Abstract
Insulator sequences guide the function of distantly located enhancer elements to the appropriate target genes by blocking inappropriate interactions. In Drosophila, five different insulator binding proteins have been identified, Zw5, BEAF-32, GAGA factor, Su(Hw) and dCTCF. Only dCTCF has a known conserved counterpart in vertebrates. Here we find that the structurally related factors dCTCF and Su(Hw) have distinct binding targets. In contrast, the Su(Hw) interacting factor CP190 largely overlapped with dCTCF binding sites and interacts with dCTCF. Binding of dCTCF to targets requires CP190 in many cases, whereas others are independent of CP190. Analysis of the bithorax complex revealed that six of the borders between the parasegment specific regulatory domains are bound by dCTCF and by CP190 in vivo. dCTCF null mutations affect expression of Abdominal-B, cause pharate lethality and a homeotic phenotype. A short pulse of dCTCF expression during larval development rescues the dCTCF loss of function phenotype. Overall, we demonstrate the importance of dCTCF in fly development and in the regulation of abdominal segmentation.
Keywords: Abdominal-B, bithorax complex, CP190, homeotic, Hox
Introduction
Eukaryotic genomes are highly organized into functional units containing individual genes or gene groups together with the corresponding regulatory elements. Regulatory elements may be separated from the transcriptional start sites by several thousands of base pairs. These functional units need to be insulated from each other in order to prevent illegitimate interactions of enhancers with other transcriptional units. Furthermore, a single regulatory element may control several genes, or several distinct regulatory elements may control the activity of a single gene in time and space. Again, a functional constraint for the appropriate regulator/gene interaction needs to be achieved. Insulator elements with enhancer blocking activity fulfill this function, such that only appropriate promoters and genes are activated (Holdridge and Dorsett, 1991; Geyer and Corces, 1992; Chung et al, 1993; Gaszner and Felsenfeld, 2006). In vertebrates, there is only one factor currently known that binds to enhancer-blocking elements and prevents the inappropriate activation by adjacent enhancers: CTC binding factor (CTCF) (Ohlsson et al, 2001). Binding sites for CTCF have been shown to be involved in gene repression (Baniahmad et al, 1990; Lobanenkov et al, 1990), in gene activation (Vostrov and Quitschke, 1997) and in enhancer blocking (Bell et al, 1999; Hark et al, 2000; Kanduri et al, 2000; Szabo et al, 2000; Lutz et al, 2003; Burke et al, 2005). Furthermore, vertebrate- and mammalian-specific functions, such as X-chromosome inactivation and control of the epigenetic DNA methylation state, seem to involve CTCF (for reviews see Lee, 2003; Lewis and Reik, 2006).
In sharp contrast to vertebrates, the genome of Drosophila is much more compacted, primarily due to shorter distances between genes. Therefore, the need for insulators to separate genes is likely greater than in vertebrates. Indeed, five different insulator binding proteins have been identified in Drosophila. These are Zw5, BEAF-32 (Zhao et al, 1995; Gaszner et al, 1999), GAGA factor (Ohtsuki and Levine, 1998; Belozerov et al, 2003), Su(Hw) (Gerasimova et al, 1995) and a Drosophila orthologue of CTCF that we have identified (Moon et al, 2005). The function of enhancer blocking has evolved such that Drosophila utilizes several proteins and probably multiple mechanisms for enhancer blocking and insulation (Kuhn et al, 2003). However, with the exception of dCTCF, none of the other known Drosophila insulator proteins have a counterpart found to be conserved in vertebrates. Su(Hw), a 12-zinc-finger factor, resembles the 11-zinc-finger protein dCTCF with respect to the overall domain structure. Detailed study of Su(Hw) has revealed that a 350-bp DNA sequence of the gypsy insulator binds a protein complex consisting of at least three components, Su(Hw), Mod(mdg4)67.2 and CP190. Su(Hw) and CP190 can bind DNA directly via their zinc-finger domains, whereas Mod(mdg4)67.2 does not bind DNA directly, but is recruited to the gypsy insulator sequence through physical interactions with Su(Hw) and CP190 (Pai et al, 2004). Several hundred endogenous binding sites for Su(Hw) are found throughout the Drosophila genome (Gerasimova and Corces, 1998; Parnell et al, 2006; Ramos et al, 2006).
Another perspective on the requirement for insulators comes from the fact that many genes are controlled by several regulatory elements needed for tissue- and cell-specific expression. For example, the three Drosophila homeotic genes of the bithorax complex (BX-C), Ultrabithorax (Ubx), abdominal A (abd-A) and Abdominal-B (Abd-B), are regulated by a cis-regulatory region comprising more than 300 kb. This region is subdivided into nine distinct regulatory domains controlling the three homeotic genes individually and specifically with respect to particular parasegments (Lewis, 1978; Sanchez-Herrero et al, 1985; Maeda and Karch, 2006). A striking feature of this complex regulatory region is the colinearity of regulatory domains controlling gene expression from the thoracic segment T3 through the abdominal segments A1–A9 (parasegments PS5–PS14). Of the eight boundaries between the nine regulatory domains, three have been functionally identified as chromatin insulators with enhancer blocking activity. These are Miscadastral pigmentation (Mcp), Frontabdominal-7 (Fab-7) and Frontabdominal-8 (Fab-8) (Maeda and Karch, 2006). The GAGA factor was shown to be required for the enhancer blocking function of Fab-7 (Schweinsberg et al, 2004). Previously we demonstrated that dCTCF binding sites are required for the function of Fab-8 (Moon et al, 2005), and that dCTCF is associated with all but one of the known or predicted insulators of BX-C (Holohan et al, 2007).
Here we addressed whether dCTCF and Su(Hw) mediate similar molecular functions and biological effects, and whether dCTCF plays a functional role in the BX-C, which is finely subdivided by regulatory domain borders. We demonstrate that the overall binding pattern of Su(Hw) is distinct from dCTCF and that, in contrast to Su(Hw), a dCTCF-null mutation causes pharate lethality and hypomorphic mutations result in a homeotic phenotype. Despite these differences, both factors interact with CP190.
Results
dCTCF binds to many interbands without overlap with Su(Hw) binding sites
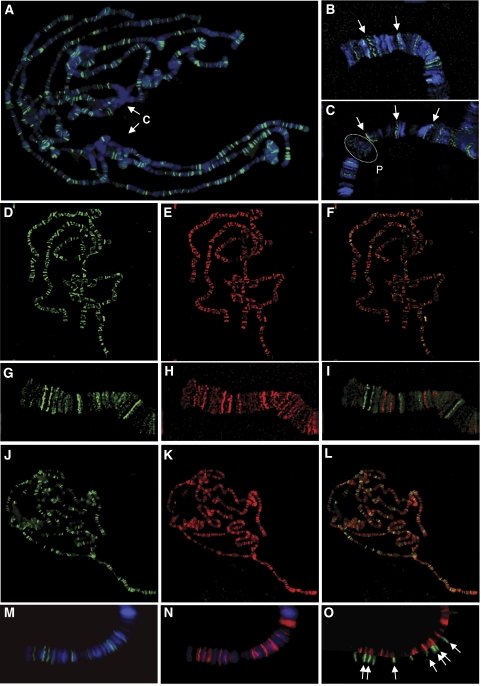
dCTCF and Su(Hw) are similar factors in that both are multi zinc-finger proteins and both mediate enhancer blocking. To test whether both factors target to identical genomic sites, or whether separate sets of target sites are accessed, we analyzed the pattern of dCTCF bound to polytene chromosomes of third instar larvae. We used two different antibodies directed against the N-terminal or the C-terminal domain of dCTCF (Moon et al, 2005). Both antibodies resulted in the same pattern, when carefully compared in independent staining experiments with each other and with GFP signals from a CTCF-eGFP line (not shown). Analysis of the overall distribution of dCTCF showed that approximately 300 to 400 dCTCF target sites (CTS) are bound (Figure 1). The heterochromatic chromocenter region is essentially free of dCTCF binding. Close inspection at higher magnification revealed that all bound CTSs are located in interband regions. In many cases, when the chromosomal quality allowed high-resolution analysis, a location of the CTS at the interband/band border or at a puff border is evident (Figure 1B and C). Next we analyzed the distribution of binding sites for Su(Hw) relative to dCTCF. The Su(Hw) antibody stains several hundreds of sites located in interbands as well. At low magnification, only few sites seem to colocalize, which may be caused by both factors bound to the same target or by low resolution of separate targets. Upon magnification and inspection of high-quality regions, no colocalization could be detected (Figure 1G–I). Therefore, we conclude that at least for the polytene chromosomes, almost all dCTCF targets are different from those of Su(Hw).
Figure 1.
dCTCF binds to several hundred loci on polytene chromosomes, different from those bound by Su(Hw), but overlapping with CP190 signals. Immunostaining of salivary gland polytene chromosomes with α-dCTCF, α-Su(Hw) and α-CP190. (A–C) dCTCF (green) detected on interband or at band/interband transitions, flanked by bands stained with Hoechst 33342 (blue). Examples are pointed out (arrows) at higher magnification for chromosomes 3L (B) and X (C). Chromocenter (‘C') of two nuclei, and the 2B puff (dashed circle) are negative for dCTCF. (D–I) α-dCTCF (green) and α-Su(Hw) (red) generate non-overlapping staining (F, I, merged). Higher magnification of the tip of 3L (G–I). (J–O) α-dCTCF (green) and α-CP190 (red) show overlapping localization. Merged picture of the whole chromosome set (L) indicates colocalization (orange signals). The enlarged view of the tip of the 3L chromosome (M–O) identifies colocalization in the split view (O, arrows).
CP190 and dCTCF binding sites overlap and both factors interact in vivo
We wondered whether dCTCF and Su(Hw) might share cofactors despite the differences in target site specificity. The protein Mod(mdg4) has been shown to be required for Su(Hw) function (Ghosh et al, 2001) and an analysis of Mod(mdg4) distribution over the polytene chromosomes revealed only a partial colocalization with dCTCF (not shown). In contrast, when we tested the binding pattern of CP190, another cofactor required for the function of Su(Hw) (Pai et al, 2004), we found that many of the CP190 sites are CTSs as well (Figure 1J–O). Indeed, analysis at high resolution revealed that the majority of CTSs are bound by CP190 (Figure 1O).
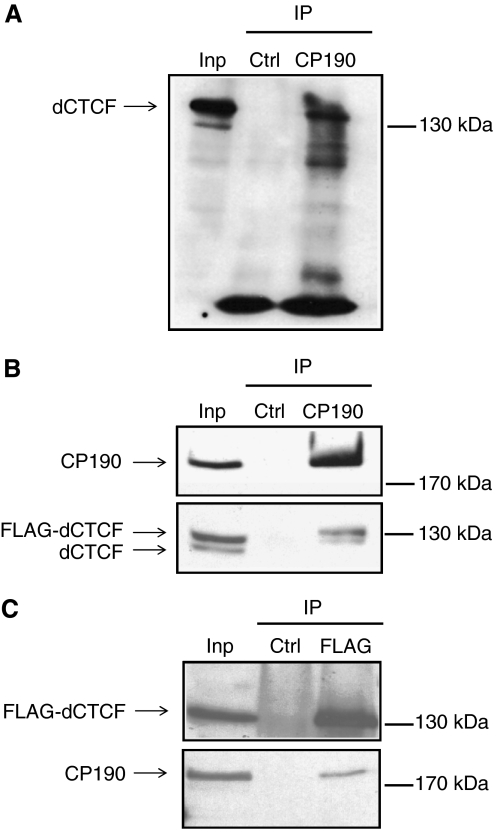
The CP190 protein contains three classical C2H2 zinc-finger motifs and an N-terminal BTB/POZ domain. Both domains could potentially be involved in chromatin binding. On the other hand, chromatin binding might be achieved by interaction with other factors, such as dCTCF. We therefore tested for a possible interaction of dCTCF with CP190 using co-immunoprecipitation. Precipitation of CP190 from Schneider cell extracts resulted in the detection of dCTCF (Figure 2A). To confirm the interaction we expressed a FLAG-dCTCF fusion protein in Schneider cells and precipitated with either an antibody against CP190 or an antibody against FLAG (Figure 2B and C). The CP190 precipitate contained endogenous dCTCF as well as FLAG-dCTCF in the same ratio as the input, suggesting that both dCTCF proteins are similarly associated with CP190. Furthermore, the reverse experiment using FLAG precipitation demonstrated that dCTCF and CP190 interact in vivo.
Figure 2.
Co-immunoprecipitation of dCTCF and CP190. (A, B) Whole-cell extracts from S2 cells were precipitated with α-CP190 antibody and the precipitates were analyzed by SDS–PAGE and subsequent Western blotting with α-CP190 or α-dCTCF. CP190 coprecipitates dCTCF (A) and FLAG-dCTCF, as well as endogenous dCTCF (B). (C) α-FLAG antibody was used in the IP, and α-CP190 or α-FLAG for detection in Western blot. Control: precipitation with mouse IgG; input: 10% in panel A, 4% of extract in panels B and C.
dCTCF mutants show a homeotic phenotype and pharate lethality
We first tested the effect of dCTCF depletion from Schneider S2 cells by RNAi knockdown (Supplementary Figure). However, we could not detect any change in cell number or cell size after dCTCF depletion. Therefore, we wondered whether dCTCF may play a role in Drosophila development.
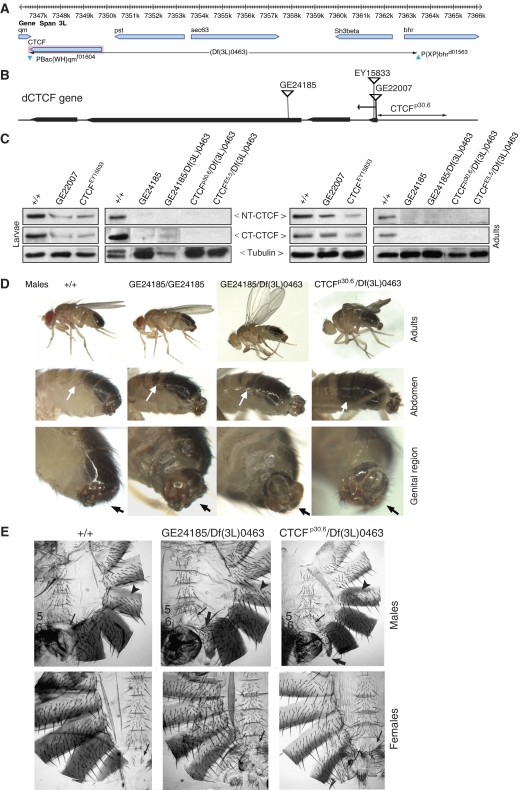
The available deficiency Df(3L)pbl-X1 does not take out dCTCF (data not shown). Therefore, we generated a 16-kb deletion (Df(3L)0463) by recombination between FRT containing P-elements (Parks et al, 2004; Thibault et al, 2004) within the quemao gene at 65F5-F6 and within the bhringi gene at 65F7 (PBac{WH}qmf01604 and P{XP}bhrd01563). The deletion is recessive lethal and takes out dCTCF, pastrel, CG8583, Sh3beta and the 5′ region of bhringi (Figure 3A). In addition, we checked three different transposon elements residing within the dCTCF locus (Figure 3B). GE22007 and CTCFEY15833, each with a P-EP or a P-element positioned 15 or 35 bp downstream of the transcription start site of dCTCF show a 50–70% reduced amount of CTCF as determined in third instar larvae and in adults (Table I; Figure 3C). The reduction in protein amount is also seen in salivary gland polytene chromosomes, where the number of CTCF-bound sites is reduced to about 50%. Trans-heterozygotes GE22007/Df(3L)0463 and CTCFEY15833/Df(3L)0463 are without phenotype (Table I). Finally, GE24185 has a P-EP element located at the nucleotide triplet coding for amino acid 158. Few F1 homozygotes showed spreading of wings by 30–45° (Table I). The GE24185/Df(3L)0463 trans-heterozygotes showed a highly penetrant mild (60°) held out wing phenotype, various homeotic transformations (see below) and sterility in both males and females. Protein extracts prepared from 10 salivary glands showed a weak band, running at ∼90kDa when tested with the C-terminal antibody, but no specific band is detectable with the N-terminal antibody (Supplementary Figure 1C). Salivary gland polytene chromosome staining showed detectable dCTCF, although at a dramatically reduced number of sites (25% of wild type) using the antibody against the C-terminus of the dCTCF protein (Table I), whereas the antibody against the N-terminus of dCTCF did not show any signal. Similarly, RNA analysis by RT–PCR showed the presence of a transcript in reduced amount (Supplementary Figure 1B). Taken together, these data suggest that insertion of the P-element results in the reduced expression of a truncated dCTCF protein that is recognized by the C-terminal-specific antibody in a Western blot, and on polytene chromosomes. This small amount appears to be highly concentrated on a subset of binding sites (see also below).
Figure 3.
dCTCF mutations cause homeotic phenotypes. (A) View of the gbrowse genome browser (FlyBase) showing CTCF and its neighboring genes, and the two FRT elements PBac{WH}qmf01604 and P{XP}bhrd01563 used to generate Df(3L)0463 (long horizontal arrow). (B) Schematic representation of the dCTCF gene; locations of exons I–IV (thick horizontal arrows), of the transposons GE22007, GE24185 and EY15833, and of the deleted fragment in CTCFp30.6 are shown. (C) Western analysis of wild type (+/+), P-element and jump-out mutants. Single fly extracts from third instar larvae and young adults were loaded on each lane and probed with N- and C-terminal-specific dCTCF antibodies. Tubulin antibody was used as control. (D) Morphological defects of the indicated dCTCF mutants. Appearance of patchy pigmentation in A4 indicates a partial transformation to A5 (white arrow). Twisting of the male genital region (black arrows). (E) Cuticle preparations of the abdomen of the indicated male mutants show a patchy appearance of pigmentation in A4 tergite (arrowhead), and small white patches in A5 tergite toward the anterior side of the segment. Loss of pigmentation in A5 indicates transformation into A4. The more compact A5 sternite in wild type has elongated into a banana shape in the mutants. This indicates transformation of A5 to A6 segment. Up to eight bristles were seen in the sixth sternite (thin arrow), and a small seventh segment (thick arrow) is present. Females show transformation of A7, with an increased number of bristles of the seventh sternite, showing a different orientation.
Table 1.
dCTCF mutants
| Genotype | dCTCF expressiona | Phenotypeb |
|---|---|---|
| P-strains | ||
| GE22007/GE22007 | W (50–70%) | wt; F2 embryos die at various stages from secondary mutations (∼5/200 survivors) |
| GE22007/Df(3L)0463 | Not determined | wt |
| EY15833/EY15833 | W (50–60%); P (50%) | wt |
| EY15833/Df(3L)0463 | Not determined | wt |
| GE24185/GE24185 | W (0%); P (25%) with C-terminal antibody | Reduced viability (66%); embryos from F2 homozygotes do not eclose; held out wings (30% of survivors); male genitalia rotated up to 30° (10%); males: A4 patchy pigment, partial loss of pigment in A5, eight bristles on sixth sternite, small seventh tergite (10–20%) |
| GE24185/Df(3L)0463 | W (0%); P with C-terminal antibody (25%) | Held out wings (100%); male and female sterile; male genitalia rotated by 40°–120° (100%); males: A4 patchy pigment, partial loss of pigment in A5 tergite, eight bristles on sixth sternite, small seventh tergite (100%); females: bristles on A7 sternite have lost orientation (100%) |
| Jump-out strains | ||
| P30.6/P30.6c | W (0%); P (0%) | Pharate lethal (100%); male genitalia rotated by 40°–90° (100%); males: A4 tergite with patchy pigmentation, A5 tergite shows a few white spots, A5 sternite is elongated into a banana shape, eight bristles on sixth sternite, small seventh tergite (100%); females: A7 sternite with up to 11 bristles pointing vertically downwards (100%) |
| P30.6/Df(3L)0463 | W (0%); P (0%) | as P30.6/P30.6 |
| P35.2/P35.2c | W (0%) | as P30.6/P30.6 |
| E5.5/E5.5d | W (0%) | as P30.6/P30.6 |
| R10.3/R10.3e | W (0%) | as P30.6/P30.6 |
|
R15.4/R15.4e |
W (0%) |
as P30.6/P30.6 |
| wt, wild type. | ||
| a(W) single fly western with the percentage of expression relative to wild type; (P) polytene chromosome staining with the percentage of the number of sites relative to wild type. | ||
| bPhenotype with percentage of penetrance within the surviving group. | ||
| cJump-out from GE22007. | ||
| dJump-out from EY15833. | ||
| eJump-out from GE24185. | ||
To create amorphic alleles by imprecise excision, we performed jump-outs from all three insertions. When homozygous, all of these mutants died at the pharate stage, with about 50% being very short lived (up to 12–24 h) but able to eclose (Table I). Immunoblot analysis of single larval protein extracts showed no dCTCF protein and polytene chromosome squashes were completely negative (not shown). Similarly, absence of dCTCF was observed when these lines were crossed to Df(3L)0463 (Figure 3C). Thus, these mutations were considered to be amorphic dCTCF alleles. For further analysis we concentrated on jump-out strain CTCFp30.6, which we molecularly characterized to carry a deletion from nucleotide −537 upstream of the transcriptional start site to position +15 (Figure 3B).
Careful inspection of the adults revealed a number of homeotic transformations in the GE24185 homozgygotes, in GE24185/Df(3L)0463 and even more severe in the amorphic mutants (Figure 3D). Some of the GE24185 homozygous males showed mild and patchy pigmentation of the abdominal segment A4, and formation of a small A7 tergite. They also showed various degrees (0–30°) of genital region rotation. This later phenotype is similar to a known homeotic mutation in the bithorax locus (Estrada et al, 2002). All GE24185/Df(3L)0463 males showed larger patches of pigmentation in A4 and formation of a small A7 tergite (Figure 3D and E). Cuticle preparations also showed a number of additional phenotypes. The dCTCFp30.6/Df(3L)0463 males have lost pigmentation in small patches toward the anterior of the A5 tergite, indicating that A5 is nominally transformed into A4 (Figure 3E). Interestingly, A5 sternite is banana shaped, which is a feature of the A6 sternite (Figure 3E). Hence, A5 shows anterior to posterior transformation, as well as posterior to anterior transformation. We also found up to eight bristles in the A6 sternite in the cuticle preparations, which are not seen in wild type. The genital region was extended out of the abdomen and rotation was more severe, from 60° to 120° (Figure 3D). Furthermore, GE24185/Df(3L)0463, as well as dCTCFp30.6/Df(3L)0463 females exhibit an A7 transformation. In the former case the bristles have lost their orientation, which in wild type is toward the mid axis. In the latter case the transformation is more apparent, as indicated by more bristles on the A7 sternite (up to 11 compared with eight in wild type), and by their vertical downward orientation (Figure 3E).
The dCTCFp30.6 phenotype can be rescued by brief dCTCF expression at the larval stage
In order to demonstrate that the phenotype seen in the various dCTCF mutants is caused by the loss of dCTCF, we generated rescue strains. First we produced genomic dCTCF transgenic flies by cloning the 5084-bp sequence spanning the dCTCF gene from a site within the quemao gene and extending to within the pastrel gene. We reasoned that the regulatory sequences for dCTCF expression in this gene dense region would most likely be framed by the two flanking genes (Figure 3A). A single integration of genomic dCTCF on the second chromosome (gCTCF) was able to rescue CTCFp30.6/Df(3L)0463, CTCFp30.6/CTCFp30.6, GE24185/Df(3L)0463 and GE24185/GE24185. We found a majority of the flies fully rescued, with a few showing various degrees of held out wings and that were unable to walk properly. The rescued flies were fertile and we could establish a breeding stock of gCTCF/gCTCF;GE24185/GE24185.
Furthermore, we tested whether a pulse of dCTCF expression during larval development would be sufficient for the rescue or whether the flies might need a continuous supply of dCTCF. We generated UASdCTCF-EGFP transgenic flies by P-element transgenesis, and established lines with insertions on the X, second and third chromosomes. On driving UASdCTCF-EGFP in wild-type background with ubiquitous drivers such as tubulin-gal4 or actin-gal4, we found early embryonic lethality (data not shown). In order to express a moderate and controlled amount of dCTCF, we used a hsp70Gal4 driver line, and gave a single heat shock (37°C) for 20 min at various stages of development. We found that a single heat shock at 24, 48 or 72 of development was sufficient to phenotypically rescue the majority of flies with a dCTCFp30.6/Df(3L)0463 or a GE24185/Df(3L)0463 genetic background. Fertility tests of the rescued flies revealed that both males and females were fertile. A few flies still showed a very mild held out wing phenotype and some had a disorganized bristle pattern on the dorsal abdomen. Heat shock at the pupal stage could not rescue all the mutant phenotypes (not shown). Thus, a short pulse of dCTCF expression is sufficient to generate phenotypes close to wild type.
Many, but not all dCTCF binding sites depend on CP190
Because we found that CP190 and dCTCF colocalize on polytene chromosomes and interact in vivo, we asked whether the overall amount of dCTCF protein might be changed in CP190-deficient third instar larvae. A Western blot analysis of both Cp1901 homozygotes (deficient in CP190) and wild-type larval extracts showed that the amount of dCTCF is reduced in Cp1901 homozygotes (Figure 4A).
Figure 4.
Many, but not all dCTCF binding sites depend on CP190. (A) Western analysis of dCTCF and tubulin in Cp1901 larval extracts. (B–F) Wild-type polytene chromosome 3L from third instar larval salivary glands stained for DNA (B), CTCF (D), CP190 (E) and the indicated merges are shown. Arrows point to all 12 CTSs as identified in panel D, counted from the tip of chromosome 3L and numbered. (G–I) Polytene 3L chromosomes from homozygote genotypes EY15833, GE24185 and CP1901, stained with DAPI and for dCTCF. Numbered arrows point to the corresponding locations as in wild type. (J) Analysis of the first 12 CTS at the 3L chromosome tip as numbered in panel D. Occupancy (+) for dCTCF and for CP190 in different genotypes. The total number of CTSs on chromosome 3 is given±the number of weak signals, which cannot be interpreted.
Next we wanted to know whether the reduced amount of dCTCF caused by the loss of CP190 affects dCTCF binding on the polytene chromosomes. We found that the total number of dCTCF labeled sites is reduced in the Cp1901 mutant, whereas the number of CP190 sites was not affected by dCTCF mutants (not shown and below). As mentioned above, the analysis of dCTCF binding in the two hypomorphic mutants CTCFEY15833/CTCFEY15833 and GE24185/GE24185 revealed that that the number of bound sites is reduced to about 50 and 25%, respectively (Table I). By close inspection of the chromosomes we found that the set of dCTCF sites missing in the CP190 or in the dCTCF mutants overlap but are not identical. This is exemplified with the first 12 CTS at the tip of chromosome 3L (Figure 4B–I). We find four types of sites: (a) those binding dCTCF only, affected by both hypomorphic dCTCF mutations, but not by CP1901 (site 6 in Figure 4G–J), (b) sites binding dCTCF only, and neither affected by hypomorphic dCTCF mutations nor by CP1901 (site 1), (c) sites binding both proteins, and dCTCF binding affected by CP1901 and dCTCF mutations (sites 2–4 and 10–12) and (d) sites colocalizing with both factors, but dCTCF binding not affected by either mutation (sites 5 and 7–9). Threshold effects caused by reduced dCTCF amounts do not seem to be problematic for this analysis. For example, sites 2, 10 and 12 are strong in wild type and are lost in the mutant, whereas weak wild-type sites (e.g., sites 1 and 5) are still bound in dCTCF mutants. This may indicate that, depending on the binding site, both factors cooperate in binding. Thus, different sites vary in their requirement for CP190/dCTCF cooperation.
dCTCF binding to the BX-C locus is lost in hypomorphic dCTCF mutants
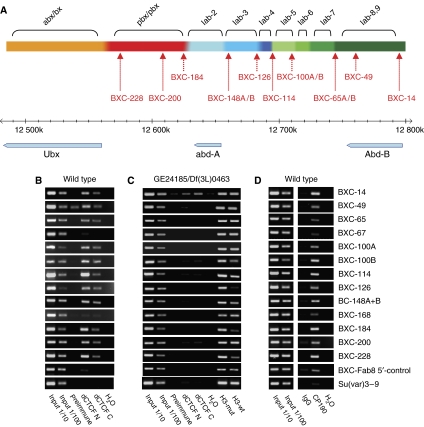
Previously we demonstrated that dCTCF binding sites are required for the function of Fab-8 (Moon et al, 2005), and that dCTCF is associated with all but one of the known or predicted insulators in embryo and Schneider cell chromatin (Holohan et al, 2007). As dCTCF loss of function mutants show a homeotic phenotype, one would predict that the dCTCF occupancy of the CTSs (Figure 5A) is lost or reduced in the case of the GE24185/Df(3L)0463 genotype, which shows a dramatic loss of dCTCF signals on polytene chromosomes (see above; Table I). Whether the BX-C locus remains bound by dCTCF cannot be analyzed on the level of salivary gland polytene chromosomes, since this region is strongly under-replicated and not clearly analyzable (Moshkin et al, 2001). Therefore, we precipitated chromatin from wild-type and GE24185/Df(3L)0463 third instar larval brains with the dCTCF antibodies (Figure 5A–C). Clearly, all 10 dCTCF binding sites within BX-C (Holohan et al, 2007) are bound by dCTCF in larval chromatin of wild-type brains (Figure 5B). Sites BXC-67 and BXC-168 are not or only weakly bound. In contrast to wild-type larval chromatin, mutant larval brains show no dCTCF occupancy except for BXC-14, where some residual dCTCF binding to chromatin can be seen (Figure 5C). A control precipitation with an antibody specific for histone H3 resulted in similar PCR signals for most of the sites in the wild-type as well in the mutant background.
Figure 5.
dCTCF binds to 13 sequences in the BX-C. (A) Summary of the dCTCF target sites (dCTSs:BXC-14–BXC-228) in the BX-C region on chromosome 3R. Coordinates are corresponding to genome release 5.1. The genomic locus with nine regulatory domains (Maeda and Karch, 2006), and the Ubx, abd-A and Abd-B genes are shown. (B) dCTCF occupies all of the CTSs in larval chromatin. ChIP was performed with chromatin from third instar larval brains with C- or N-terminal dCTCF-specific antibodies. Negative controls were preimmune serum or a non-binding sequence (Fab8 5′-control). BXC-67 and BXC-168 are only bound in vitro (Holohan et al, 2007). (C) ChIP with mutant strain GE24185/Df(3L)0463 chromatin from larval brains. All dCTS but BXC-14 are negative for dCTCF-specific precipitation. Control precipitation of histone H3 shows precipitation with either chromatin from wild type (H3-wt) or mutant (H3-mut) extracts. (D) CP190 is present at all dCTSs of the BX-C. Same extracts as in panel B were precipitated with CP190-specific antibody.
As we have found an interaction and colocalization of dCTCF and CP190 (Figures 1, 2 and 4), we asked whether CP190 might be present at dCTCF-sites within the BX-C locus. We performed ChIP analysis with a CP190-specific antibody (Figure 5D). Clearly, all of the CTSs are positive for CP190. In contrast, a negative control sequence (Su(var)3–9) did not show any CP190 association, supporting the specificity of the CP190 ChIP experiment. Therefore we conclude that dCTCF and CP190 do occupy similar target sites in the BX-C.
The correct function of the BX-C locus requires dCTCF
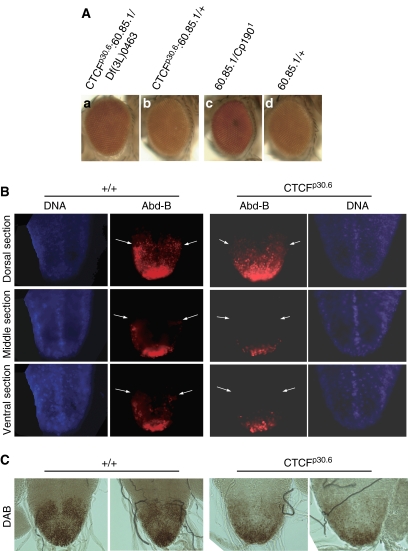
Previously we have shown that the Fab-8 sequence harbors two adjacent dCTCF binding sites, and that the mutation of these sites causes loss of enhancer blocking in a reporter assay. With the dCTCF mutants now available, we wanted to confirm that Fab-8 enhancer blocking requires dCTCF. The enhancers previously used for this analysis were only active in embryonic tissues. As dCTCF is provided as a maternal component (Moon et al, 2005), we decided to use a reporter system where expression could be analyzed in the adult tissue. We chose the Fab-8 enhancer blocking line 60.85.1 (kindly provided by P Schedl), which is similar to nine other lines showing enhancer blocking activity of Fab-8 (Barges et al, 2000). The line harbors the Fab-8 boundary inserted in between the white enhancer and the mini-white reporter on the third chromosome. This line shows weak enhancer blocking, resulting in reduced eye pigment (Figure 6Ad). Any relief from enhancer blocking should increase eye color. We found that a low, but significant relief in enhancer blocking is seen in null CTCF background (CTCFp30.6:60.85.1/Df(3L)0463) (Figure 6Aa and b). All trans-heterozygote females (17 individuals) showed an increase in eye pigmentation compared with nearly all heterozyogtes (49 of 51 showed weaker pigmentation). Next we analyzed whether a mutation in Cp190 has an effect on Fab-8 enhancer blocking. We found that a single copy of Cp1901 is sufficient to impair enhancer blocking (Figure 6Ac and d). All of the Cp1901 heterozygote flies showed the increase in eye pigmentation. Thus, Fab-8-mediated enhancer blocking requires both dCTCF and CP190.
Figure 6.
dCTCF affects Fab-8 insulator function and Abd-B expression. (A) Enhancer blocking assay using the Fab8 reporter line 60.85.1 (Barges et al, 2000) in dCTCF and Cp190 mutant females. 60.85.1 shows more pigmentation in null dCTCF background (a) compared with the heterozygous CTCF mutation (b). In the presence of one copy of CP1901, 60.85.1 shows higher amount of pigmentation (c) compared with wild-type background (d). (B) Abd-B antibody detects expression in wild-type 3rd instar larval nerve cord in dorsal, middle and ventral optical sections. The expression is reduced in CTCFp30.6 homozygotes. Arrows mark the anterior Abd-B expression border in wild type and a comparable region in the mutant. (C) Similar as in panel B, except that Abd-B staining was visualized by DAB reaction, which can be controlled more accurately to reflect the antigen amount.
Given that dCTCF binds to 10 sites in the bithorax locus at known or predicted insulators (see above), and since dCTCF has an effect on the Fab-8 insulator (Moon et al, 2005), we wondered whether the expression of Abd-B might be altered in the absence of dCTCF. Abd-B is expressed in various tissues and is important for organogenesis (Lovegrove et al, 2006). We expected that the high amount of maternal dCTCF (Moon et al, 2005) might be functional in early stages of development of amorphic dCTCF mutants, and therefore analyzed the expression at later stages. Abd-B immunostaining in the larval nerve cord revealed an expression pattern in parasegments PS10–PS14 (Figure 6B) that is very similar to the expression pattern as seen by in situ hybridization (Schmitt et al, 2005). The expression in homozygote CTCFp30.6 is clearly reduced when compared with wild type as seen in fluorescent images (Figure 6B). We also performed Abd-B staining followed by DAB reaction to control saturation of signals, which can occur with fluorescent dyes. Samples with the same reaction times are shown, and they show a similar reduction of Abd-B staining in dCTCF mutants (Figure 6C). Therefore, within the potential effects mediated by dCTCF at the many binding sites throughout the genome, Abd-B within the bithorax locus is one of the functional dCTCF targets.
Discussion
Insulator elements with enhancer blocking activity establish independent regulatory domains. An analysis of binding sites (CTS) for the enhancer blocking factor dCTCF on salivary gland polytene chromosomes resulted in the identification of several hundred sites bound by dCTCF. All of these sites are found in interbands, and when inspected more precisely are often at the borders of interbands next to bands. Interbands harbor active housekeeping genes or regulatory regions of inactive genes, whereas bands contain the bodies of inactive genes (Demakov et al, 2004). Interbands and bands differ in chromatin composition and modification (Ebert et al, 2006). Thus, there is a clear border between interbands and bands. Any factors generating functional chromatin boundaries would be expected to be localized to the interband/band transition. This is not only the case for dCTCF, as a similar location has been found for Su(Hw) (Pai et al, 2004). Also, BEAF-32 and Zw5 are located in interbands at hundreds of binding sites throughout the genome (Zhao et al, 1995; Cuvier et al, 1998; Blanton et al, 2003; Parnell et al, 2006; Ramos et al, 2006; Roy et al, 2006).
The obvious question was whether dCTCF has a redundant function and therefore similar targets as the other Drosophila enhancer blocking factors. We could not detect a significant colocalization of dCTCF with either BEAF-32 (data not shown) or with Su(Hw) on polytene chromosomes. This may provide an explanation of how an organism with a small genome, such as Drosophila, can prevent promiscuous enhancer interaction with any nearby gene. Apparently, an elaborate system of different enhancer blockers and barrier factors fulfills the insulation of regulatory units.
The biochemical composition and function of insulator complexes involving Su(Hw) have been studied in detail. The best studied binding site is the gypsy transposon with a 350-bp sequence containing 12 binding sites for Su(Hw) (Geyer and Corces, 1992). A functional complex of Su(Hw), Mod(mdg4)67.2, CP190, and possibly other factors has been documented (Capelson and Corces, 2005; Lei and Corces, 2006). Although there is no colocalization of Su(Hw) with dCTCF on polytene chromosomes, and only partial colocalization with Mod(mdg4) (data not shown), we were interested in examining whether CP190 plays a role in dCTCF function. We have shown that vertebrate CTCF is a centrosomal factor during mitosis and a nuclear protein during interphase (Zhang et al, 2004), and that CP190 (centrosome binding protein) is associated with centrosomes as well. CP190 is essential for viability, but is not required for cell division (Butcher et al, 2004). CP190 knockdown in Schneider cells has no effect, whereas a null mutation in flies leads to pharate lethality. Here we have seen a similar phenotype after dCTCF depletion in Schneider cells and pharate lethality in flies as well. The centrosomal function of CP190 is not required for the insulator activity in the context of Su(Hw) bound to gypsy (Pai et al, 2004). The localization of CP190 on polytene chromosomes overlaps with sites bound by Su(Hw) or by Mod(mdg4)67.2. In addition, CP190 is found at loci devoid of Su(Hw) or Mod(mdg4)67.2, suggesting that other factors might recruit CP190 to these sites (Pai et al, 2004). There is a significant overlap in dCTCF with CP190 binding sites. A functional dependence is seen, as at many sites binding of dCTCF depends on CP190. Although there is an overall reduction in the dCTCF amount observed in the CP190 mutant, differences in dCTCF occupancy in dCTCF and CP190 mutants indicate a discrimination between CP190-independent and -dependent sites. Furthermore, the previously characterized insulator Fab-8 (Barges et al, 2000) is impaired in the absence of dCTCF (Moon et al, 2005) and by the reduction of CP190.
Another perspective on the requirement of insulators comes from the fact that many genes are controlled by several regulatory elements that are required for tissue and cell-specific expression. A prominent example is the Drosophila BX-C. This is one of two Hox gene clusters, which contain regulator genes controlling development. The BX-C is responsible for the correct specification of the posterior thorax segment (T3) and all of the abdominal segments (for review see Maeda and Karch, 2006). Within BX-C, only three protein coding genes, Ubx, abd-A and Abd-B, are responsible for the segment-specific development of organs and tissues. On the other hand, nine separate groups of many mutations are affecting segment-specific functions (Lewis, 1978). The borders of some of these domains are genetically defined by elements Fab-6, Fab-7, Fab-8 and by Mcp (Hagstrom et al, 1996; Barges et al, 2000; Gruzdeva et al, 2005; Mihaly et al, 2006). Proteins involved in such a functional separation are the GAGA factor in case of the Fab-7 element (Schweinsberg et al, 2004), and dCTCF for the Fab-8 sequence (Moon et al, 2005). Recently we have demonstrated that six of the BX-C domain junctions are bound by dCTCF (Holohan et al, 2007). Consequently, if these would contribute to boundary function, gene activity within this locus should be changed. Indeed, we find a homeotic phenotype and a reduced expression of Abd-B in larval nerve cord. If dCTCF plays a central role in separating the different regulator domains in the BX-C and elsewhere in the genome, it is difficult to predict the dCTCF phenotype. The situation could be complicated as the three BX-C genes are controlling realizator genes as well as other regulators (Pearson et al, 2005; Jeong et al, 2006; Lovegrove et al, 2006). Furthermore, individual BX-C genes repress others, for example Abd-B as well as the miRNA iab-4 and bxd expression repress Ubx (Struhl and White, 1985; Choi et al, 2000; Ronshaugen et al, 2005; Petruk et al, 2006). In addition, other factors, such as CP190 and perhaps additional unknown factors may contribute to the enhancer blocking function of dCTCF. For all of the CTS in the BX-C, we find dCTCF and CP190 binding. Although both factors clearly interact as seen by co-immunoprecipitation, CP190 may contact other DNA-bound factors as well, or may be directly targeted to chromatin (Pai et al, 2004).
Thus, dCTCF shares several biochemical and functional features with Su(Hw), but is clearly targeted to dCTCF-specific sites. Overall, we show that dCTCF is important for fly development, and has important functions in the regulation of abdominal segmentation.
Materials and methods
Fly strains, transgenic flies and plasmid constructs
Fly crosses were maintained at 25°C on standard medium. Hsp70Gal, actin-Gal4 and tubulin-Gal4 were from Bloomington stock center. P(EPgy2)CTCFEY15833 was from the Genome Disruption project and supplied by H Bellen. PBac{WH}qmf01604 and P{XP}bhrd01563 were provided by the Drosophila Stock Collection at Harvard Medical School. GE24185 and GE22007 were purchased from GenExcel. Cp1901 was provided by WGF Whitfield and JW Raff. For jump-outs from GE22007, EY15833 and GE24185 we screened 59, 131 and 169 lines in the background of Df(3L)0463. From these, 8, 2 and 7 lines showed lethality at pharate stage. At least one pharate lethal line from each P-element was chosen for further analysis. As both GE22007 and GE24185 have secondary mutations (Table I), we performed complementation assays between the different jump-out lines. Crossing females from dCTCFp30.6or dCTCFp35.2 (both generated from GE22007) to males from dCTCFE5.5 (from EY15833), dCTCFR15.4 or dCTCFR10.3 (both generated from GE24185) resulted in offspring with pharate lethality in all cases (not shown), verifying that the phenotype is caused by the loss of CTCF.
UASdCTCF-EGFP and gCTCF transgenic lines were generated with single inserts on different chromosomes. Heat shock was given by immersing the tubes in a 37°C water bath. The Fab-8 reporter stock was kindly provided by P Schedl. The eye color was determined at identical times and photographed: 12 h after eclosion for the dCTCF mutants and controls, and 48 h for the CP190 mutant and control.
For plasmid construction see Supplementary data.
Co-immunoprecipitation, Western blot and chromatin immunoprecipitation
Schneider cells were stably transfected with pRm-HA-FLAG-dCTCF. A single cell clone was induced with 500 μM CuSO4 for 24 h. Extract preparation and immunoprecipitation were performed as described (Eckey et al, 2007). CP190-specific antibody (clone Bx63; Frasch et al, 1986; Whitfield et al, 1995) or FLAG-M2 antibody (Sigma) was used for precipitation. SDS–PAGE and Western blot were performed as previously described (Moon et al, 2005). α-Tubulin (DHSB) was used at 1:2000, and α-CP190 at 1:2000. Chromatin immunoprecipitation was essentially as described (Holohan et al, 2007), except that larval brain tissue was used. For details see Supplementary data.
Immunohistochemistry
Third instar larval brains were stained with α-Abd-B (DSHB) following standard procedures. Hoechst 33342 was used to stain DNA. DAB (Sigma, Deisenhofen) staining was performed as described (Putz et al, 2005). Care was taken to perform the reaction for the same amount of time in both wild type and mutants. For salivary gland polytene chromosomes, flies were grown on semi-defined medium (Flyfood-Bloomington) at 18°C. Polytene chromosome staining was performed as described (Saumweber et al, 1990). To detect dCTCF we used either α-dCTCF N- or C-terminal antisera. CP190 was detected with the Bx63 monoclonal antibody (Frasch et al, 1986; Whitfield et al, 1988). Su(Hw) was detected with a rat antibody kindly provided by V Corces. A Zeiss Apotome or a Deltavsion image restoration microscope was used to capture images, and Photoshop was used to process images.
Abdominal cuticle preparations
Flies (3–4 days old) were killed in ethanol and incubated in 10% KOH solution for 1h at 65°C. Adult abdominal cuticles were cut along the dorsal midline and mounted with a drop of Hoyer's medium (Jeong et al, 2006). The cuticles were flattened with a coverslip and incubated for 3 h at 65°C.
Supplementary Material
Supplementary Figure 1
Supplementary materials
Acknowledgments
We thank Hugo J Bellen, Victor Corces, Rainer Dorn, Ian Duncan, Jordan W Raff, Paul Schedl, Jean-Paul Vincent and William GF Whitfield for materials, Francois Karch and Henrik Gyurkovics for their help in the cuticle interpretation, and Ernesto Sánchez-Herrero, Jishy Varghese and Manish Jaiswal for advice. We thank Eva Günther for help in cloning and Nadine Müller and Leni Schäfer-Pfeiffer for technical assistance. We are thankful to Development Studies Hybridoma Bank for supplying the anti-Abd-B and anti-Tubulin antibodies, Bloomington stock center and Drosophila Stock Collection at Harvard Medical School for providing fly reagents and Drosophila Genomics Resource Center for pCasper4 vector. This work was supported by grants from the Deutsche Forschungsgemeinschaft.
References
- Baniahmad A, Steiner C, Kohne AC, Renkawitz R (1990) Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell 61: 505–514 [DOI] [PubMed] [Google Scholar]
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F (2000) The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127: 779–790 [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Belozerov VE, Majumder P, Shen P, Cai HN (2003) A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J 22: 3113–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton J, Gaszner M, Schedl P (2003) Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev 17: 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke LJ, Zhang R, Bartkuhn M, Tiwari VK, Tavoosidana G, Kurukuti S, Weth C, Leers J, Galjart N, Ohlsson R, Renkawitz R (2005) CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J 24: 3291–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RD, Chodagam S, Basto R, Wakefield JG, Henderson DS, Raff JW, Whitfield WG (2004) The Drosophila centrosome-associated protein CP190 is essential for viability but not for cell division. J Cell Sci 117: 1191–1199 [DOI] [PubMed] [Google Scholar]
- Capelson M, Corces VG (2005) The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell 20: 105–116 [DOI] [PubMed] [Google Scholar]
- Choi SH, Oh CT, Kim SH, Kim YT, Jeon SH (2000) Effects of Polycomb group mutations on the expression of Ultrabithorax in the Drosophila visceral mesoderm. Mol Cell 10: 156–161 [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Cuvier O, Hart CM, Laemmli UK (1998) Identification of a class of chromatin boundary elements. Mol Cell Biol 18: 7478–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demakov S, Gortchakov A, Schwartz Y, Semeshin V, Campuzano S, Modolell J, Zhimulev I (2004) Molecular and genetic organization of Drosophila melanogaster polytene chromosomes: evidence for two types of interband regions. Genetica 122: 311–324 [DOI] [PubMed] [Google Scholar]
- Ebert A, Lein S, Schotta G, Reuter G (2006) Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res 14: 377–392 [DOI] [PubMed] [Google Scholar]
- Eckey M, Hong W, Papaioannou M, Baniahmad A (2007) The nucleosome assembly activity of NAP1 is enhanced by Alien. Mol Cell Biol 27: 3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Casares F, Busturia A, Sanchez-Herrero E (2002) Genetic and molecular characterization of a novel iab-8 regulatory domain in the Abdominal-B gene of Drosophila melanogaster. Development 129: 5195–5204 [DOI] [PubMed] [Google Scholar]
- Frasch M, Glover DM, Saumweber H (1986) Nuclear antigens follow different pathways into daughter nuclei during mitosis in early Drosophila embryos. J Cell Sci 82: 155–172 [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Gaszner M, Vazquez J, Schedl P (1999) The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer–promoter interaction. Genes Dev 13: 2098–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG (1995) A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597 [DOI] [PubMed] [Google Scholar]
- Geyer PK, Corces VG (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865–1873 [DOI] [PubMed] [Google Scholar]
- Ghosh D, Gerasimova TI, Corces VG (2001) Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J 20: 2518–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P (2005) The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol Cell Biol 25: 3682–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P (1996) Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev 10: 3202–3215 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Holdridge C, Dorsett D (1991) Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol 11: 1894–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R (2007) CTCF genomic binding sites in Drosophila and the organisation of the Bithorax complex. PLoS Genet 3: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB (2006) Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125: 1387–1399 [DOI] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10: 853–856 [DOI] [PubMed] [Google Scholar]
- Kuhn EJ, Viering MM, Rhodes KM, Geyer PK (2003) A test of insulator interactions in Drosophila. EMBO J 22: 2463–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT (2003) Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr Biol 13: R242–R254 [DOI] [PubMed] [Google Scholar]
- Lei EP, Corces VG (2006) RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet 38: 936–941 [DOI] [PubMed] [Google Scholar]
- Lewis A, Reik W (2006) How imprinting centres work. Cytogenet Genome Res 113: 81–89 [DOI] [PubMed] [Google Scholar]
- Lewis EB (1978) A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH (1990) A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5: 1743–1753 [PubMed] [Google Scholar]
- Lovegrove B, Simoes S, Rivas ML, Sotillos S, Johnson K, Knust E, Jacinto A, Hombria JC (2006) Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Curr Biol 16: 2206–2216 [DOI] [PubMed] [Google Scholar]
- Lutz M, Burke LJ, LeFevre P, Myers FA, Thorne AW, Crane-Robinson C, Bonifer C, Filippova GN, Lobanenkov V, Renkawitz R (2003) Thyroid hormone-regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. EMBO J 22: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda RK, Karch F (2006) The ABC of the BX-C: the bithorax complex explained. Development 133: 1413–1422 [DOI] [PubMed] [Google Scholar]
- Mihaly J, Barges S, Sipos L, Maeda R, Cleard F, Hogga I, Bender W, Gyurkovics H, Karch F (2006) Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development 133: 2983–2993 [DOI] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, Burke LJ, Renkawitz-Pohl R, Ohlsson R, Zhou J, Renkawitz R, Lobanenkov V (2005) CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin YM, Alekseyenko AA, Semeshin VF, Spierer A, Spierer P, Makarevich GF, Belyaeva ES, Zhimulev IF (2001) The bithorax complex of Drosophila melanogaster: underreplication and morphology in polytene chromosomes. Proc Natl Acad Sci USA 98: 570–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M (1998) GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev 12: 3325–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG (2004) The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748 [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS et al. (2004) Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet 36: 288–292 [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Kuhn EJ, Gilmore BL, Helou C, Wold MS, Geyer PK (2006) Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol Cell Biol 26: 5983–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W (2005) Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6: 893–904 [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A (2006) Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz M, Kesper DA, Buttgereit D, Renkawitz-Pohl R (2005) In Drosophila melanogaster, the Rolling pebbles isoform 6 (Rols6) is essential for proper Malpighian tubule morphology. Mech Dev 122: 1206–1217 [DOI] [PubMed] [Google Scholar]
- Ramos E, Ghosh D, Baxter E, Corces VG (2006) Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics 172: 2337–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC (2005) The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev 19: 2947–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Tan YY, Hart CM (2006) A genetic screen supports a broad role for the Drosophila insulator proteins BEAF-32A and BEAF-32B in maintaining patterns of gene expression. Mol Genet Genomics 277: 273–286 [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E, Vernos I, Marco R, Morata G (1985) Genetic organization of Drosophila bithorax complex. Nature 313: 108–113 [DOI] [PubMed] [Google Scholar]
- Saumweber H, Frasch M, Korge G (1990) Two puff-specific proteins bind within the 2.5 kb upstream region of the Drosophila melanogaster Sgs-4 gene. Chromosoma 99: 52–60 [DOI] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R (2005) Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev 19: 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F (2004) The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168: 1371–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, White RA (1985) Regulation of the Ultrabithorax gene of Drosophila by other bithorax complex genes. Cell 43: 507–519 [DOI] [PubMed] [Google Scholar]
- Szabo P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR (2000) Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 10: 607–610 [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D et al. (2004) A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Vostrov AA, Quitschke WW (1997) The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. evidence for a role in transcriptional activation. J Biol Chem 272: 33353–33359 [DOI] [PubMed] [Google Scholar]
- Whitfield WG, Chaplin MA, Oegema K, Parry H, Glover DM (1995) The 190 kDa centrosome-associated protein of Drosophila melanogaster contains four zinc finger motifs and binds to specific sites on polytene chromosomes. J Cell Sci 108 (Pt 11): 3377–3387 [DOI] [PubMed] [Google Scholar]
- Whitfield WG, Millar SE, Saumweber H, Frasch M, Glover DM (1988) Cloning of a gene encoding an antigen associated with the centrosome in Drosophila. J Cell Sci 89 (Pt 4): 467–480 [DOI] [PubMed] [Google Scholar]
- Zhang R, Burke LJ, Rasko JE, Lobanenkov V, Renkawitz R (2004) Dynamic association of the mammalian insulator protein CTCF with centrosomes and the midbody. Exp Cell Res 294: 86–93 [DOI] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK (1995) Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81: 879–889 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary materials