Abstract
TSG101 and ALIX both function in HIV budding and in vesicle formation at the multivesicular body (MVB), where they interact with other Endosomal Sorting Complex Required for Transport (ESCRT) pathway factors required for release of viruses and vesicles. Proteomic analyses revealed that ALIX and TSG101/ESCRT-I also bind a series of proteins involved in cytokinesis, including CEP55, CD2AP, ROCK1, and IQGAP1. ALIX and TSG101 concentrate at centrosomes and are then recruited to the midbodies of dividing cells through direct interactions between the central CEP55 ‘hinge' region and GPP-based motifs within TSG101 and ALIX. ESCRT-III and VPS4 proteins are also recruited, indicating that much of the ESCRT pathway localizes to the midbody. Depletion of ALIX and TSG101/ESCRT-I inhibits the abscission step of HeLa cell cytokinesis, as does VPS4 overexpression, confirming a requirement for these proteins in cell division. Furthermore, ALIX point mutants that block CEP55 and CHMP4/ESCRT-III binding also inhibit abscission, indicating that both interactions are essential. These experiments suggest that the ESCRT pathway may be recruited to facilitate analogous membrane fission events during HIV budding, MVB vesicle formation, and the abscission stage of cytokinesis.
Keywords: abscission, CEP55, cytokinesis, ESCRT pathway, midbody
Introduction
TSG101(yeast Vps23p) and ALIX/AIP1(Bro1p) both function at the endosome to help sort membrane proteins into vesicles that bud into the lumen to create multivesicular bodies (MVBs) (Hurley and Emr, 2006; Gill et al, 2007). One important function of this pathway is to target membrane proteins for degradation, which occurs when MVBs fuse with lysosomes and thereby expose the internal vesicles and their contents to the action of lysosomal lipases and hydrolases. TSG101 functions as a subunit of the heterotetrameric ESCRT-I complex (Endosomal Sorting Complex Required for Transport-I), together with three other MVB pathway proteins: VPS28, VPS37, and MVB12 (Chu et al, 2006; Curtiss et al, 2007; Kostelansky et al, 2007; Morita et al, 2007; Oestreich et al, 2007). ALIX also functions in the MVB pathway, where it can interact with several different proteins and complexes, including TSG101/ESCRT-I (Martin-Serrano et al, 2003; Strack et al, 2003; von Schwedler et al, 2003; Odorizzi, 2006). Both TSG101 and ALIX can also bind directly to retroviral Gag proteins, including HIV-1 Gag, and facilitate late stages of virus budding (Demirov and Freed, 2004; Morita and Sundquist, 2004; Bieniasz, 2006). In some contexts, ALIX and TSG101 can substitute for one another in the release of infectious virions, indicating that they can perform similar (or complementary) roles in virus budding (Fisher et al, 2007; Usami et al, 2007).
TSG101/ESCRT-I and ALIX both function together with other ESCRT pathway members, including ESCRT-II, ESCRT-III and VPS4. Although mechanistic details are lacking, current models hold that the ESCRT pathway mediates the protein sorting and membrane fission events required for release of cargo-filled vesicles and viruses. Importantly, both MVB vesicles and retroviruses bud away from (rather than into) the cytoplasm, implying that the cytoplasmic ESCRT machinery may help mediate membrane fission events from inside the neck of the budding virus/vesicle. The family of related ESCRT-III proteins appears to play a particularly critical role in this process, by assembling into membrane-bound lattices associated with sites of vesicle formation (Babst et al, 2002; Lin et al, 2005). Once assembled on the membrane, the ESCRT-III subunits are bound and remodeled by the action of the VPS4 ATPases, which redistributes the ESCRT machinery back into the cytoplasm (Babst et al, 1998).
Another important cellular process in which a thin membrane tubule must be resolved from within is the final abscission step of cytokinesis (reviewed in McCollum, 2005). During abscission, the thin microtubule-filled midbody that connects the dividing cells is severed to release two discrete daughter cells. Abscission is mediated by proteins of the Flemming body, a dense proteinaceous ring that occupies a central position within the midbody. Many midbody proteins have been identified (Skop et al, 2004), and recent studies have revealed that one such protein, CEP55, performs important roles in organizing the Flemming body and in recruiting a series of late-acting protein required for abscission (Fabbro et al, 2005; Martinez-Garay et al, 2006; Zhao et al, 2006).
Cytokinesis proceeds through a series of sequential, but coupled stages that ultimately lead to abscission. Initially, a cleavage furrow is created through constriction of the actomyosin contractile ring. A number of factors help control contractile ring assembly and constriction, including IQGAP1 and Rho-associated kinases such as citron and ROCK1 (reviewed in Machesky, 1998; Matsumura, 2005). The centrosome also appears to play critical roles in helping to regulate the different stages of cytokinesis, and a series of proteins, including centriolin, centrin, and CEP55, concentrate at centrosomes during most of the cell cycle, but then migrate to Flemming bodies during cytokinesis (reviewed in Doxsey et al, 2005). Cleavage furrow ingression eventually creates a thin midbody that connects the dividing cells and the Flemming body forms at the center of the midbody. Some Flemming body components are present throughout furrow ingression, whereas others are delivered late, including cellular membranes, endocytic factors, and secretory vesicles and their associated fusion machinery (McCollum, 2005). Once all of these components are properly assembled and activated, abscission occurs and the two daughters separate completely.
As part of our continuing effort to characterize the functions of human TSG101/ESCRT-I and ALIX, we performed a series of proteomic-style experiments aimed at identifying cellular binding partners. These experiments revealed that both ESCRT-I and ALIX bind a series of proteins that localize to centrosomes and midbodies, and function in cytokinesis. When we began this work, there were already several observations linking the ESCRT pathway with centrosomes and cytokinesis, although the potential implications of these links were not widely appreciated. First, Xie et al (1998) reported that TSG101/ESCRT-I can localize to centrosomes and midbodies, and that TSG101 downregulation leads to mitotic abnormalities. Second, Spitzer et al (2006) demonstrated that elc/tsg101 mutants in Arabidopsis exhibited high levels of multinucleate cells, and the authors suggested that this might reflect a cytokinesis defect arising from misregulation of the microtubule cytoskeleton, although this defect was not characterized further. Third, we reported that EAP20/ESCRT-II also concentrates at centrosomes, but did not characterize a centrosomal function for the ESCRT-II complex (Langelier et al, 2006). Fourth, Jin et al (2005) reported that EAP30/ESCRT-II negatively regulates maturation of the meiotic spindle pole body (centrosome) in Schizosaccharomyces pombe, although the authors did not note that EAP30 is a component of ESCRT-II. Finally, Furukawa and co-workers used affinity purification/mass spectrometry methods to show that CEP55 (called ‘C10orf3') binds both TSG101 and ALIX (called ‘PDCD6IP'), but the role of CEP55 in cytokinesis was not yet known and the authors apparently did not recognize that PDCD6IP corresponds to ALIX (Sakai et al, 2006). Subsequently, while our work was in progress, Carlton and Martin-Serrano (2007) reported that both ESCRT-I and ALIX localize to the Flemming body and are required for abscission in human cells. We have therefore interpreted our observations in the light of their discovery, and proceeded to characterize the roles of ESCRT-I, ALIX, and other ESCRT pathway components in the abscission step of cytokinesis.
Results
Identification of ALIX and ESCRT-I binding partners involved in cytokinesis
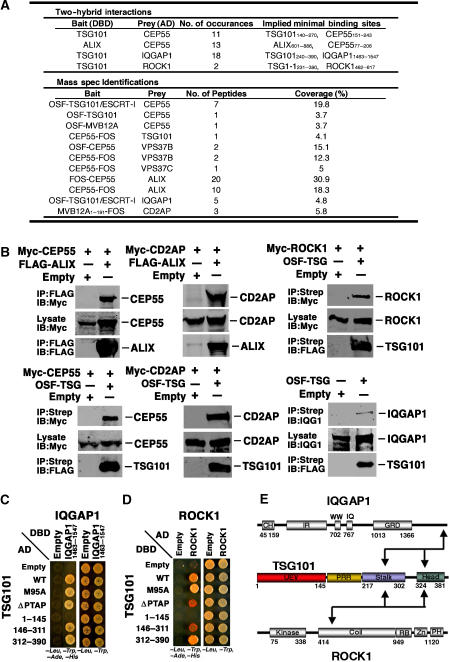
Extensive yeast two-hybrid and One-STrEP-tagged affinity purification/mass spectrometry experiments were performed to identify potential binding partners for ESCRT-I and ALIX. Remarkably, these experiments identified more than 10 proteins previously implicated in centrosome and midbody function. Four of these proteins, CEP55, ROCK1, IQGAP1, and CD2AP, were selected for further study because each (1) localizes to the midbody, (2) functions in cytokinesis/abscission, and (3) was identified in at least two independent proteomics screens (summarized in Figure 1A and caption). Our initial surveys indicated that three of these proteins, ROCK1, IQGAP1, and CD2AP, associated with ESCRT-I, and that CEP55 associated with both ESCRT-I and ALIX. Moreover, CD2AP was recently reported as an ALIX binding partner (Usami et al, 2007), indicating that this protein also binds both ESCRT-I and ALIX.
Figure 1.
Cellular binding partners of ESCRT-I and ALIX. (A) Proteomic identification of ESCRT-I and ALIX binding partners. Upper panel: summarized data from yeast two-hybrid screens of AD prey libraries for DBD-TSG101 and DBD-ALIX binding partners. Binding sites were inferred from the minimal overlapping regions of bait and prey fragments that gave positive interactions. Lower panel: proteins identified as ESCRT-I or ALIX binding partners in affinity purification/mass spectrometric experiments (and absent in control purifications). Bait proteins were expressed as One-STrEP-FLAG (OSF, N-terminal) or FLAG-One-STrEP (FOS, C-terminal) fusions, and OSF-TSG101 was coexpressed with untagged versions of other ESCRT-I subunits (VPS28 and VPS37A-D) in samples labeled ‘TSG101/ESCRT-I'. (B) Cytokinesis proteins co-precipitate with ALIX and ESCRT-I. ‘Bait' FLAG-ALIX and OSF-TSG101 or empty vector controls were tested for co-precipitation with overexpressed Myc-tagged ‘prey' proteins or with endogenous IQGAP1 (lower right). Western blots show (1) prey protein levels in soluble lysates (middle panels, Lysate, IB: anti-Myc), (2) bait proteins bound to anti-FLAG (FLAG-ALIX) or StrepTactin (OSF-TSG/ESCRT-I) matrices (lower panels, IP: anti-FLAG or Strep), or (3) prey proteins co-precipitated onto the matrix (upper panels, IB: anti-Myc or anti-IQGAP1). Note that untagged versions of the three other ESCRT-I subunits (VPS37B, MVB12A, and VPS28) were always coexpressed with OSF-TSG101, and that others have also reported TSG101-CEP55, ALIX-CEP55, and ALIX-CD2AP interactions (Sakai et al, 2006; Usami et al, 2007). (C) Yeast two-hybrid mapping of the TSG101 binding sites for IQGAP11463–1547. TSG101-AD fusions (rows 2–6) or control AD constructs (row 1, Empty) were coexpressed together with IQGAP11463–1547-DBD (column 2) or control DBD constructs (column 1, Empty) and tested for positive yeast two-hybrid interactions (left) or co-transformation (control, right). Note that IQGAP11463-1547 shows positive interactions with both the stalk and head regions of TSG101. (D) Yeast two-hybrid mapping of the TSG101 binding sites for ROCK1. The experiment is analogous to that in panel C, except that TSG101 constructs were expressed as DBD fusions and ROCK1 was expressed as an AD fusion. Note that ROCK1 shows positive interactions with both the stalk and core regions of TSG101. (E) Summary of TSG101 interactions with IQGAP1 and ROCK1. Domain abbreviations: CH, Calponin Homology; IR, IQGAP-Specific Repeat; GRD, GAP-Related Domain; UEV, Ubiquitin E2 Variant; PRR, Proline-Rich Region; Coil, predicted coiled-coil region; RB, Rho-Binding region; Zn, Zinc finger; PH, Plexstrin Homology Domain.
Five of the six relevant TSG101/ESCRT-I and ALIX interactions were initially verified by demonstrating that Myc-tagged candidate proteins bound immobilized FLAG-ALIX or OSF-TSG101/ESCRT-I but not control matrices (Figure 1B). The sixth interaction, IQGAP1-TSG101/ESCRT-I, could not be verified in this manner because Myc-IQGAP1 exhibited high background binding to the control matrix. In this case, preferential co-immunoprecipitation of endogenous IQGAP1 by OSF-TSG101/ESCRT-I was documented, albeit at levels only modestly above background. Finally, because the two CD2AP interactions were underrepresented in our initial proteomic screens, we further verified that CD2AP-ALIX and CD2AP-ESCRT-I co-immunoprecipitation reactions were robust and specific in both directions (Supplementary Figures S1A and B). Taken together, these experiments demonstrate that TSG101/ESCRT-I and ALIX interact with a series of proteins that function in cytokinesis.
ROCK1 and IQGAP1 interact with the TSG101/ESCRT-I PRR/stalk and headpiece
ROCK1 and IQGAP1 have both been implicated in contractile ring assembly and activation (Machesky, 1998; Matsumura, 2005), and may therefore belong to a similar functional class of ESCRT-I binding proteins. As shown in Figures 1C and D, both ROCK1 and IQGAP11465–1547 exhibited positive two-hybrid interactions with TSG101, and these assays were used to map their TSG101 binding requirements. TSG101 constructs were designed to span the N-terminal UEV domain (TSG1–145), the central proline-rich (PRR) and ‘stalk' regions (TSG101146–311), and the C-terminal ‘headpiece' (TSG101312–390) (Kostelansky et al, 2007), and also to test requirements for the TSG101 PTAP element (ΔPTAP) and PTAP binding activity (M95A) (Pornillos et al, 2002; Kostelansky et al, 2007). ROCK1 and IQGAP11465–1547 associated specifically with both the TSG101 PRR/stalk and headpiece, indicating that both proteins must make multiple contacts with TSG101/ESCRT-I. The TSG101 stalk and headpiece make a complex series of interactions with other ESCRT-I subunits (Kostelansky et al, 2007; Morita et al, 2007), and we therefore did not attempt to map the IQGAP1 and ROCK1 binding sites more precisely. Nevertheless, our experiments show that both IQGAP1 and ROCK1 associate with the core of TSG101/ESCRT-I (Figure 1E).
ALIX and TSG101 bind CEP55
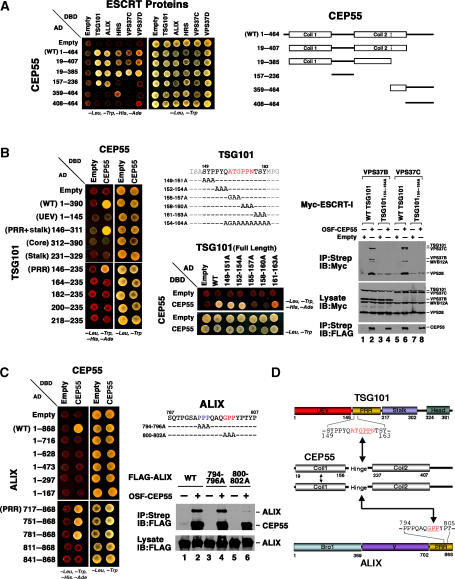
We focused our studies on CEP55 because our initial proteomics screens indicated that this protein bound both TSG101 and ALIX, and because CEP55 localizes to Flemming bodies and is required for abscission (Fabbro et al, 2005; Martinez-Garay et al, 2006; Zhao et al, 2006). The specificity and reciprocity of the CEP55–ESCRT-I and CEP55–ALIX interactions were confirmed in a series of co-immunoprecipitation and directed yeast two-hybrid experiments, which established that (1) OSF-CEP55 specifically co-immunoprecipitated endogenous TSG101 and ALIX (Supplementary Figure S2A), (2) OSF-CEP55 co-immunoprecipitated the entire ESCRT-I complex (not just TSG101) (Supplementary Figure S2B), and (3) CEP55 exhibited yeast two-hybrid interactions with several additional TSG101 binding proteins in the MVB pathway proteins, including HRS, VPS37C, and VPS37D (Figure 2A).
Figure 2.
CEP55 interactions with ESCRT-I and ALIX. (A) Yeast two-hybrid identification of interactions between CEP55 and proteins of the ESCRT pathway. CEP55-AD fusions or control AD constructs (Empty) were coexpressed together with TSG101-, ALIX-, HRS-, VPS37C- or VPS37D-DBD fusions, or control DBD constructs (Empty), and tested for positive yeast two-hybrid interactions (left) or co-transformation (control, right). CEP55 did not interact with other human ESCRT proteins in this assay (not shown). The different CEP55 constructs are summarized on the right. (B) Mapping of the CEP55 binding site on TSG101. Left panel: yeast two-hybrid mapping experiments showing that CEP55-DBD constructs bind the proline-rich region (PRR) of TSG101-AD, and that binding requires TSG101 residues 146–163. Middle panels: further yeast two-hybrid mapping experiments showing that the CEP55 binding site maps to TSG101 residues 155–163. Right panels: co-precipitation experiments confirming that OSF-CEP55 co-precipitates wild-type (WT) ESCRT-I complexes that contain either VPS37B or VPS37C subunits, but not analogous ESCRT-I complexes with a mutant TSG101 binding site (TSG101154–164A). Note that Myc-tagged versions of all four ESCRT-I subunits were coexpressed in these experiments. (C) Mapping of the CEP55 binding site on ALIX. Left panel: yeast two-hybrid mapping experiments showing that CEP55-DBD constructs bind the proline-rich region (PRR) of ALIX-AD, and that binding requires ALIX residues 781–810. Middle panels: co-precipitation experiments showing that OSF-CEP55 specifically co-precipitates WT FLAG-ALIX and FLAG-ALIX794–796A (control mutant), but not the binding site mutant FLAG-ALIX800–802A. (D) Summary of CEP55 interactions with TSG101, ALIX, and itself. Mapped binding residues are highlighted in red and the GPP motifs within the two CEP55 binding sites are underlined.
CEP55 contains two predicted coiled-coil ‘arms' separated by a ‘hinge' (Figure 2A) and the N-terminal arm region reportedly mediates homo-oligomerization (Martinez-Garay et al, 2006; Zhao et al, 2006). Consistent with these observations, we found that two-hybrid constructs corresponding to full-length CEP55 or to just the two arms and the central hinge (CEP5519–385) self-associated in two-hybrid assays (Supplementary Figure S3). Importantly, both ALIX and TSG101 bound the CEP55 hinge alone (Figure 2A), although enhanced ALIX binding was observed for fragments that included arm 1 (Figure 2A and data not shown). In contrast, strong HRS, VPS37C, and VPS37D binding was only detected for longer CEP55 fragments that spanned both arms and the hinge.
Two-hybrid and co-immunoprecipitation assays were used to map the CEP55 binding sites on TSG101/ESCRT-I and ALIX (Figures 2B–D). As shown in Figure 2B, CEP55 bound central fragments of TSG101, including the proline-rich region (left panel, PRR, TSG101146–235). Alanine scanning mutations through the TSG101 PRR revealed that the CEP55 binding site spanned TSG101 residues 155–163, because binding was eliminated by the 158PPN160/AAA mutation and reduced by the adjacent 157ATG157/GAA and 161TSY163/AAA mutations (Figure 2B, middle panels). This CEP55 binding site was confirmed in co-immunoprecipitation experiments showing that Ala/Gly mutations across the TSG101 The 154QATGPPNTSYM164 sequence (TSG101154–164A; see Figure 2B) abrogated co-immunoprecipitation of OSF-CEP55 with Myc-tagged ESCRT-I complexes (Figure 2B, right panels, compare lanes 2 and 6 to lanes 4 and 8). These data are in excellent agreement with those of Carlton and Martin-Serrano (2007) mapping the CEP55 binding site to TSG101 residues 158–162.
Analogous yeast two-hybrid and co-immunoprecipitation experiments were used to characterize and map the CEP55–ALIX interaction (Figure 2C). Two-hybrid mapping experiments indicated that the hinge region of CEP55 bound ALIX residues 781–810 (Figures 2A and C, lower left panel). ALIX constructs containing mutations that spanned each of the two Pro–Pro tracts in this region were tested for CEP55 binding, which revealed that the ALIX 800GPP802/AAA mutation specifically abrogated CEP55 binding (Figure 2C, right panels). Hence, the CEP55 binding sites on both TSG101 and ALIX were centered about ‘GPP' sequence elements, suggesting that the CEP55 hinge may recognize a common core sequence motif in both proteins. TSG101 and ALIX mutants that lacked CEP55 binding activities still supported HIV-1 budding, however, arguing against a role for CEP55 in virus release (Supplementary data and Supplementary Figure S4).
ALIX and ESCRT-I colocalize with CEP55 at centrosomes and Flemming bodies
We next examined the possibility that ALIX and ESCRT-I might function together with CEP55 in the abscission step of cytokinesis. CEP55 concentrates at centrosomes during the G2/M phase of the cell cycle, and then migrates to the midbody to function in abscission during cytokinesis (Fabbro et al, 2005; Martinez-Garay et al, 2006; Zhao et al, 2006). We therefore examined the localization of tagged ALIX and TSG101/ESCRT-I at different stages of the HeLa cell cycle (Figure 3). In non-dividing cells, exogenous ALIX and ESCRT-I were both distributed throughout the cytoplasm, including on endosomal membranes (Welsch et al, 2006). Strikingly, however, both ALIX and TSG101 were also highly enriched together with CEP55 on centrosomes whenever these structures were clearly visible (Figure 3B, rows 2 and 3) (Xie et al, 1998). Centrosomal localization was also confirmed for the endogenous ALIX protein (Supplementary Figure S5, lower row). Thus, both ALIX and ESCRT-I concentrate at the centrosomes of non-dividing cells.
Figure 3.
CEP55, ALIX, and TSG101/ESCRT-I colocalize at centrosomes and Flemming bodies. (A) Triple labeled immunofluorescence and DIC images showing that FLAG-CEP55 (0.2 μg DNA), GFP-TSG101/ESCRT-I (0.2 μg GFP-TSG101, VPS28, VPS37B, and MVB12A DNA), or GFP-ALIX (0.5 μg DNA) colocalize at the midbodies (arrowheads) of dividing HeLa cells. Microtubule staining (white, α-Tubulin) is also shown for reference in columns 1 and 5. Expanded and merged views of the midbodies from the lower pair of cells in each image are shown in column 5. (B) Double labeled immunofluorescence and DIC images showing that α-Tubulin, FLAG-CEP55, or CEP55-Myc (0.2 or 0.5 μg DNA), Myc-TSG101/ESCRT-I (0.2 μg Myc-TSG101, VPS28, VPS37B, and MVB12A DNA), and FLAG-ALIX (0.5 μg DNA) colocalize at the centrosomes of non-dividing cells (arrowheads). (C) Triple-labeled immunofluorescence and DIC images showing that GFP-CEP55 (0.5 μg DNA) localizes to midbodies (arrowheads) whereas OSF-TSG101/ESCRT-I (0.2 μg GFP-TSG101154–164A, VPS28, VPS37B, and MVB12A DNA) and FLAG-ALIX800-802A (0.5 μg DNA) proteins that cannot bind CEP55 are not recruited to midbodies. Microtubule staining (white, α-Tubulin) is also shown for reference.
The distributions of TSG101/ESCRT-I and ALIX in dividing cells were even more striking, as both tagged proteins and endogenous ALIX localized to Flemming bodies, together with CEP55 (Figure 3A; Supplementary Figure S5, upper row). Importantly, however, TSG101/ESCRT-I and ALIX mutants that were unable to bind CEP55 failed to localize to Flemming bodies, although these mutants did not affect the midbody localization of CEP55 (Figure 3C). Similarly, siRNA depletion of endogenous CEP55 blocked the accumulation of ALIX and TSG101/ESCRT-I at Flemming bodies (Supplementary Figures S6A and B, and S7). In contrast, TSG101 depletion did not affect CEP55 localization, and ALIX depletion did not affect CEP55 or TSG101 localization (Supplementary Figures S6C–E). Hence, CEP55 appears to recruit ALIX and TSG101/ESCRT-I to Flemming bodies, where all three proteins are positioned to function together in abscission.
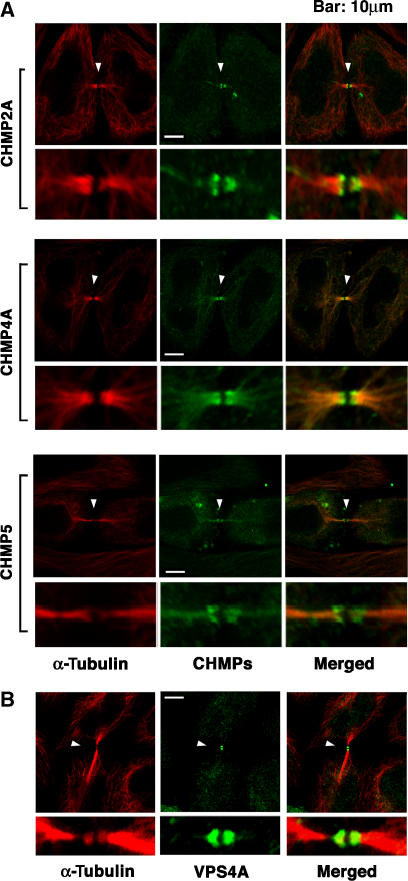
CHMP/ESCRT-III and VPS4 concentrate at midbodies
Both ESCRT-I and ALIX function together with other ESCRT factors, particularly the ESCRT-III and VPS4 proteins, to facilitate the vesiculation events required for virus budding and MVB biogenesis (Hurley and Emr, 2006; Gill et al, 2007). We therefore tested whether representative proteins from three of the seven different ESCRT-III families were also recruited to the Flemming bodies during cytokinesis. As shown in Figure 4A, C-terminally FLAG-tagged CHMP2A, CHMP4A, and CHMP5 proteins all localized to the midbodies of dividing cells. Moreover, all three proteins were localized into two distinct rings, one on either side of the Flemming body. Hence, ESCRT-III proteins were also recruited to midbodies where they formed distinctive double ring assemblies.
Figure 4.
ESCRT-III Proteins and VPS4A Concentrate at Flemming bodies. (A) Double labeled immunofluorescence images showing that CHMP2A-FLAG, CHMP4A-FLAG, and CHMP5-FLAG (each 0.5 μg DNA) concentrate in double ring structures (arrowheads) at the midbodies of dividing HeLa cells. Expanded views are shown below each image. Note that these ESCRT-III constructs were employed because they exhibited no (CHMP2A-FLAG and CHMP4A-FLAG) or minimal (CHMP5-FLAG) effects on HIV budding (von Schwedler et al, 2003). Microtubules were also stained for reference (red, α-Tubulin). (B) Double-labeled immunofluorescence images showing that endogenous VPS4A concentrates in double ring structures (arrowheads) within very thin midbodies of dividing HeLa cells. Microtubules were also stained (red, α-Tubulin). Examples of wider midbodies that lacked VPS4A staining are shown in Supplementary Figure S8A, and VPS4 localization is quantified in Supplementary Figure S8B.
The localization of VPS4A was also examined, and we found that endogenous VPS4A also formed double-ring structures at the midbodies (Figure 4B). In this case, however, VPS4A localization was only observed when the midbodies were particularly thin, suggesting that VPS4A may only be recruited at the final stages of cytokinesis (see Supplementary Figure S8A). Quantitative analyses revealed that endogenous VPS4A was present at 36% of identifiable midbodies (Supplementary Figure S8B). The efficiency of midbody localization was not affected by TSG101 depletion (35%), but was reduced by ALIX depletion (22%) and eliminated by CEP55 depletion (3%), indicating that ALIX contributes to VPS4 midbody localization and CEP55 is required. Taken together, these experiments demonstrate that many different ESCRT pathway members, including ALIX, ESCRT-I, ESCRT-III subunits, and VPS4A, are recruited to midbodies where they could all function in abscission.
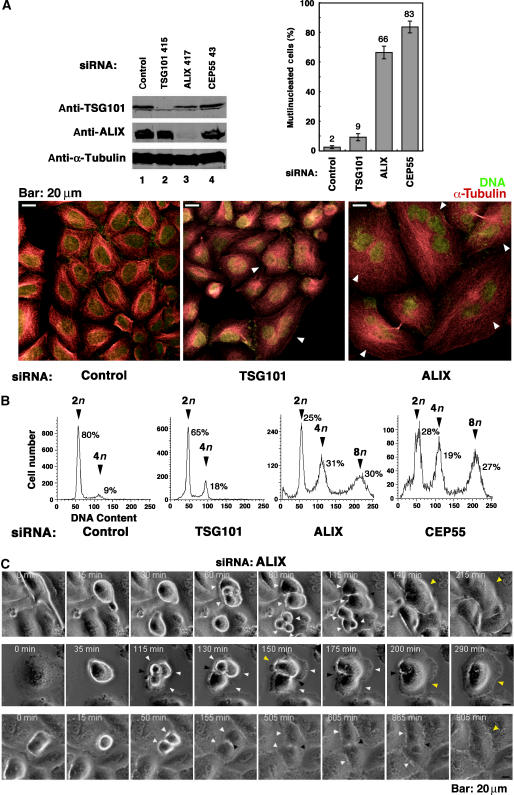
ALIX and ESCRT-I are required for efficient cytokinesis
To test whether TSG101/ESCRT-I and ALIX are actually required for abscission, we examined cell division in cells depleted of endogenous TSG101 and ALIX (Figure 5). Replicating HeLa cells depleted of TSG101 appeared relatively normal, albeit with a moderate enrichment of multinuclear cells (Figure 5A, lower central panel). In contrast, cells depleted of ALIX were highly aberrant, with most of the cells exhibiting multiple nuclei and/or unusual intercellular connections that appeared to be the remnants of arrested midbodies (Figure 5A, lower right panel). These observations were quantified both by counting multinuclear cells (Figure 5A, upper right) and by using both FACS analyses to measure the relative numbers of cells with 2n, 4n and 8n DNA contents (Figure 5B). As shown in Figure 5B, 9% of HeLa cells treated with an irrelevant control siRNA had 4n DNA contents, and cells with 8n DNA contents were essentially undetectable (<1%). TSG101 depletion doubled the number of cells with a 4n DNA content (18%), but did not detectably increase the numbers of cells with 8n DNA contents. More strikingly, ALIX depletion increased the percentages of cells with 4n and 8n DNA contents to 31 and 30%, respectively. Similarly, direct microscopic quantification also showed that TSG101 depletion modestly increased the percentage of multinuclear cells (from 2 to 9%), whereas ALIX depletion dramatically increased the percentage of multinuclear cells (to 66%). As expected, CEP55 depletion also resulted in a very high percentage of multinuclear cells in both assays, consistent with previous reports that CEP55 performs an important function in cytokinesis (Fabbro et al, 2005; Martinez-Garay et al, 2006; Zhao et al, 2006).
Figure 5.
ALIX and ESCRT-I are required for efficient cytokinesis and abscission. (A) Effects of siRNA depletion of TSG101 and ALIX on cell division. Upper left panel: Western blot showing efficient siRNA depletion of endogenous TSG101 (lane 2) and ALIX (lane 3). Controls show protein levels in HeLa cells treated with an irrelevant siRNA against luciferase (lane 1) and cells depleted of CEP55 (lane 4). Numbering denotes the first nucleotide of the siRNA target site within the gene. α-Tubulin loading controls are shown below. Lower panels: immunofluorescent images of control-treated (left), TSG101-depleted (middle), and ALIX-depleted (right) HeLa cells. Microtubules (red, α-Tubulin) and nuclei (SYTOX green) were stained for reference, and arrowheads highlight multinuclear cells. Upper right panel: quantification of multinucleated HeLa cells following depletion of TSG101, ALIX or CEP55, or treatment with the control siRNA. Measurements were performed in triplicate (n=200 cells each) and error bars denote standard deviations. (B) FACS analyses of the DNA content of cells depleted of TSG101, ALIX, CEP55, or treated with a control siRNA. Peaks corresponding to 2n, 4n, and 8n DNA contents are labeled, and the integrated peak volumes are provided. (C) ALIX is required for successful completion of cytokinesis and abscission. Time-lapse phase-contrast imaging of cell division following ALIX depletion. White arrowheads highlight cells that are starting to divide, black arrowheads highlight arrested midbody structures, and yellow arrowheads highlight cells that have recoalesced to produce multinucleated cells following aborted cytokinesis. The figure illustrates that cells depleted of ALIX either abort cleavage furrow formation at an early stage of cell division to form a multinucleated cell (row 1), or arrest late in cytokinesis and remain tethered via thin membranes before eventually recoalescing to form multinucleated cells (rows 2 and 3). Elapsed times are provided in each panel. Movies showing time-lapse images of the cytokinesis blocks induced by ALIX depletion are provided in Supplementary Figure S9.
Time-lapse images of ALIX-depleted cells were also collected to confirm that the dramatic increases in the number of multinucleated cells resulted from defects in cytokinesis. Previous studies have shown that cytokinesis defects can induce (1) ‘early' defects in cell division, where the invaginating cleavage furrow retracts quickly, thereby creating a single cell with two nuclei or (2) ‘late' defects, where the furrow closes but the daughters fail to undergo abscission and remain linked through stable midbodies until they again eventually recoalesce into a single cell with two nuclei (Wheatley and Wang, 1996; Canman et al, 2000; Straight and Field, 2000). Both of these phenotypes were observed frequently, and for three different siRNA constructs against ALIX (Figure 5C and data not shown). Examples of ‘early' cytokinesis defects are shown in the top panel of Figure 5C, and in the corresponding movie provided as Supplementary Figure S9B. In this case, the cleavage furrow in the featured cell began to form (80 min), but then rapidly reversed (115 min) to create a single cell with two nuclei (140 min). Examples of dividing cells arrested late in cytokinesis are shown in the lower two panels of Figure 5C, and in the corresponding movies provided as Supplementary Figures S9C and D. The second example was particularly striking because the cleavage furrow formed early (50 min), but the cells remained connected through an unresolved midbody for hours (50–865 min) until finally recoalescing into a single cell with two nuclei (905 min).
ALIX interactions required for cytokinesis
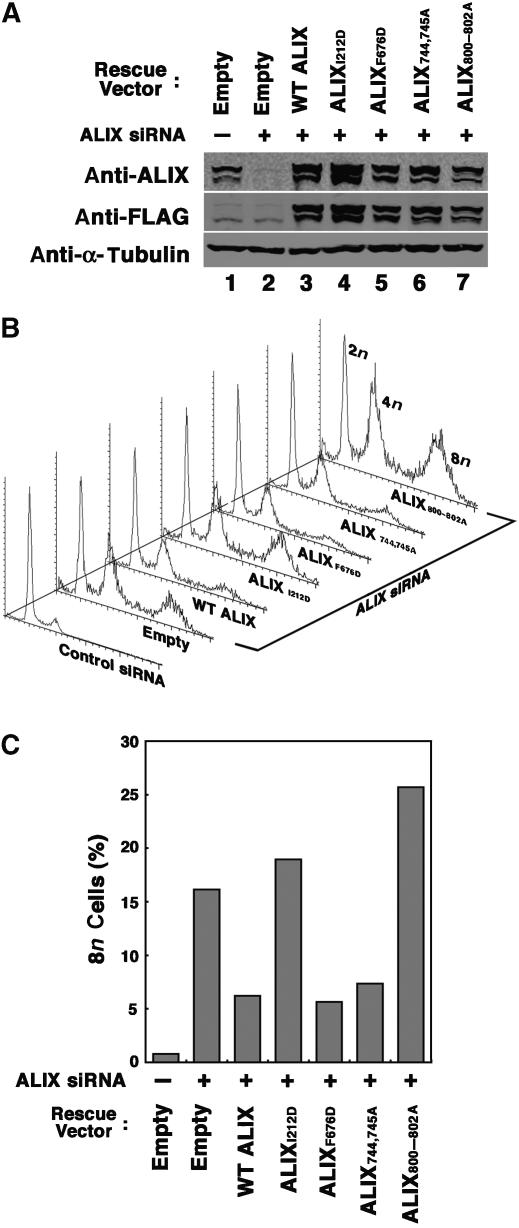
The specificity of the ALIX depletion phenotype was confirmed in experiments that tested the ability of siRNA-resistant ALIX expression constructs to rescue the cytokinesis defects induced by depletion of endogenous ALIX (Figure 6). As expected, ALIX depletion again induced substantial accumulation of HeLa cells with 8n DNA content (16%) as compared with cells treated with an siRNA control (<1%). However, this defect was corrected to a significant extent by re-expression of the WT ALIX protein from an siRNA-resistant construct (6%). We believe that our inability to rescue the replication phenotype completely stems from the technical challenges of simultaneously obtaining very high transfection efficiencies for both the siRNA and the ALIX constructs, avoiding cellular toxicity, and obtaining comparable expression levels for the endogenous and exogenous ALIX proteins. Nevertheless, rescue of the cytokinesis defect was significant and reproducible (n=11), demonstrating that the cytokinesis defects were caused by ALIX depletion and not by off target or other nonspecific effects.
Figure 6.
Requirements for different ALIX interactions in cytokinesis. (A) Western blot showing ALIX depletion/re-expression. Upper panel: ALIX levels (anti-ALIX detection) in cells depleted of endogenous ALIX (lanes 2–7) or treated with a control siRNA (lane 1). Cells in lanes 3–7 were cotransfected with vectors expressing WT FLAG-ALIX (lane 3), or the designated FLAG-ALIX mutants. Middle panel: same samples probed with an anti-FLAG antibody. Lower panel: α-Tubulin loading controls. (B) Stacked FACS profiles showing the effects of ALIX depletion/re-expression on cell polyploidy. Samples are the same as in panel A. (C) Percentages of cells with 8n contents following ALIX depletion/re-expression. The plot shows the relative integrated 8n peak volumes in the FACS profiles from panel B. Note that although siRNA-resistant WT ALIX did not fully rescue the polyploid phenotype, the partial rescue phenotypes shown here were highly reproducible (n=11).
ALIX interacts with a series of different proteins, including CEP55, CHMP4/ESCRT-III, CD2AP, and viral late domains of the YPXL class, and we therefore tested whether mutations that blocked these interactions also inhibited the ability of ALIX to support cytokinesis (Fisher et al, 2007). As shown in Figures 6B and C, ALIX mutants that could not bind CEP55 (ALIX800–802A) or CHMP4/ESCRT-III (ALIXI212D) also failed to rescue the cytokinesis defects in cells that lacked endogenous ALIX. Indeed, these mutant ALIX proteins reproducibly enhanced the cytokinesis block even further, perhaps because they function as dominant inhibitors. In contrast, ALIX proteins with inactivating mutations in the YPXL (ALIXF676D) or the CD2AP (ALIX744,745A) binding sites supported cytokinesis. Thus, ALIX must not with both CEP55 and CHMP4/ESCRT-III to function in cytokinesis, but not with CD2AP nor with an endogenous YPXL-containing protein.
VPS4 overexpression inhibits cytokinesis
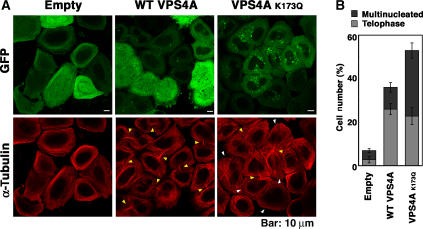
Inhibition of VPS4 ATPase activity induces the formation of enlarged, aberrant endosomal structures called Class E compartments. These structures trap the ESCRT machinery on their surfaces (e.g., see Supplementary Figure S10), reducing their availability to participate in other functions. As shown in Figure 7, expression of the dominant-negative VPS4K173Q-GFP construct resulted in a dramatic increase in the percentage of cells that were arrested in telophase and that had multiple nuclei. Indeed, even overexpression of WT VPS4-GFP, which induces a mild Class E phenotype, substantially increased the percentage of cells arrested (or delayed) in telophase. We therefore conclude that normal levels of VPS4 ATPase activity are also required for efficient cytokinesis. Carlton and Martin-Serrano (2007) similarly reported that a dominant-negative VPS4A construct moderately inhibited cytokinesis, and the more dramatic effects seen in our experiments may simply reflect differences in protein expression levels.
Figure 7.
VPS4 protein overexpression inhibits cytokinesis. (A) Double labeled immunofluorescence images showing cells expressing VPS4-GFP, VPS4AK173Q-GFP, or GFP (top row), and co-stained for microtubules (α-Tubulin, red, bottom row). Examples of multinuclear cells and cells in telophase are indicated by yellow and white arrowheads, respectively. (B) Percentages of cells with multiple nuclei (dark bars) or in telophase (light bars). Three fields of 200 cells each were counted, and error bars denote standard deviations.
Discussion
Our experiments confirm and extend the remarkable discovery of Carlton and Martin-Serrano (2007) that ESCRT-I and ALIX localize to Flemming bodies, are required for efficient abscission, and can also influence the efficiency of furrow ingression and midbody formation. Specifically, we have shown that (1) TSG101/ESCRT-I and ALIX interact with at least four other proteins that function in cytokinesis (CEP55, CD2AP, ROCK1, and IQGAP1), (2) TSG101/ESCRT-I and ALIX concentrate at Flemming bodies during the final stages of cytokinesis, and (3) depletion of ALIX and (to a lesser extent) ESCRT-I inhibits both early and late stages of cytokinesis. Thus, ESCRT-I and ALIX are required for (at least) three important membrane fission processes: MVB biogenesis, virus budding, and cytokinesis.
The less penetrant cytokinesis phenotypes seen for TSG101/ESCRT-I versus ALIX could, in principle, reflect incomplete depletion of TSG101. We disfavor this explanation, however, because TSG101 depletion was efficient and reduced HIV-1 budding 50-fold or more (Figure 5A; Supplementary Figure S4A). Hence, it is more likely that TSG101/ESCRT-I is simply less essential than ALIX for HeLa cell cytokinesis. This situation is similar to the marked differences in the degree to which different lentiviruses rely on TSG101 versus ALIX for viral budding (e.g., see Fisher et al, 2007).
Importantly, our experiments also indicate that most of the known ESCRT pathway is likely to function in abscission. The following observations are in support of this idea: (1) CEP55 exhibits two-hybrid interactions with multiple ESCRT pathway components, including HRS/ESCRT-0 and TSG101/ESCRT-I, VPS37/ESCRT-I, and ALIX, (2) three different ESCRT-III subunits and VPS4A are all recruited to Flemming bodies, (3) dominant inhibitory ESCRT-III and VPS4 constructs inhibit cytokinesis (Carlton and Martin-Serrano, 2007, and this work), and (4) an ALIX mutant that cannot recruit the CHMP4 class of ESCRT-III proteins fails to support cytokinesis (Figure 6). The role of ESCRT-II in cytokinesis remains an open question, however, because ESCRT-II subunits localize to centrosomes (Jin et al, 2005; Langelier et al, 2006), but we observed only very minor cytokinesis defects upon depletion of the ESCRT-II subunit EAP20 (data not shown).
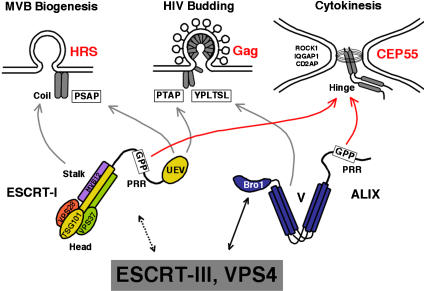
As illustrated in Figure 8, distinct adaptor proteins recruit the ESCRT machinery to the different membrane sites for MVB vesicle formation (e.g., HRS and its binding partners), virus budding (e.g., HIV-1 Gag), and cytokinesis (CEP55). Importantly, CEP55 is not generally required for ESCRT pathway function, since the protein is dispensable for virus budding (Supplementary Figure S4). However, CEP55 recruits both TSG101/ESCRT-I and ALIX to midbodies by binding to GPP-based sequence motifs within the proline-rich regions of both ALIX and TSG101 (Figure 3; Supplementary Figures S6 and S7; Carlton and Martin-Serrano, 2007). These interactions must then help recruit additional ESCRT machinery because ALIX depletion reduces (but does not eliminate) the concentration of VPS4 at midbodies and CEP55 depletion eliminates VPS4 midbody localization completely (Supplementary Figure S8).
Figure 8.
ESCRT pathway functions. The schematic model illustrates how ESCRT-I, ALIX, and other ESCRT pathway proteins may be recruited by different adaptor proteins (red) to perform similar roles in the terminal membrane fission events of MVB biogenesis, virus budding, and cytokinesis.
The dimeric CEP55 protein forms a central ring within the Flemming body (Zhao et al, 2006), and some of our images also showed ring-like ESCRT protein assemblies (not shown). ESCRT-I and ALIX typically formed a single (or two unresolvable) central ring(s), whereas ESCRT-III and VPS4 typically formed two distinct rings, one at either end of the Flemming body. This issue requires further study, but one possibility is that the ESCRT assembly has two-fold symmetry, with ESCRT-III and VPS4 rings flanking central rings of ESCRT-I and ALIX that assemble on a CEP55 template. Alternatively, ESCRT-III and VPS4 may join the Flemming body at a late stage of abscission, as the structure is separating.
CEP55 trafficking is notable because the protein is concentrated at centrosomes until the onset of prophase when it is released by phosphorylation and then moves sequentially to spindle pole regions (late prophase), to the mitotic spindle (metaphase), to the spindle midzone (anaphase), and then ultimately assembles into a ring within the Flemming body via interactions with the MKLP2 subunit of centralspindlin (cytokinesis) (Fabbro et al, 2005; Martinez-Garay et al, 2006; Zhao et al, 2006). Thus, CEP55 is present early at the cleavage furrow, and could therefore recruit at least a subset of the ESCRT machinery early in cytokinesis. However, the most important known CEP55 function is to orchestrate the final stages of abscission late in cytokinesis (Fabbro et al, 2005; Martinez-Garay et al, 2006; Zhao et al, 2006). In the absence of CEP55, a series of late-acting abscission factors fail to concentrate at Flemming bodies, including the Aurora B, MKL2, Plk1, PRC1, and ECT2 (Zhao et al, 2006), and now the ESCRT machinery (Carlton and Martin-Serrano, 2007, and this work).
TSG101/ESCRT-I and/or ALIX also interact with three other proteins involved in cytokinesis, CD2AP, ROCK1, and IQGAP, although the functional implications of these interactions remain to be determined. CD2AP (CD2-Associated Protein) is best characterized as an adaptor protein that functions in various stages of endocytic protein trafficking and actin remodeling, and whose depletion causes glomerular disease (reviewed in Wolf and Stahl, 2003). Reductions in CD2AP levels lead to defects in both MVB biogenesis (Kim et al, 2003) and abscission (Monzo et al, 2005), implying that CD2AP can function in both processes. Notably, Monzo et al (2005) have reported that CD2AP localizes to Flemming bodies and that CD2AP depletion induces abscission defects similar to those described here. We have confirmed this observation (not shown), but the cytokinesis defects in HeLa cells depleted of CD2AP were modest (resembling those seen for TSG101/ESCRT-I depletion), and the ALIX-CD2AP interaction was not absolutely required for cytokinesis (Figure 6). Similarly, mice lacking CD2AP in most tissues are viable, implying that CD2AP cannot be absolutely essential for cytokinesis in most cell types, although this viability could simply reflect functional redundancy with CIN85 and/or other paralogs (Grunkemeyer et al, 2005). Nevertheless, our data show that CD2AP associates with both TSG101/ESCRT-I and ALIX, and all three proteins are now functionally implicated in both MVB biogenesis and cytokinesis.
We also found that both IQGAP1 and ROCK1 bound the stalk and headpiece of TSG101/ESCRT-I. Both of these proteins regulate cytoskeletal remodeling, and both appear to function during actomyosin ring assembly and contraction (Machesky, 1998; Matsumura, 2005), and possibly also during abscission. The evidence for ROCK1 involvement in abscission is not yet strong, but a related Rho-kinase, citron, clearly functions at the midbody during abscission (e.g., Madaule et al, 1998), and was also detected as a TSG101/ESCRT-I binding protein in our original proteomics screens (not shown). The evidence for IQGAP1 involvement in abscission is stronger, as IQGAP1 localizes to the midbody of mammalian cells (Skop et al, 2004) and Dictyostelium mutants lacking the IQGAP1 homolog, GAPA, form midbodies but then arrest before completing abscission (Adachi et al, 1997). Interestingly, IQGAP1 is also present within HIV-1 particles (Chertova et al, 2006) and binds directly to the Gag protein of murine leukemia viral and facilitates virion release (Leung et al, 2006). These observations provide additional links between retrovirus budding and cytokinesis, and raise the intriguing possibility that virus budding, like cytokinesis, may require actomyosin remodeling or contraction. Furthermore, proteomic analyses (Skop et al, 2004) have shown that midbodies also contain at least four additional proteins that have previously been implicated in retrovirus budding: Nedd4 (reviewed in Morita and Sundquist, 2004), Annexin II (Ryzhova et al, 2006), AP-2 (Puffer et al, 1998), and endophilins (Wang et al, 2003), suggesting additional possible parallels between these processes.
The precise function and generality of the ESCRT machinery in eukaryotic cell cytokinesis remains to be determined. For example, ESCRT proteins are not essential for Saccharomyces cerevisiae replication (Hurley and Emr, 2006; Gill et al, 2007), indicating that the ESCRT machinery is not universally required for cytokinesis. Furthermore, our preliminary results indicate that even mammalian cells may differ in their requirements for cytokinesis, as HEK 293T cells continue to divide following CEP55 depletion (not shown). Further experiments will therefore be required to test whether the ESCRT pathway plays a fundamental role in the division of all mammalian cells, and whether this role is indirect (e.g., by functioning in vesicle trafficking or membrane protein degradation) or direct (e.g., by participating in the actual abscission process). Nevertheless, it is striking that the three best-characterized functions for the ESCRT machinery all involve systems in which a thin membrane tubule must be resolved through an internal membrane fission event (see Figure 8). We therefore speculate that the ESCRT pathway is a modular membrane fission machine that can be recruited to perform analogous functions in a series of different biological processes.
If true, this model implies that the ESCRT machinery can mediate membrane fission reactions at midbodies (which are roughly 2 μm in diameter) and at MVB vesicles and retroviruses (which are roughly 100 nm in diameter). We envision that the resolution to this apparent paradox may come from the stepwise nature of cytokinesis. In the first step, furrow ingression is driven by contraction of the actomyosin ring to produce a midbody that is approximately 2 μm in diameter. The Flemming body then forms, and it has been proposed that it recruits secretory vesicles that fuse to create membrane ‘nets' that help constrict the midbody further (Gromley et al, 2005). We speculate that this process may reduce the size of the midbody pore(s) to the 100-nm scale, and at that point the cytoplasmic ESCRT machinery could then mediate membrane fission from within those narrowed pores, thereby completing abscission and releasing the two daughter cells.
Materials and methods
Cell cultures
HeLa, 293T, and COS7 cells were maintained in DMEM supplemented with 10% FCS. For immunofluorescence experiments, HeLa cells were seeded onto coverslips and cotransfected at 50–60% confluence.
Expression vectors and plasmids
Expression vectors used in this study are summarized in Supplementary Table S2.
Co-immunoprecipitation and Western blotting assays
Experimental details of co-immunoprecipitation and Western blotting experiments are provided in the caption to Supplementary Figure S1A and in Supplementary Table S4.
Yeast two-hybrid assays
Automated yeast two-hybrid screens were performed as described previously (Bartel and Fields, 1997; Garrus et al, 2001), using cDNA prey libraries from macrophages and from breast and prostate cancer cell lines. Genes emerging from the screens were recloned (Supplementary Table S3) and tested for protein interactions using the Matchmaker GAL4 Yeast Two Hybrid 3 system (Clontech) (Langelier et al, 2006).
Immunofluorescent imaging
HeLa cells were transfected with 10 nM siRNA and/or plasmids expressing the designated combination of epitope-tagged or GFP-fused proteins. Except where noted, cells were fixed 18 h post-transfection with 3% paraformaldehyde in PBS or ice-cold methanol (for cases involving γ-tubulin staining). Confocal immunofluorescence images were acquired using Fluoview software on a FV300 IX81 Olympus microscope. Images are single confocal slices, except where noted. Antibodies and concentrations are provided in Supplementary Table S4.
FACS analyses of cellular DNA contents
HeLa cells from six-well plates were collected by trypsin treatment, and half were resuspended in propidium iodide (PI) solution (50 μg of PI per milliliter of PBS, 0.1% Triton X-100, 0.25 mg RNase/ml, 30 min, 4°C). PI-positive cells were counted with a FACScan fluorescence-activated cell sorter (FACScan, BD Bioscience), and peak volumes associated with each DNA content were analyzed using CellQuest software. The other half of the cells were lysed (50 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, protease inhibitors) and TSG101 and ALIX proteins were detected by Western blotting.
Time-lapse microscopy
siRNA-transfected HeLa cells were cultured in a 37°C microscope chamber (Oko-Lab) with 5% CO2 and observed by phase contrast with an Olympus IX81 microscope (× 10 objective). Images were acquired every 5 min with MetaMorph v6.2r6 software (Molecular Devices Corp).
Rescue of cytokinesis defects in cells depleted of endogenous ALIX
For cell cycle analyses, HeLa cells in six-well plates were transfected with ALIX expression vectors (2 μg/well, Lipofectamine LTX, Invitrogen) and siRNA (10 nM, Lipofectamine RNAi MAX, Invitrogen), following the time course as mentioned here: t=0, cells seeded at 4 × 105 cells/well; t=24 h, ALIX vector; t=32 h, media changed+ALIX vector; t=44 h, cells trypsin treated and reseeded with 2- to 8-fold dilution; t=50 h, siRNA; t=56 h, media changed+siRNA; t=68 h, media changed+siRNA; t=98 h, cells harvested and analyzed.
Supplementary Material
Supplementary Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9A
Supplementary Figure S9B
Supplementary Figure S9C
Supplementary Figure S9D
Supplementary Figure S10
Acknowledgments
We thank Jez Carlton and Juan Martin-Serrano for sharing their data on the roles of ESCRT-I and ALIX in cytokinesis, before publication, the Taplin Biological Mass Spectrometry Facility at Harvard Medical School for protein ID data, and the ProNet group of Myriad Genetics Inc. for identification of yeast two-hybrid interactions. Funding was provided by NIH AI051174 and AI45405 (to WIS), and the American Foundation for AIDS Research (to VS).
References
- Adachi H, Takahashi Y, Hasebe T, Shirouzu M, Yokoyama S, Sutoh K (1997) Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis. J Cell Biol 137: 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann D, Estepa-Sabal E, Meerloo T, Emr S (2002) Escrt-III. An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell 3: 271–282 [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD (1998) The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J 17: 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel PL, Fields BN (1997) The Yeast Two Hybrid Screen. New York: Oxford Press Inc. [Google Scholar]
- Bieniasz PD (2006) Late budding domains and host proteins in enveloped virus release. Virology 344: 55–63 [DOI] [PubMed] [Google Scholar]
- Canman JC, Hoffman DB, Salmon ED (2000) The role of pre- and post-anaphase microtubules in the cytokinesis phase of the cell cycle. Curr Biol 10: 611–614 [DOI] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J (2007) Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316: 1908–1912 [DOI] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW Jr, Sowder II RC, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE (2006) Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol 80: 9039–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu T, Sun J, Saksena S, Emr SD (2006) New component of ESCRT-I regulates endosomal sorting complex assembly. J Cell Biol 175: 815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss M, Jones C, Babst M (2007) Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol Biol Cell 18: 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Freed EO (2004) Retrovirus budding. Virus Res 106: 87–102 [DOI] [PubMed] [Google Scholar]
- Doxsey S, McCollum D, Theurkauf W (2005) Centrosomes in cellular regulation. Annu Rev Cell Dev Biol 21: 411–434 [DOI] [PubMed] [Google Scholar]
- Fabbro M, Zhou BB, Takahashi M, Sarcevic B, Lal P, Graham ME, Gabrielli BG, Robinson PJ, Nigg EA, Ono Y, Khanna KK (2005) Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell 9: 477–488 [DOI] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP (2007) Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128: 841–852 [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107: 55–65 [DOI] [PubMed] [Google Scholar]
- Gill DJ, Teo H, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL (2007) Structural studies of phosphoinositide 3-kinase-dependent traffic to multivesicular bodies. Biochem Soc Symp 74: 47–57 [DOI] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ (2005) Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 123: 75–87 [DOI] [PubMed] [Google Scholar]
- Grunkemeyer JA, Kwoh C, Huber TB, Shaw AS (2005) CD2-associated protein (CD2AP) expression in podocytes rescues lethality of CD2AP deficiency. J Biol Chem 280: 29677–29681 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Emr SD (2006) The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct 35: 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Mancuso JJ, Uzawa S, Cronembold D, Cande WZ (2005) The fission yeast homolog of the human transcription factor EAP30 blocks meiotic spindle pole body amplification. Dev Cell 9: 63–73 [DOI] [PubMed] [Google Scholar]
- Kim JM, Wu H, Green G, Winkler CA, Kopp JB, Miner JH, Unanue ER, Shaw AS (2003) CD2-associated protein haploinsufficiency is linked to glomerular disease susceptibility. Science 300: 1298–1300 [DOI] [PubMed] [Google Scholar]
- Kostelansky MS, Schluter C, Tam YY, Lee S, Ghirlando R, Beach B, Conibear E, Hurley JH (2007) Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell 129: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, Hill CP, Kaplan J, Ward D, Sundquist WI (2006) Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol 80: 9465–9480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Yueh A, Appah FS Jr, Yuan B, de los Santos K, Goff SP (2006) Interaction of Moloney murine leukemia virus matrix protein with IQGAP. EMBO J 25: 2155–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI (2005) Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem 280: 12799–12809 [DOI] [PubMed] [Google Scholar]
- Machesky LM (1998) Cytokinesis: IQGAPs find a function. Curr Biol 8: R202–R205 [DOI] [PubMed] [Google Scholar]
- Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S (1998) Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature 394: 491–494 [DOI] [PubMed] [Google Scholar]
- Martinez-Garay I, Rustom A, Gerdes HH, Kutsche K (2006) The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics 87: 243–253 [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Yaravoy A, Perez-Caballero D, Bieniasz PD (2003) Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA 100: 12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura F (2005) Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol 15: 371–377 [DOI] [PubMed] [Google Scholar]
- McCollum D (2005) Cytokinesis: breaking the ties that bind. Curr Biol 15: R998–R1000 [DOI] [PubMed] [Google Scholar]
- Monzo P, Gauthier NC, Keslair F, Loubat A, Field CM, Le Marchand-Brustel Y, Cormont M (2005) Clues to CD2-associated protein involvement in cytokinesis. Mol Biol Cell 16: 2891–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sundquist WI (2004) Retrovirus budding. Annu Rev Cell Dev Biol 20: 395–425 [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Alam S, Eckert D, Gygi SP, Sundquist WI (2007) Identification of the human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host & Microbe 2: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G (2006) The multiple personalities of Alix. J Cell Sci 119: 3025–3032 [DOI] [PubMed] [Google Scholar]
- Oestreich AJ, Davies BA, Payne JA, Katzmann DJ (2007) Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol Biol Cell 18: 646–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Alam SL, Davis DR, Sundquist WI (2002) Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol 9: 812–817 [DOI] [PubMed] [Google Scholar]
- Puffer BA, Watkins SC, Montelaro RC (1998) Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J Virol 72: 10218–10221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryzhova EV, Vos RM, Albright AV, Harrist AV, Harvey T, Gonzalez-Scarano F (2006) Annexin 2: a novel human immunodeficiency virus type 1 Gag binding protein involved in replication in monocyte-derived macrophages. J Virol 80: 2694–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai M, Shimokawa T, Kobayashi T, Matsushima S, Yamada Y, Nakamura Y, Furukawa Y (2006) Elevated expression of C10orf3 (chromosome 10 open reading frame 3) is involved in the growth of human colon tumor. Oncogene 25: 480–486 [DOI] [PubMed] [Google Scholar]
- Skop AR, Liu H, Yates J III, Meyer BJ, Heald R (2004) Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305: 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, Bechtold N, Herzog M, Muller S, Hanisch FG, Hulskamp M (2006) The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133: 4679–4689 [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG (2003) AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114: 689–699 [DOI] [PubMed] [Google Scholar]
- Straight AF, Field CM (2000) Microtubules, membranes and cytokinesis. Curr Biol 10: R760–R770 [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG (2007) Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol 81: 6614–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI (2003) The protein network of HIV budding. Cell 114: 701–713 [DOI] [PubMed] [Google Scholar]
- Wang MQ, Kim W, Gao G, Torrey TA, Morse HC III, De Camilli P, Goff SP (2003) Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production. J Biol 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Habermann A, Jager S, Muller B, Krijnse-Locker J, Krausslich HG (2006) Ultrastructural analysis of ESCRT proteins suggests a role for endosome-associated tubular-vesicular membranes in ESCRT function. Traffic 7: 1551–1566 [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Wang Y (1996) Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol 135: 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Stahl RA (2003) CD2-associated protein and glomerular disease. Lancet 362: 1746–1748 [DOI] [PubMed] [Google Scholar]
- Xie W, Li L, Cohen SN (1998) Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc Natl Acad Sci USA 95: 1595–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WM, Seki A, Fang G (2006) Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell 17: 3881–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9A
Supplementary Figure S9B
Supplementary Figure S9C
Supplementary Figure S9D
Supplementary Figure S10