably high affinity as a monomer of the DDR2-DS domain. In addition, it is consistent with the previous collagen binding and tyrosine phosphorylation studies, in which the DS domain bound to collagen type II but not type IV (Shrivastava et al, 1997; Vogel et al, 1997; Leitinger et al, 2004). This indicates that the prepared DDR2-DS domain possesses the same binding properties as the native protein. It should be noted that nonspecific binding of the DDR2-DS domain to either type of collagen was not observed under the conditions of the SPR experiments. Therefore, it is likely that the effect of nonspecific binding in the TCS experiments should be negligible.
Collagen-binding site of the DDR2-DS domain
The TCS experiments revealed that Trp52, Ser53, Cys73, Val78, Arg105, Ile112, Glu113, Phe114, Met174, Asn175, Val176, and Cys177, which are located in the L1, L3, L4, and L6 loops on the ‘top' of the DDR2-DS domain, are in the proximity of the fibrillar collagen (Figure 3C). The mutagenesis study revealed that Asp69 in the L2 loop is also another interacting residue. The lack of TCS information for Asp69 is presumably due to its amide proton being remote (more than 7 Å) from any of the collagen protons, which could happen in the case of a long-range electrostatic interaction involving the side-chain terminus. In contrast, Ser53, Ile112, Phe114, Met174, and Asn175, which were detected in the TCS experiments, exhibited no significant loss in the affinity in the mutagenesis studies. The affinity depends on both energetically critical hot-spot residues and a surrounding seal of contacting residues, which may establish the correct solvation environment by occluding bulk solvent from the hot spot (Bogan and Thorn, 1998). This might be the reason why the residues mentioned above have little effect on the collagen-binding affinity.
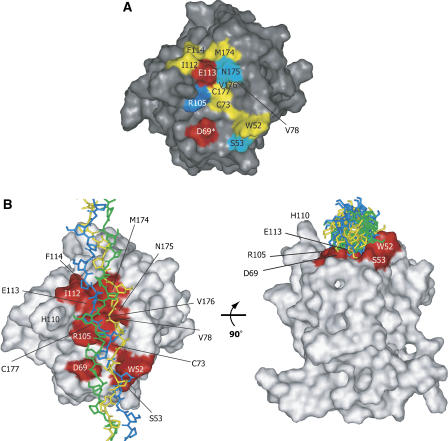
Altogether, we conclude that the collagen-binding site of the DDR2-DS domain is composed of the following residues: Trp52, Ser53, Asp69, Cys73, Val78, Arg105, Ile112, Glu113, Phe114, Met174, Asn175, Val176, and Cys177 (Figure 4A).
Figure 4.
The properties of the collagen-binding trench. (A) Surface representation of the binding interface of the DDR2-DS domain. The residues at the collagen-binding interface are colored according to their side-chain properties: hydrophilic residues are shown in cyan; hydrophobic residues are yellow; acidic residues are red; and the basic residue is blue. An asterisk (*) indicates that the residue was identified by the mutation, but did not display TCS signal intensity reduction. (B) The proposed model of the DDR2-DS domain–triple-helical peptide, obtained by using the PatchDock software (Schneidman-Duhovny et al, 2003) (http://bioinfo3d.cs.tau.ac.il/PatchDock). The model peptide of collagen with the sequence (GPO)12 (PDB code 2CLG; Chen et al, 1991) was used as the ligand, with each of the 20 conformers of the DDR2-DS domain as the receptor. The solution with the highest score among the top 20 solutions that satisfied the NMR and mutagenesis data was chosen. The collagen-binding residues of the DDR2-DS domain are colored red. The three model peptides of collagen are shown as stick models colored green, blue, and yellow, respectively.
The binding site determined in the present study forms a trench with a curved surface, which seems to accommodate the triple helix of collagen (Figures 3C and 4A). Therefore, the model peptide of collagen, (GPO)12 (O=hydroxyproline) (PDB code 2CLG) (Chen et al, 1991), was docked to the DDR2-DS domain (Figure 4B). The model shows that the width and depth of the trench are suitable to accommodate the triple helix in a reasonable orientation, which is consistent with the TCS experiments and the mutagenesis study.
As shown in Figure 4B, His110 appears to play a role in the collagen recognition, since His110 is located at the apex of the most prominent loop, L4, on the trench. However, both the TCS experiments and mutagenesis studies indicated that His110 is not involved in collagen recognition. It is likely that L4, which includes the glycine/alanine-rich region (107AGGHG111), is somewhat flexible, because fewer inter-residue NOEs were observed in this region, leading to the less converged structure (Figure 2A). Therefore, the flexibility might result in the lack of the interaction between His110 and collagen.
It should be noted that the collagen triple helix might bend upon binding, in order to achieve maximum complementarity of the binding to the trench of the DDR2-DS domain, which exhibits a curved surface. Such bending of the triple helix has been observed in the integrin–collagen structure (Emsley et al, 2000, 2004).
Ligand recognition by DS domains
The present results for the DDR2-DS domain show that the collagen binds to the loop region, where the L1, L2, L3, L4, and L6 loops form a trench suitable to accommodate the triple helix of the collagen fibril (Figures 2B and 4B). This result agrees well with the previous observation where the loop chimeras of L1, L2, and L4 lost the ability to bind the collagen, and the chimera of L3 somewhat reduced affinity (Leitinger, 2003).
The DDR2-DS domain displays the short L1 loop with eight residues, which is far apart from L4 to facilitate the binding of collagen. Furthermore, the disulfide bond between Cys73 and Cys177 makes the trench wide and hydrophobic, and stabilizes the overall fold (Figure 2B). In addition, the DDR2-DS domain has both electropositive and negative residues, Asp69, Arg105, and Glu113, which might play a role in determining the specificity of the target sequence on collagen (see below).
The DS domains are known to recognize a variety of ligands, such as sugars, lipids, and proteins (Baumgartner et al, 1998). To date, a number of crystal structures of DS domains have been reported, including sugar-recognizing lectins (Ito et al, 1994; Gaskell et al, 1995; Bianchet et al, 2002), the C2 domains of F5 and F8 (Macedo-Ribeiro et al, 1999; Pratt et al, 1999), and the Npn-1 b1 domain (Lee et al, 2003; Vander Kooi et al, 2007). Although these DS domains share remarkable similarity in their overall fold, the differences in the loop region consisting of L1, L2, L3, L4, and L6 lead to the recognition of a wide variety of ligands, as noted in the literature (Fuentes-Prior et al, 2002). The DS domains of the blood coagulation factors, F5 and F8, bind to membranes containing phosphatidyl-L-serine (PLS). They possess long hydrophobic loops, which might be suitable for insertion into the lipid bilayer. In addition, the hydrophilic and charged residues in the deep pocket formed by the loops seem to contribute to the recognition of the hydrophilic moiety of PLS (Macedo-Ribeiro et al, 1999; Srivastava et al, 2001).
The structures of the lectin DS domains in complex with a sugar molecule were also reported (Ito et al, 1994; Gaskell et al, 1995; Bianchet et al, 2002). In each of their loop regions, the residues such as Arg, Glu, and His directly form hydrogen bonds and/or electrostatic interactions with the ligand molecule. Therefore, the DDR2-DS domain is different from the other DS domains, in that the combination of hydrophobic and charged residues on the widely opened loops enables the specific recognition of collagen.
Specific binding of the DDR2-DS domain to collagen
The present study has revealed the collagen-binding interface and its properties on the DDR2-DS domain (Figure 4A). The axis of the triple helix may become oriented along the trench upon binding (Figure 4B). Therefore, the DDR2-binding site on collagen should be complementary to the revealed collagen-binding interface on the DDR2-DS domain.
As shown in Figure 4B, the binding site of the DDR2-DS domain, which spans almost the same length as two to three triplets in collagen, has two aromatic residues, Trp52 and Phe114, and the charged residues Asp69, Arg105, and Glu113. Notably, the negatively charged residue, Glu113, exists at the bottom of the trench and is surrounded by hydrophobic residues, which probably enhance the contribution of the electrostatic interaction with collagen. The other charged residues, Asp69 and Arg105, could also participate in electrostatic interactions. Therefore, the DDR2-binding sequences on collagen might be the two to three triplets, which include at least one negative residue, one or two positive residues, and hydrophobic residues, which could interact with aromatic residues on the DDR2-DS domain. Both negative and positive residues comprise only 8% of the total sequence of collagen type II. Based on the properties of the collagen-binding interface, the recognition sequence of DDR2 may be rare in the collagen type II, which is consistent with the previous result that DDR2 binds to only the D2 period of the collagen type II (Agarwal et al, 2002; Leitinger et al, 2004).
Some collagen sequences recognized by other collagen-binding proteins have been identified. Among them, CNA recognizes (GPO)n (O=hydroxyproline) segments, which are frequently found in collagen, through mainly hydrophobic interactions (Zong et al, 2005). On the other hand, the I domain from the α2 subunit of integrin contains key electrostatic interactions as well as a surrounding hydrophobic surface in its collagen-binding site (Emsley et al, 2000), similar to the DDR2-DS domain, and recognizes the specific sequences GXOGER (X=Phe, Leu, Arg, Met) and GLOGEN, which appear only three times in collagen type III (Knight et al, 2000; Kim et al, 2005; Raynal et al, 2006).
The collagen sequence recognized by the DDR2-DS domain remains to be identified. The structural basis of the collagen recognition by the DDR2-DS domain provided here should be useful to predict the DDR2-binding sequences on collagen, which are justified by the binding study, for example, using synthetic peptides containing the predicted collagen sequence.
Materials and methods
Sample preparation
A cDNA fragment containing the DS domain (residues 26–186) of human DDR2 was amplified by PCR, and was cloned into the pPICZαA plasmid (Invitrogen). The resultant plasmid was transformed into the P. pastoris X-33 strain. A Mut+ transformant was grown in buffered minimal medium (100 mM potassium phosphate, pH 6, yeast nitrogen base (0.34%, w/v), biotin (4 × 10−5%, w/v), and carbon and nitrogen sources, as described below). The cells were initially grown the cells in medium containing 1% (w/v) ammonium chloride and 1% (w/v) glycerol, and then protein production was induced by transferring the cells to medium containing 0.5% methanol as the carbon source. Uniformly 15N-labeled and 13C/15N-labeled proteins were prepared by initially growing the cells in medium containing 0.2% (w/v) 15N-ammonium chloride, and 0.5% (w/v) glucose or 0.5% (w/v) 13C-glucose supplemented with 0.1% (w/v) Celtone-N or -CN powder, and then inducing protein production by transferring the cells to medium containing methanol or 13C-methanol as a carbon source. Methanol-induced cultures were grown for 4 days at 20°C, with the daily addition of 0.5% (v/v) methanol or 13C-methanol. Similarly, uniformly 2H/15N-labeled proteins were prepared by initially growing the cells using 99% 2H2O-buffered minimal medium, containing 0.2% (w/v) 15N-ammonium chloride and 0.5% (w/v) 2H6-labeled glucose supplemented with 0.1% (w/v) Celtone-DN powder, and then inducing protein production by growing the cells in 99% 2H2O medium, containing 0.5% (v/v) 2H4-labeled methanol as a carbon source, for 8 days, with the daily addition of 0.5% (v/v) 2H4-labeled methanol (Morgan et al, 2004). The culture supernatant was concentrated and subjected to HiTrap DEAE FF (Amersham Biosciences) ion-exchange chromatography, followed by Mono-Q (Amersham Biosciences) ion-exchange chromatography, and HiLoad Superdex 75 pg (Amersham Biosciences) size-exclusion chromatography. Fibrillar collagen was obtained as follows: collagen type II from bovine nasal septum (Sigma) was dissolved in 0.1 M acetic acid, pH 4, and dialyzed against 10 mM phosphate buffer, pH 7. We confirmed that no free triple-helical collagen existed in the supernatant fraction of the NMR sample by an SDS–PAGE analysis. All mutants were generated according to the QuikChange™ (Stratagene) protocol.
NMR spectroscopy
All NMR spectra (0.1–0.4 mM protein in 100 mM NaCl, 10 mM sodium phosphate, pH 7, 95% H2O, 5% D2O) were recorded at 25 or 37°C with a Bruker Avance 500 or Avance 600 spectrometer equipped with a cryogenic probe. The buffer was replaced with 100% D2O for the 2D-NOESY, 13C-NOESY-HSQC, HCCH-TOCSY, and HCCH-COSY spectra, and for the 1H–15N HSQC spectrum for hydrogen–deuterium exchange. Sequential assignments of the backbone resonances of the DDR2-DS domain were achieved using the following triple resonance experiments recorded on a uniformly 15N and 13C-labeled sample in H2O buffer: HNCA, HNCACB, CBCA(CO)NH, HNCO, and HN(CA)CO. Side-chain resonances were assigned on the basis of triple resonance experiments, 15N-TOCSY-HSQC, HBHA(CO)NH, H(CCO)NH, (H)C(CO)NH, HCCH-TOCSY, and HCCH-COSY (Clore and Gronenborn, 1994). All spectra were processed by XWIN-NMR and analyzed by Sparky (Goddard and Kneller, 2004).
In the TCS experiments, the molar ratio of the 2H and 15N-labeled DDR2-DS domain to collagen type II was set to 1:0.3, and the pH was 7, to detect the resonance of the DDR2-DS domain in the free state. The proton content of the solvent was lowered to 20%, to suppress the spin diffusion caused by the exchangeable amide protons in the DDR2-DS domain. The TCS experiments were performed using the pulse scheme, as described (Takahashi et al, 2000). The saturation frequency was set at 1.2 p.p.m., and the maximum radiofrequency amplitude was 0.17 kHz for WURST-2 (the adiabatic factor Q0=1). The measurement time was 24 h, with a relaxation delay of 3.5 s and a saturation time of 0.6 s. To evaluate the effect of the residual aliphatic protons within the DDR2-DS domain, TCS experiments were also carried out under the same conditions as those mentioned above, but without the collagen. All NMR spectra were processed and analyzed with XWIN-NMR/Sparky.
Structure calculation
Approximate interproton distances were obtained from the 2D-NOESY, 13C-NOESY-HSQC, and 15N-NOESY-HSQC spectra described above. The mixing time was 150 ms for all NOESY experiments. A series of 1H–15N HSQC spectra was acquired on a sample freshly dissolved in 2H2O, to identify the slowly exchanging amides. Amides that had not exchanged after 1 h and were located in regions of defined secondary structure, based on the NOE data, were restrained to form HN–CO hydrogen bonds, using the distance restraints of 2.7–3.0 Å for O–N and 1.8–2.0 Å for O–HN, respectively. For structure calculations, 70 restraints were used for 35 backbone hydrogen bonds. The torsion angle φ values were calculated using the JHNHA value, based on the HNHA.
CYANA v. 2.1 was used to compute seven cycles, each with 200 conformers (Herrmann et al, 2002). Input data and structure calculation statistics are summarized in Table I. The accuracy of the NMR models may be assessed based on the criteria for successful structure calculation using the program CYANA, which have been defined by Jee and Guntert (2003). The 20 lowest-energy (total energy) structures were chosen for the final structural ensemble. A Ramachandran plot of the final 20 structures, calculated using PROCHECK v. 3.4.4, showed 73.9, 25.0, 1.0, and 0.0% of the residues in the most favored, additional allowed, generously allowed, and disallowed regions, respectively (Laskowski et al, 1993). The structure coordinates have been deposited in the Protein Data Bank, with the accession number 2Z4F.
SPR assays
SPR measurements were performed using a BIAcore 2000 instrument (Biacore AB). Collagen types II and IV (from human placenta; Sigma) were immobilized on the sensor chip CM5 as described (Sasaki et al, 1997), resulting in a signal of 3500–4500 resonance units. The binding assay was performed in running buffer (100 mM NaCl, 10 mM sodium phosphate, pH 7) at a flow rate of 10 μl min−1, using serial dilutions of the DDR2-DS domain in the 1.2–75 μM range. The dissociation constants (Kd) were obtained from the steady-state curve fitting analysis, using BIAevaluation 3.0 (Biacore AB) and Origin v. 5.0 (Microcal Software). The dissociation rates were also estimated from the simulation analysis using BIAsimulation (Biacore AB).
Acknowledgments
This work was supported by a grant from the Japan New Energy and Industrial Technology Development Organization (NEDO).
References
- Agarwal G, Kovac L, Radziejewski C, Samuelsson SJ (2002) Binding of discoidin domain receptor 2 to collagen I: an atomic force microscopy investigation. Biochemistry 41: 11091–11098 [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Hofmann K, Chiquet-Ehrismann R, Bucher P (1998) The discoidin domain family revisited: new members from prokaryotes and a homology-based fold prediction. Protein Sci 7: 1626–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchet MA, Odom EW, Vasta GR, Amzel LM (2002) A novel fucose recognition fold involved in innate immunity. Nat Struct Biol 9: 628–634 [DOI] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS (1998) Anatomy of hot spots in protein interfaces. J Mol Biol 280: 1–9 [DOI] [PubMed] [Google Scholar]
- Chen JM, Kung CE, Feairheller SH, Brown EM (1991) An energetic evaluation of a ‘Smith' collagen microfibril model. J Protein Chem 10: 535–552 [DOI] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM (1994) Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol 239: 349–363 [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ (2004) Structure of the integrin alpha2beta1-binding collagen peptide. J Mol Biol 335: 1019–1028 [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC (2000) Structural basis of collagen recognition by integrin alpha2beta1. Cell 101: 47–56 [DOI] [PubMed] [Google Scholar]
- Ferri N, Carragher NO, Raines EW (2004) Role of discoidin domain receptors 1 and 2 in human smooth muscle cell-mediated collagen remodeling: potential implications in atherosclerosis and lymphangioleiomyomatosis. Am J Pathol 164: 1575–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, Fujikawa K, Pratt KP (2002) New insights into binding interfaces of coagulation factors V and VIII and their homologues lessons from high resolution crystal structures. Curr Protein Pept Sci 3: 313–339 [DOI] [PubMed] [Google Scholar]
- Gaskell A, Crennell S, Taylor G (1995) The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure 3: 1197–1205 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG (2004) SPARKY 3. San Francisco: University of California [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319: 209–227 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Wang LH, Torres R, Zhao H, Olaso E, Eng FJ, Labrador P, Klein R, Lovett D, Yancopoulos GD, Friedman SL, Lin HC (2002) Discoidin domain receptor 2 interacts with Src and Shc following its activation by type I collagen. J Biol Chem 277: 19206–19212 [DOI] [PubMed] [Google Scholar]
- Ito N, Phillips SE, Yadav KD, Knowles PF (1994) Crystal structure of a free radical enzyme, galactose oxidase. J Mol Biol 238: 794–814 [DOI] [PubMed] [Google Scholar]
- Jee J, Guntert P (2003) Influence of the completeness of chemical shift assignments on NMR structures obtained with automated NOE assignment. J Struct Funct Genomics 4: 179–189 [DOI] [PubMed] [Google Scholar]
- Juliano RL, Haskill S (1993) Signal transduction from the extracellular matrix. J Cell Biol 120: 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadler KE, Holmes DF, Trotter JA, Chapman JA (1996) Collagen fibril formation. Biochem J 316 (Part 1): 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Xu Y, Xu X, Keene DR, Gurusiddappa S, Liang X, Wary KK, Hook M (2005) A novel binding site in collagen type III for integrins alpha1beta1 and alpha2beta1. J Biol Chem 280: 32512–32520 [DOI] [PubMed] [Google Scholar]
- Knight CG, Morton LF, Peachey AR, Tuckwell DS, Farndale RW, Barnes MJ (2000) The collagen-binding A-domains of integrins alpha(1)beta(1) and alpha(2)beta(1) recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J Biol Chem 275: 35–40 [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- Labrador JP, Azcoitia V, Tuckermann J, Lin C, Olaso E, Manes S, Bruckner K, Goergen JL, Lemke G, Yancopoulos G, Angel P, Martinez C, Klein R (2001) The collagen receptor DDR2 regulates proliferation and its elimination leads to dwarfism. EMBO Rep 2: 446–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lee CC, Kreusch A, McMullan D, Ng K, Spraggon G (2003) Crystal structure of the human neuropilin-1 b1 domain. Structure 11: 99–108 [DOI] [PubMed] [Google Scholar]
- Leitinger B (2003) Molecular analysis of collagen binding by the human discoidin domain receptors, DDR1 and DDR2. Identification of collagen binding sites in DDR2. J Biol Chem 278: 16761–16769 [DOI] [PubMed] [Google Scholar]
- Leitinger B, Hohenester E (2007) Mammalian collagen receptors. Matrix Biol 26: 146–155 [DOI] [PubMed] [Google Scholar]
- Leitinger B, Steplewski A, Fertala A (2004) The D2 period of collagen II contains a specific binding site for the human discoidin domain receptor, DDR2. J Mol Biol 344: 993–1003 [DOI] [PubMed] [Google Scholar]
- Macedo-Ribeiro S, Bode W, Huber R, Quinn-Allen MA, Kim SW, Ortel TL, Bourenkov GP, Bartunik HD, Stubbs MT, Kane WH, Fuentes-Prior P (1999) Crystal structures of the membrane-binding C2 domain of human coagulation factor V. Nature 402: 434–439 [DOI] [PubMed] [Google Scholar]
- Morgan WD, Lock MJ, Frenkiel TA, Grainger M, Holder AA (2004) Malaria parasite-inhibitory antibody epitopes on Plasmodium falciparum merozoite surface protein-1(19) mapped by TROSY NMR. Mol Biochem Parasitol 138: 29–36 [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Miyazawa M, Sakakura M, Terasawa H, Takahashi H, Shimada I (2002) Determination of the interface of a large protein complex by transferred cross-saturation measurements. J Mol Biol 318: 245–249 [DOI] [PubMed] [Google Scholar]
- Nishida N, Sumikawa H, Sakakura M, Shimba N, Takahashi H, Terasawa H, Suzuki EI, Shimada I (2003) Collagen-binding mode of vWF-A3 domain determined by a transferred cross-saturation experiment. Nat Struct Biol 10: 53–58 [DOI] [PubMed] [Google Scholar]
- Okuyama K, Wu G, Jiravanichanun N, Hongo C, Noguchi K (2006) Helical twists of collagen model peptides. Biopolymers 84: 421–432 [DOI] [PubMed] [Google Scholar]
- Olaso E, Ikeda K, Eng FJ, Xu L, Wang LH, Lin HC, Friedman SL (2001) DDR2 receptor promotes MMP-2-mediated proliferation and invasion by hepatic stellate cells. J Clin Invest 108: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaso E, Labrador JP, Wang L, Ikeda K, Eng FJ, Klein R, Lovett DH, Lin HC, Friedman SL (2002) Discoidin domain receptor 2 regulates fibroblast proliferation and migration through the extracellular matrix in association with transcriptional activation of matrix metalloproteinase-2. J Biol Chem 277: 3606–3613 [DOI] [PubMed] [Google Scholar]
- Pratt KP, Shen BW, Takeshima K, Davie EW, Fujikawa K, Stoddard BL (1999) Structure of the C2 domain of human factor VIII at 1.5 Å resolution. Nature 402: 439–442 [DOI] [PubMed] [Google Scholar]
- Raynal N, Hamaia SW, Siljander PR, Maddox B, Peachey AR, Fernandez R, Foley LJ, Slatter DA, Jarvis GE, Farndale RW (2006) Use of synthetic peptides to locate novel integrin alpha2beta1-binding motifs in human collagen III. J Biol Chem 281: 3821–3831 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Gohring W, Mann K, Maurer P, Hohenester E, Knauper V, Murphy G, Timpl R (1997) Limited cleavage of extracellular matrix protein BM-40 by matrix metalloproteinases increases its affinity for collagens. J Biol Chem 272: 9237–9243 [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Polak V, Shatsky M, Halperin I, Benyamini H, Barzilai A, Dror O, Haspel N, Nussinov R, Wolfson HJ (2003) Taking geometry to its edge: fast unbound rigid (and hinge-bent) docking. Proteins 52: 107–112 [DOI] [PubMed] [Google Scholar]
- Shimada I (2005) NMR techniques for identifying the interface of a larger protein–protein complex: cross-saturation and transferred cross-saturation experiments. Methods Enzymol 394: 483–506 [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Radziejewski C, Campbell E, Kovac L, McGlynn M, Ryan TE, Davis S, Goldfarb MP, Glass DJ, Lemke G, Yancopoulos GD (1997) An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Mol Cell 1: 25–34 [DOI] [PubMed] [Google Scholar]
- Srivastava A, Quinn-Allen MA, Kim SW, Kane WH, Lentz BR (2001) Soluble phosphatidylserine binds to a single identified site in the C2 domain of human factor Va. Biochemistry 40: 8246–8255 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Nakanishi T, Kami K, Arata Y, Shimada I (2000) A novel NMR method for determining the interfaces of large protein–protein complexes. Nat Struct Biol 7: 220–223 [DOI] [PubMed] [Google Scholar]
- Vander Kooi CW, Jusino MA, Perman B, Neau DB, Bellamy HD, Leahy DJ (2007) Structural basis for ligand and heparin binding to neuropilin B domains. Proc Natl Acad Sci USA 104: 6152–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel W, Gish GD, Alves F, Pawson T (1997) The discoidin domain receptor tyrosine kinases are activated by collagen. Mol Cell 1: 13–23 [DOI] [PubMed] [Google Scholar]
- Vogel WF (2001) Collagen-receptor signaling in health and disease. Eur J Dermatol 11: 506–514 [PubMed] [Google Scholar]
- Vogel WF, Abdulhussein R, Ford CE (2006) Sensing extracellular matrix: an update on discoidin domain receptor function. Cell Signal 18: 1108–1116 [DOI] [PubMed] [Google Scholar]
- Wall SJ, Werner E, Werb Z, DeClerck YA (2005) Discoidin domain receptor 2 mediates tumor cell cycle arrest induced by fibrillar collagen. J Biol Chem 280: 40187–40194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lu H, Liu X, Deng Y, Sun T, Li F, Ji S, Nie X, Yao L (2002) Functional analysis of discoidin domain receptor 2 in synovial fibroblasts in rheumatoid arthritis. J Autoimmun 19: 161–168 [DOI] [PubMed] [Google Scholar]
- Xu L, Peng H, Wu D, Hu K, Goldring MB, Olsen BR, Li Y (2005) Activation of the discoidin domain receptor 2 induces expression of matrix metalloproteinase 13 associated with osteoarthritis in mice. J Biol Chem 280: 548–555 [DOI] [PubMed] [Google Scholar]
- Yang K, Kim JH, Kim HJ, Park IS, Kim IY, Yang BS (2005) Tyrosine 740 phosphorylation of discoidin domain receptor 2 by Src stimulates intramolecular autophosphorylation and Shc signaling complex formation. J Biol Chem 280: 39058–39066 [DOI] [PubMed] [Google Scholar]
- Zong Y, Xu Y, Liang X, Keene DR, Hook A, Gurusiddappa S, Hook M, Narayana SV (2005) A ‘Collagen Hug' model for Staphylococcus aureus CNA binding to collagen. EMBO J 24: 4224–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]