Abstract
Hepatitis B virus (HBV) is a major human pathogen causing about 750 000 deaths per year. The virion consists of a nucleocapsid and an envelope formed by lipids, and three integral membrane proteins. Although we have detailed structural insights into the organization of the HBV core, the arrangement of the envelope in virions and its interaction with the nucleocapsid is elusive. Here we show the ultrastructure of hepatitis B virions purified from patient serum. We identified two morphological phenotypes, which appear as compact and gapped particles with nucleocapsids in distinguishable conformations. The overall structures of these nucleocapsids resemble recombinant cores with two α-helical spikes per asymmetric unit. At the charged tips the spikes are contacted by defined protrusions of the envelope proteins, probably via electrostatic interactions. The HBV envelope in the two morphotypes is to some extent variable, but the surface proteins follow a general packing scheme with up to three surface protein dimers per asymmetric unit. The variability in the structure of the envelope indicates that the nucleocapsid does not firmly constrain the arrangement of the surface proteins, but provides a general template for the packing.
Keywords: capsid-envelope-interaction, cryo-electron microscopy, envelope protein, hepatitis B virus (HBV), virus structure
Introduction
The human hepatitis B virus (HBV) is a remarkably small, enveloped, DNA-containing pararetrovirus causing acute and chronic liver infections (Seeger and Mason, 2000). About 1/3 of the world population have had contact with HBV, and about 2/3 of all liver cancers are attributable to an HBV infection (Chan and Sung, 2006). The unique replication strategy of HBV and related viruses involves the cytoplasmic encapsidation of the HBV pregenomic RNA into a nucleocapsid (core), followed by reverse transcription of this RNA into a partially double-stranded DNA. DNA synthesis is associated with the maturation of the nucleocapsid, triggering the specific interaction with HBV envelope proteins at the ER compartment (Bruss, 2004). This process leads to specific envelopment, budding into the ER and release of the virion from the infected hepatocyte. In contrast to many other enveloped viruses, the secretion of viral envelope proteins is independent of the interaction with mature nucleocapsids and proceeds autonomously (Standring et al, 1986), resulting in a huge excess of empty envelope protein containing shells (subviral particles) in the serum of infected patients.
Most of our structural understanding of the organization of HBV comes from electron microscopy and X-ray crystallography of subviral structures. Escherichia coli-derived core protein (HBc), devoid of any HBV-specific nucleic acid forms large (240 subunits, T=4 arrangement) and small (180 subunits, T=3 arrangement) core particles (Crowther et al, 1994; Böttcher et al, 1997; Conway et al, 1997; Wynne et al, 1999). In virions, mainly the large nucleocapsid particles are found (Roseman et al, 2005; Dryden et al, 2006). The building blocks of the cores are HBc-dimers with a spike in the center that is composed of a four helix bundle to which each monomer contributes two helices. In T=4 particles, dimers occupy two spatially different surroundings called AB-dimer (closest to the five-fold axes) and CD-dimer (closest to the three-fold axes) (Zlotnick et al, 1996).
The envelope of HBV is formed by three different surface proteins (HBs) termed L- (large), M- (middle) and S- (small), with a ratio of approximately 1:1:4 (Heermann et al, 1984). They share the C-terminal S-domain, which contains four predicted transmembrane helices involved in membrane anchorage and dimerization (Seeger and Mason, 2000). The M-protein has a 55-amino-acid-long extension (PreS2) to the S-domain and is not required for envelopment and infectivity. The L-protein has in addition to the PreS2-domain a further N-terminal extension called PreS1 (108 or 119 amino acids long). PreS1 of the L-protein plays a crucial role in envelopment of the capsid (Bruss, 1997) and specific receptor interaction with hepatocytes (Glebe and Urban, 2007). In order to fulfill this dual role, L- adopts two alternate topologies with PreS (PreS1+PreS2) either located at the inside or the outside of the viral membrane (Bruss et al, 1994). Remarkably, structural analysis of the HBV preS1-domain revealed that it is intrinsically unfolded (unpublished data).
It is still a matter of debate how the envelope and the capsid interact in virions. Recent image reconstructions of virions suggested a non-icosahedral packing of HBs in the envelope and failed to identify any specific interactions to the capsid (Dryden et al, 2006). Our work on the structure of infectious virions confirms the variability of HBs packing, but more importantly reveals distinct rules for the packing of HBs in the envelope by showing specific interactions between HBc and HBs.
Results and discussion
Gross architecture of HBV
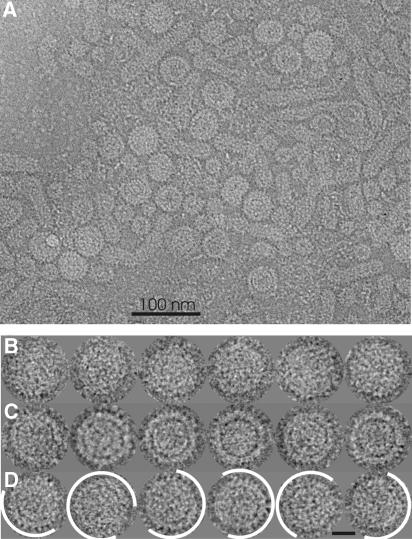
For electron cryo-microscopy we purified viral particles from a high-titer serum of a chronically infected patient by size-exclusion chromatography and isopycnic sucrose density gradient centrifugation. This procedure has been shown to preserve the infectivity of the virions. The final preparation contained virions at a concentration of >5 × 1012 genome equivalents per ml and also filamentous and spherical subviral particles (Figure 1A). The virions could be subdivided into two previously undescribed morphological phenotypes as follows: in compact particles (Figure 1B), the border between the nucleocapsid and the envelope is hardly cognizable in projection. In gapped particles (Figure 1C), the envelope is clearly demarcated from the nucleocapsid by a rim of lower electron density. Gapped and compact particles occurred with similar incidence and could not be separated by their buoyant density (data not shown). Not all particles had a spherical shape, some had filamentous appendices and in some gapped particles the envelope seemed to peel off from the nucleocapsid. For image processing we selected 457 regular compact and 113 gapped particles (Figure 1B and C). With an outer diameter of 45 nm they were indistinguishable in size. This becomes evident when looking at the minor fraction of chimeric particles with a mixture of both phenotypes (Figure 1D). In total, approximately 30% of virions were selected for analysis.
Figure 1.
Micrograph and gallery of HBV embedded in vitrified buffer. (A) Part of a micrograph (inverted contrast) recorded with a Philips CM120 Biotwin. (B) Gallery of selected particles with compact morphology. (C) Gallery of selected particles with gapped morphology. (D) Gallery of particles with mixed morphology. Areas consistent with the gapped morphology are delineated in white. Particles appear bright against darker background. The length of the bar in panel D equals 20 nm.
The averaged centered virions had a radial density distribution (Supplementary Figure 1, dotted lines), with the main maximum at a radius of 13.5 nm, the same radius as the protein shell of recombinant T=4 capsids. Inside the capsid, in close proximity to the protein shell, a density shoulder (radius=10.5 nm) coincided with a density maximum ascribed to partly ordered RNA in recombinant capsids (Crowther et al, 1994). At the outer radii, an asymmetric peak corresponded to the envelope of the virions. In gapped particles the maximum of this peak was shifted toward larger radii (18.9 nm compared with 18.1 nm), indicating a larger distance between the nucleocapsid and the inner surface of the envelope.
3D-image reconstructions were calculated for compact and gapped virions (Figure 2). Plots of the radial density distribution of the 3D-maps (Supplementary Figure 1, solid lines) showed that despite the tight packing of surface proteins (Heermann et al, 1984), the envelope of the virus had the profile of a classical lipid bilayer.
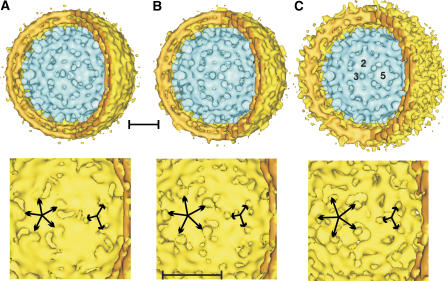
Figure 2.
Surface representations of compact particles (A) A-map and (B) B-map, and gapped particles (C). The A- and B-maps of the compact particles were derived from iterative alignment, supervised classification and reconstruction using two reference maps (see Supplementary data). The A-, B- and C-maps were reconstructed from 114, 106 and 26 particles, respectively. In the upper row a third of the envelope is cut away to reveal the structure of the nucleocapsid inside. For all three maps the Fourier-Shell-correlation dropped to 0.5 at 1/22 Å−1 (see Supplementary Figure 3). For representation the maps were low-pass-filtered to this spatial frequency. The view is centered on a local three-fold axes. In panel C the positions of a strict two-fold, three-fold and five-fold axes are labeled. The lower row shows a view onto the inner surface of the envelope centered at a local three-fold axis. The protrusions around the three-fold (short arrows) and five-fold axes (long arrows) that contact the spikes of the nucleocapsid are marked. The length of the scale bars equals 10 nm. The threshold for calculating the surface representation of the envelope of the gapped particles was adjusted to compensate for the reduced density in this area (see density plots in Supplementary Figure 1).
3D reconstruction and refinement strategy
Initially, 3D-image reconstructions were independently calculated over the whole particle radius for the complete set of regularly shaped gapped and compact virions, respectively. Both types of virions showed the known T=4 architecture of the core shell, but in all 3D-maps the spikes of the nucleocapsid displayed less density and a smaller diameter (see Supplementary Figure 2, lanes 1–3) than expected from image reconstructions of E. coli-derived cores at comparable resolution and similar radii (Crowther et al, 1994). The observed reduced density indicated either wrong orientations or conformational variability in the spike domain. Support for the latter came from our observation that during iterative refinement, reconstructions of the compact particles resulted in split spikes of the CD-dimers. This is consistent with the averaging of particles with spikes in diverse but defined positions. Therefore, we separated different conformations computationally by aligning particle images to projections of two different reference maps (for details, see Supplementary data and Supplementary Figure 2). The particle images were assigned to the one or the other conformation according to the reference map in which the phase residuals of the cross common lines were lower. The resulting two maps, designated A- and B- (Figure 2A and B, EMDB accession IDs: EMD-1400 and EMD-1401), showed spikes that were more solid. Furthermore, the particle images correlated better with the respective references, indicating an overall improvement in the representation of the raw data by the calculated models. Gapped particles were also sorted into two classes following the same strategy as for the compact particles. However, this did not lead to a general improvement (see Supplementary Figure 2) and only one of the two resulting reconstructions showed a good representation of the spikes, whereas in the other map the spikes almost disappeared. Accordingly, only one reference was used in further iterative refinement and only the 25% best-correlating particles were included in the three-dimensional image reconstruction. This led to a clear improvement of the representation of the spikes (C-map; Figure 2C, EMDB accession ID: EMD-1402). However, the density of the envelope was comparatively weak, indicating less order than in compact particles. The reduced peak separation of the lipid bilayer in gapped particles, as well as the smearing of the density towards higher radii, suggested that this lack of order was caused by variability in the distance between the nucleocapsid and the envelope. This variation together with the smaller number of particles resulted in a rather noisy representation of the envelope (Figure 2C).
A-, B- and C-maps showed solid spikes in the nucleocapsid, which were arranged in a T=4 packing, as observed earlier for virion derived capsids (Roseman et al, 2005) and nucleocapsids in virions (Dryden et al, 2006). Major differences between the maps emerged in the position of the dimer axes of the spikes and in the interaction between spikes and envelope (Figure 3). The positions of the centers of the spikes in the B- and C-maps were similar to that in recombinant cores (Crowther et al, 1994; Böttcher et al, 1997; Conway et al, 1997; Wynne et al, 1999), but deviated in the A-map with a clear displacement of the CD-dimers (Figure 3H). The displacement in the A-map resembled the distortions to the spikes induced by conformational stress to the tips of the spikes in recombinant cores (Böttcher et al, 2006). Such stress could occur upon binding of HBs that is tightly arranged in an independently packed lattice. Consistent with this idea is that nucleocapsids without envelope (Roseman et al, 2005) are lacking conformational variability in the spike positions.
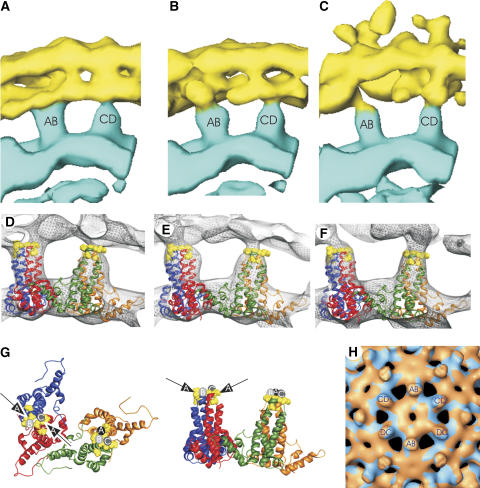
Figure 3.
Spike organization and core envelope contacts. (A) A-map, (B) B-map and (C) C-map show side views of the two types of spikes (labeled as AB and CD spike) in contact to the envelope. The envelope is shown in yellow, the lower part of the nucleocapsid in blue. The radial assignment of color is the same in panels A, B and C. The two dimers in the asymmetric unit (AB-dimer, red, blue; CD-dimer, green, orange, 1QGT; Wynne et al, 1999) fitted into the densities of the A-map (D) and B-map (E) of compact virions and in the C-map (F) of gapped particles. Negatively charged amino acids at the tips of the spikes are shown in yellow. (G) Ribbon diagram of the two dimers in the asymmetric unit (1QGT). The observed contact areas are highlighted by arrows and circles. Contacts in the A-map are colored black, in the B map medium gray and in the C-map light gray. (H) shows a superposition of the nucleocapsid of the A map (blue) and B map (red). In the six spikes closest to the two-fold axis the AB- and CD-dimers are labeled. The change in surface color indicates a relative lateral displacement of the CD-spikes (approximately 4–6 Å). The position of the centers of the AB-spikes in the two maps is identical at the current resolution (less than 3 Å displacement). For spherical slices see Supplementary Figure 3.
The tips of the spikes contacted the envelope via protrusions emerging from the inner surface of the envelope (Figure 2, arrows). This indicated that the contact was formed between core spikes and HBs, rather than between spikes and lipid membrane as suggested earlier (Dryden et al, 2006). The exact positions of the contact area on the spike, the size of the contact area and the length of the protrusion (shortest in the A-map and longest in the C-map) varied between the three reconstructions, which demonstrated a high degree of plasticity of the envelope capsid contact. The variability of the connections became only apparent when the particles were sorted according to their conformational state. However, despite careful sorting, we cannot completely exclude that the reconstructions still include small fractions of particles in mixed conformations similar to the ones shown in Figure 1D. Inclusion of such particles would add noise to the reconstruction. In order to minimize this effect, we included only about 25% of best-correlating particles in the final maps, assuming that particles in mixed conformations would correlate less than particles in uniform conformations.
In the B- and C-maps the nucleocapsid and the inner surface of the envelope were further apart than in the A-map (radial density plot; Supplementary Figures 1 and 3). It is conceivable that these somewhat larger gaps are related to more flexible interactions of the two lattices, which reduced the conformational stress and therefore permitted the relaxed conformation of the nucleocapsid.
The AB-dimers, whose positions remained constant in all three maps, formed the more intimate contacts with the envelope. In the A-map, HBs bound from two sides to the periphery of the tips of the AB-spikes (Figure 3D and G). In the B- and C-maps, the AB-spikes were contacted from only one side (Figure 3E–G). However, the direction from which the spikes were contacted did not superimpose in the three maps. The binding sites to the envelope coincided with high density of negative charges in the core at the tip of the spike (E77, D78) and at the start of the descending helix (D83) (Figure 3D–G, yellow spheres) (Wynne et al, 1999). The residues in the nucleocapsid, which were part of the contact area in all three maps were D78 and P79 in monomer A and D78 in monomer B. The other residues in the contact area with the envelope varied with 6–10 amino acids distributed across both HBc monomers contributing to the interface. Three to five of these residues were negatively charged (Table I).
Table 1.
Amino acids in the core protein involved in envelope contactsa
 |
The CD-dimers were contacted by HBs at a central part of the tips of the spike (Figure 3) with D78 of both HBc monomers being part of the interface in all three maps. The contact area with the envelope was less extensive than in the AB-dimers. Five to seven amino acids of HBc, of which 3–4 were negatively charged, contributed to the interaction site. In the B-and C-map the contact sites were mainly located on the C-monomer whereas in the A-map the contact site was predominantly on the D-monomer.
The different insertion areas and the different dimensions of the contacts between envelope and nucleocapsid suggest an involvement of different binding regions of HBs. This is in agreement with the two reported capsid-binding sites in the PreS-domain (Bruss, 1997; Poisson et al, 1997) and in the first cytosolic loop of the S-domain (Poisson et al, 1997; Löffler-Mary et al, 2000) as well as with the proposed biphasic binding behavior of the L-protein to recombinant cores (Tan et al, 1999). The cytosolic loop contains a cluster of three arginine residues (R73, R78 and R79), which could mediate the interaction with the negative charges at the tips of the spikes. A peptide comprising this part of the cytosolic loop of S- (aa 56–80) competitively inhibits immunoprecipitation of capsids (Poisson et al, 1997), suggesting that this fragment binds to the major antigenic site at the tips of the spikes (aa 78–82; Salfeld et al, 1989), which overlaps with the observed contact site in our maps. There are also several basic amino acid residues in the PreS-domain, among them three arginines (R88, R92 and R102 in subtype ayw). The elimination of the latter two leads to a reduced affinity of the L-protein to recombinant cores, with a substitution of R92 showing the strongest effect (Tan et al, 1999). Furthermore, double point mutations in this region block virion formation (Bruss, 1997). It is also notable that binding of a positively charged peptide to the tips of the spikes (Böttcher et al, 1998) blocks virion assembly through competition with the L-interaction (Dyson and Murray, 1995). These findings together with our structural observations indicate that binding of HBs to the nucleocapsid is mediated by specific types of electrostatic interactions and not by non-specific binding to the lipid membrane that occurs during virion maturation in duck HBV(Mabit and Schaller, 2000).
All image reconstructions of the different virions (A-, B- and C-map) identify the tips of the spikes as the major interface between nucleocapsid and envelope. Interestingly, the tips of the spikes are also the sites where pronounced differences between virion-derived and recombinant cores occur (Roseman et al, 2005), demonstrating the conformational flexibility of this area. Structural changes in this region may alter the charge distribution in such a way that the affinity of the nucleocapsid to the envelope enables control of envelopment. However, the observed contact sites at the tips of the spikes do not match with sites at the base of the spike necessary for envelopment and virion secretion (Ponsel and Bruss, 2003). At these sites we do not observe stable envelope contacts although at the current resolution we cannot decide if the base of the spike is directly involved in the interaction with the envelope. The same mutational screen (Ponsel and Bruss, 2003) also showed that the exchange of a single negatively charged residue at the tips of the spikes is insufficient to interfere with envelopment. This would be consistent with the idea that multiple charged residues contribute with similar efficiency to the electrostatic interaction with HBs, but that not all charges are required at the same time for effective binding. Ponsel and Bruss also tested Asp78, which we identified as being part of the interaction site in all four types of HBc monomers in all three maps. Unfortunately, mutation of Asp78 abolished nucleocapsid formation completely, which left the importance of this residue for envelope binding still elusive. Another important amino acid, which is part of the binding site between HBc and envelope in all three maps, is Pro79 of the A- monomers. Mutation of this residue leads to reduced virus secretion in the mutational screen (Ponsel and Bruss, 2003), which highlights the possible role of Pro79 in envelope binding.
Organization of HBs in the envelope
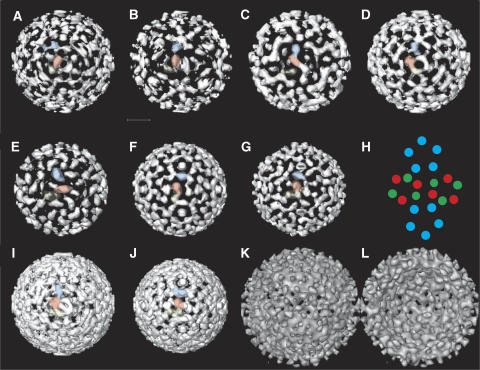
The maps of the compact particles and the map of the gapped particles represent the most common structural arrangement where a majority of contacts in the particles are similar. The total range of variability between individual particles is still not clear. However, even within the same virion, different patterns of interaction can occur (Figure 1D). There are also a few particles with more substantial asymmetry, protrusion or ‘noses'. In order to investigate the packing of HBs in the envelope it is necessary to analyze individual particles. In exceptional cases the icosahedral symmetry enables the calculation of meaningful 3D-volumes from a single particle projection. Therefore, we calculated single particle volumes and classified them according to the pattern in the capsid and the envelope shell (gapped and compact particles separately). We selected single particle volumes, which grouped into classes with the typical arrangement of the spikes in the capsid region. This ensured that further analysis was based on particles with correctly determined orientations and a meaningful single particle volume. From particles of this subset, which grouped again into the same classes in a subsequent classification of the equatorial slices, we calculated new volumes (4–6 particles contributed to each). These volumes were used as references in an iterative refinement process of supervised classification of the particle projections. For analysis of the packing of HBs we focused on the transmembrane area of the envelope, where the four-helix footprint is expected to form compact denser regions in the membrane. We observed in all except one map a comparable overall packing of three denser regions in the asymmetric unit (Figure 4A–D, F and G). The average footprint of these regions was ca. 25 × 45 Å, which is compatible with a HBs-dimer (7–8 transmembrane helices) and the footprint of two adjacent denser regions (ca. 20 × 36 Å) in the membrane part of the small 22 nm HBs-spheres (EMD-1158; Gilbert et al, 2005; Supplementary Figure 6). Therefore, we assume that the observed elongated densities are HBs-dimers. The general packing of HBs-dimers showed 180 elongated densities with five around each 5-fold axis and six around each 3-fold axis (Figure 4H, red and green), of which three were weaker in some reconstructions (Figure 4H, green). Considering that the two spikes in the asymmetric unit provided only two potential binding sites for HBs-dimers it is conceivable that the third HBs-dimer in the asymmetric unit is not bound to the spikes. This is likely to cause a higher lateral mobility, which would be consistent with the observed lower density in the icosahedral reconstructions.
Figure 4.
Organization of HBs in the membrane. (A–D) Transmembrane region of compact particles (EMD-1399, EMD-1403, EMD-1404, EMD 1405); (E–G) transmembrane region of gapped particles (EMD-1406, EMD-1407, EMD-1408). Densities in the asymmetric unit are color-coded: blue, closest to the five-fold axes, red, closest to the two-fold axes and green close to the three-fold axes but weaker in some reconstructions. The common building principle is shown in (H). (I) Superposition of maps (A–D) (compact particles); (J) superposition of maps (E–G) (gapped particles). The superposition shows that without separating the particles according to their HBs-packing, an almost featureless envelope region is observed. (K, L) Envelope region of panel B, which had the lowest phase residuals in cross common lines of the compact maps after refinement. (K) Envelope seen from the outside and (L) envelope seen from the inside (see Supplementary Figure 6 for other envelopes of the other particle reconstructions).
Assuming that the elongated densities corresponded to HBs-dimers, a maximum of 360 HBs-molecules would contribute to the envelope, if all equivalent positions were fully occupied. The transmembrane regions of the HBs-dimers, thus account for about 26–40% of the total area of the membrane region, which is similar to the packing density found in the small HBs-spheres (Gilbert et al, 2005).
Surprisingly, little density is observed in the three-dimensional maps of the compact and gapped particles in the 20 Å-wide-gap between nucleocapsid and envelope (Figure 2; Supplementary Figure 4). However, this gap should not only contain the 120 resolved spikes of the nucleocapsid (aa 63–94), but also the 47 amino acids per cytosolic loop I (33–79, according to assignment in UniProt Q60GL2HBV) and another five amino acids per cytosolic loop II (200–204, according to assignment UniProt Q60GL2HBV) plus the N-terminal domains of the L-HBs with inverted topology (probably one in 12 HBs molecules, given an L:M:S ratio of 1:1:4 and 50% of L-protein molecules bearing preS inside; Heermann et al, 1984; Bruss et al, 1994). This adds up to a total of about 32 000 residues (ca. 3.5 MDa) in the gap. The calculated volume of the gap (width=20 Å; medium radius=165 Å) is only 6.7 × 106 Å3, which could accommodate approximately 5.7 MDa (0.844 Da/Å3). Accordingly, 61% of the gap should be filled with protein (51% of the volume which is not accounted for by spikes should be filled with HBs). Even if these residues were completely disordered, at this high occupancy they should generate a dense diffuse background. However, most of the gap appears surprisingly empty, suggesting that most of the cytosolic loops are probably packed as a thin layer mainly on the inner surface of the envelope. In agreement with this idea, radial density slices of the virions show more density between the tips of the spikes at outer radii (Supplementary Figure 4, especially radius 16.8 nm) than does a map of a capsid without an envelope.
Although most of the volumes analyzed followed the same building pattern, the exact position and orientation of the HBs-dimers varied as seen in the superposition of the maps (Figure 4I and J). In order to test whether there is a defined spatial relation between the arrangement of the surface protein and the lattice of the core protein, we used the envelope region as reference in iterative refinement as suggested by Dryden et al (2006). In contrast to the findings of Dryden and co-workers, we found that in some of the maps the typical arrangement of the capsid was maintained even after three iterations (see Supplementary Figure 5B and F), highlighting the fact that at least the particles in these maps have a specific spatial relation between HBs and HBc. Therefore, the packing of HBs in the envelope is not completely variable, with pentamers inserted at random positions, as suggested (Dryden et al, 2006), but is more likely to be substantially biased toward a common icosahedral building principle. However, this building principle allows for some degree of variability of the orientation of individual HBs in the envelope, which would give rise to none icosahedral components in the particle projections that would add noise to the reconstructions. The comparable size and shape of the transmembrane cross-sections indicated that the majority of HBs in any given particle followed the given icosahedral building principle of its class. In this respect the maps can be regarded as representations of prototypes of particles rather than the representation of the structure of an individual particle.
Considering that the approximate position of the HBs-dimers in the envelope are defined, but not their orientations, the packing of HBs in the envelope appears to be controlled by the binding interaction with the spikes. In agreement with this idea we find that one side of the well-preserved densities (Figure 4, red and blue) attributable to HBs-dimers, is always close to the potential binding site at the tips of a spikes.
The structural basis and the functional implications of the morphological variation between the gapped and compact virions need further clarification. A simple explanation could be a variation in the lipid distribution that may result in envelopes with somewhat larger or smaller radii (Supplementary Figure 1). This leads to variations in the distance between nucleocapsid and envelope, which might be compensated by some plasticity in the binding region of HBs that forms larger and smaller protrusions, but maintains similar interactions with the nucleocapsid (Figure 2). Whether the change in morphology is linked to a change in the topology of the PreS-domain remains unclear. However, if this would be the case, the appearance of partially gapped particles (Figure 1D) would suggest a locally cooperative switching of PreS to the outside. An interesting observation along these lines is that the most infectious material that we purified so far from cell cultures showed almost exclusively gapped particles (data not shown) highlighting the significance of this species.
Materials and methods
Purification of HBV
Viral particles from a 204 ml sample of a high-titer serum donated by a chronic HBV carrier were 34 × concentrated by pelleting through a 20% (w/w) sucrose cushion at 83 000 g in a Beckmann SW28 rotor for 22 h, 10°C. The pellets were resuspended overnight in 20 mM TrisCl (pH 7.4), 140 mM NaCl, 1 mM EDTA under gentle shaking and then subjected to FPLC-directed size-exclusion chromatography (Superose-6 prep grade by Amersham Biosciences, bed height 56 cm, column diameter 26 mm). Fractions showing a peak of viral DNA (18 ml in total) were then centrifuged through a discontinuous sucrose density gradient (2 ml 60% w/w, 4 ml 45% w/w, 4 ml 35% w/w, 5 ml 25% w/w, 5 ml 15% w/w) at 100 000 g in a Beckmann SW28 rotor for 20 h, 10°C. The virion peak fraction contained >5 × 1010 viral genome equivalents per ml, and was further concentrated for EM analysis by about 100-fold in a microcon YM-30 filter device (Millipore).
Electron microscopy
Virus suspensions were frozen with a homemade freezing apparatus. The sample (2 μl) was applied to a perforated carbon grid, which was automatically blotted for 1–2 s from both sides with filter paper (Whatman No. 1). Then the grid was automatically plunged into liquid ethane cooled by a surrounding bath of liquid nitrogen. The ethane was heated to prevent freezing. Grids were transferred to a Philips CM120-Biotwin microscope (LaB6-filament, operated at 100 kV) using a Gatan transfer holder. Micrographs were recorded under low-dose conditions at a calibrated magnification of 50 000. Micrographs (Kodak SO-163) were developed in full-strength D19 for 10 min at room temperature.
Image processing
Micrographs were scanned with a Zeiss-Scai scanner at a pixel size of 21 μm (0.42 nm at specimen level). Particles were selected interactively for a regular, circular outline avoiding damaged particles and particles with an irregular morphology. Gapped particles and compact particles (see Results) were processed separately. Particle orientations were determined using cross common lines against projections of a reference map (Böttcher et al, 1997) (for details see Supplementary data). Reconstructions were computed from particle-transforms multiplied by the contrast transfer function (ctf) determined for each micrograph using ctffind2 (Mindell and Grigorieff, 2003). In the final maps the amplitudes were corrected (divided by (Σ(ctf2)+0.1) to restore proper representation of the density distribution. For iterative refinement of orientations, new reference projections were calculated from the current best map. We used different references for the iterative refinement (see Supplementary data) to test the convergence of the data. For separating different conformations, particles were aligned to multiple sets of reference projections and were grouped into the set with which they showed the lowest phase residuals in cross-common lines. Maps were reconstructed from particles in the same group.
For analyzing the variable organization of the envelope, three-dimensional maps (volumes) were calculated from individual particle images, using the orientations determined in the refinement of the capsid region. Spherical shells of capsid and envelope region were calculated and mounted side by side for classification using IMAGIC V (van Heel et al, 1996). For further processing, only those individual 3D volumes that grouped into classes showing the typical T=4 arrangement of spikes in the capsid were selected. Equatorial slices of these volumes were then further reclassified using IMAGIC V. Particles, which grouped into the same classes in the two classifications steps (typically 4–6 particles) were used to calculate combined three-dimensional particle volumes. These three-dimensional maps then served as the first references in iterative refinement and supervised classification. Supervised classification and determination of orientations was based on phase residuals of cross common lines (see above). For each class, the best new volumes were calculated from the 15 particles with, the lowest phase residuals and the final result from this analysis are presented in Figure 4.
Supplementary Material
Supplementary Information
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Acknowledgments
We are grateful to Ralf Bartenschlager who constantly supported our work. We appreciate Stefanie Held for excellent technical assistance. We also appreciate the fruitful discussions with WH Gerlich, RA Crowther and R Henderson. This work received financial support from the ‘Deutsche Forschungsgemeinschaft' (DFG, UR/72-1-4) to SU.
References
- Böttcher B, Tsuji N, Takahashi H, Dyson MR, Zhao S, Crowther RA, Murray K (1998) Peptides that block hepatitis B virus assembly: analysis by cryomicroscopy, mutagenesis and transfection. EMBO J 17: 6839–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher B, Vogel M, Ploss M, Nassal M (2006) High plasticity of the hepatitis B virus capsid revealed by conformational stress. J Mol Biol 356: 812–822 [DOI] [PubMed] [Google Scholar]
- Böttcher B, Wynne SA, Crowther RA (1997) Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386: 88–91 [DOI] [PubMed] [Google Scholar]
- Bruss V (1997) A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol 71: 9350–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss V (2004) Envelopment of the hepatitis B virus nucleocapsid. Virus Res 106: 199–209 [DOI] [PubMed] [Google Scholar]
- Bruss V, Lu X, Thomssen R, Gerlich WH (1994) Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J 13: 2273–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HL, Sung JJ (2006) Hepatocellular carcinoma and hepatitis B virus. Semin Liver Dis 26: 153–161 [DOI] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC (1997) Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature 386: 91–94 [DOI] [PubMed] [Google Scholar]
- Crowther RA, Kiselev NA, Böttcher B, Berriman JA, Borisova GP, Ose V, Pumpens P (1994) Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77: 943–950 [DOI] [PubMed] [Google Scholar]
- Dryden KA, Wieland SF, Whitten-Bauer C, Gerin JL, Chisari FV, Yeager M (2006) Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol Cell 22: 843–850 [DOI] [PubMed] [Google Scholar]
- Dyson MR, Murray K (1995) Selection of peptide inhibitors of interactions involved in complex protein assemblies: association of the core and surface antigens of hepatitis B virus. Proc Natl Acad Sci USA 92: 2194–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert RJ, Beales L, Blond D, Simon MN, Lin BY, Chisari FV, Stuart DI, Rowlands DJ (2005) Hepatitis B small surface antigen particles are octahedral. Proc Natl Acad Sci USA 102: 14783–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebe D, Urban S (2007) Viral and cellular determinants involved in hepadnaviral entry. World J Gastroenterol 13: 22–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heermann KH, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich WH (1984) Large surface proteins of hepatitis B virus containing the pre-s sequence. J Virol 52: 396–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler-Mary H, Dumortier J, Klentsch-Zimmer C, Prange R (2000) Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology 270: 358–367 [DOI] [PubMed] [Google Scholar]
- Mabit H, Schaller H (2000) Intracellular hepadnavirus nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J Virol 74: 11472–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P (1997) Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology 228: 115–120 [DOI] [PubMed] [Google Scholar]
- Ponsel D, Bruss V (2003) Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol 77: 416–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman AM, Berriman JA, Wynne SA, Butler PJ, Crowther RA (2005) A structural model for maturation of the hepatitis B virus core. Proc Natl Acad Sci USA 102: 15821–15826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salfeld J, Pfaff E, Noah M, Schaller H (1989) Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J Virol 63: 798–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Mason WS (2000) Hepatitis B virus biology. Microbiol Mol Biol Rev 64: 51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standring DN, Ou JH, Rutter WJ (1986) Assembly of viral particles in Xenopus oocytes: presurface-antigens regulate secretion of the hepatitis B viral surface envelope particle. Proc Natl Acad Sci USA 83: 9338–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan WS, Dyson MR, Murray K (1999) Two distinct segments of the hepatitis B virus surface antigen contribute synergistically to its association with the viral core particles. J Mol Biol 286: 797–808 [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- Wynne SA, Crowther RA, Leslie AG (1999) The crystal structure of the human hepatitis B virus capsid. Mol Cell 3: 771–780 [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Cheng N, Conway JF, Booy FP, Steven AC, Stahl SJ, Wingfield PT (1996) Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry 35: 7412–7421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6