Abstract
The activity state of cofilin, which controls actin dynamics, is driven by a phosphorylation–dephosphorylation cycle. Phosphorylation of cofilin by LIM-kinases results in its inactivation, a process supported by 14-3-3ζ and reversed by dephosphorylation by slingshot phosphatases. Here we report on a novel cellular function for the phosphorylation–dephosphorylation cycle of cofilin. We demonstrate that muscarinic receptor-mediated stimulation of phospholipase D1 (PLD1) is controlled by LIM-kinase, slingshot phosphatase as well as 14-3-3ζ, and requires phosphorylatable cofilin. Cofilin directly and specifically interacts with PLD1 and upon phosphorylation by LIM-kinase1, stimulates PLD1 activity, an effect mimicked by phosphorylation-mimic cofilin mutants. The interaction of cofilin with PLD1 is under receptor control and encompasses a PLD1-specific fragment (aa 585–712). Expression of this fragment suppresses receptor-induced cofilin–PLD1 interaction as well as PLD stimulation and actin stress fiber formation. These data indicate that till now designated inactive phospho-cofilin exhibits an active cellular function, and suggest that phospho-cofilin by its stimulatory effect on PLD1 may control a large variety of cellular functions.
Keywords: cofilin, LIM-kinase1, phospholipase D, slingshot, 14-3-3
Introduction
The dynamic nature of actin filament assembly/disassembly and its cellular organization is regulated by several actin-binding and -regulatory proteins, including the actin-depolymerizing factor (ADF)/cofilin family (Bamburg, 1999; Bamburg and Wiggan, 2002; Pollard and Borisy, 2003; DesMarais et al, 2005). Members of the ADF/cofilin family (hereafter referred to as cofilin) are now considered to be pivotal regulators of actin filament dynamic-dependent processes, including cell motility, neuronal pathfinding, membrane dynamics, establishment of cell polarity, cell division and apoptosis, and their dysfunction seems to contribute to the progression of diseases as diverse as cancer, Alzheimer's dementia and ischemic kidney disease (Bamburg and Wiggan, 2002; Chua et al, 2003; Wang et al, 2006). Cofilin binds to both G- and F-actin, but due to a higher affinity for ADP-bound subunits, the off-rate of actin monomers from the pointed end of actin filaments is increased; in addition, cofilin severs actin filaments and thus directly generates free actin barbed ends (Condeelis, 2001; DesMarais et al, 2005).
The cofilin–actin interaction is tightly controlled by phosphocycling. Phosphorylation of cofilin at serine 3, located in the actin-binding domain, by the LIM (Lin-11/Isl-1/Mec-3) and the TES (testicular protein) kinases results in its inactivation, and the Unphosphorylatable S3A cofilin mutant as well as the phosphorylation-mimic cofilin mutants (S3D cofilin and S3E cofilin) have been widely used to define the role of cofilin in cellular responses (Bamburg, 1999; Huang et al, 2006). The two LIM-kinases, LIM-kinase1 and LIM-kinase2, are expressed in most tissues (Foletta et al, 2004; Acevedo et al, 2006) and are activated by Rho GTPases, through their effectors, Rho-kinase and p21-activated protein kinases (Edwards and Gill, 1999; Kaibuchi et al, 1999). Recently, interaction of LIM-kinase1 and cofilin with the scaffold protein 14-3-3ζ has been reported (Birkenfeld et al, 2003), leading to accumulation of inactive, phosphorylated cofilin (Gohla and Bokoch, 2002). Dephosphorylation of cofilin results in its reactivation, and is catalyzed by the cofilin-specific phosphatases of the slingshot family (Niwa et al, 2002; Kaji et al, 2003; Ohta et al, 2003) and chronophin (Gohla et al, 2005; Huang et al, 2006). The activity of the slingshot phosphatases is apparently also controlled by multiple signaling pathways, including Ca2+, cyclic AMP and phosphatidylinositol 3-kinase (for recent review see Huang et al, 2006). It has been also shown that the slingshot phosphatase 1L not only dephosphorylates cofilin but also LIM-kinase1, resulting in its inactivation, and that the phosphatase activity is regulated by F-actin and 14-3-3ζ (Nagata-Ohashi et al, 2004; Soosairajah et al, 2005).
Phospholipase D (PLD) enzymes, PLD1 splice variants and PLD2, hydrolyze phosphatidylcholine of cell membranes to phosphatidic acid (PA), in response to stimuli, and are considered to be involved in a large variety of early and late cellular responses, including calcium mobilization, secretion, superoxide production, endocytosis, exocytosis, vesicle trafficking, glucose transport, mitogenesis and apoptosis (Exton, 2002). The rise in cellular PA, in particular by PLD1, has also been reported to induce stress fiber formation in suitable cell types (Ha and Exton, 1993; Cross et al, 1996; Kam and Exton, 2001; Porcelli et al, 2002; Komati et al, 2005). As PA-activatable phosphatidylinositol-4-phosphate 5-kinase (PIP 5-kinase) generates phosphatidylinositol 4,5-bisphosphate (PIP2) (reviewed in Oude Weernink et al, 2004a), known to act as a PLD cofactor and to associate with a plethora of actin-binding proteins that regulate actin dynamics (Exton, 2002; Yin and Janmey, 2003; Hilpela et al, 2004), such mechanisms may act in concert to promote actin cytoskeleton reorganization. PLD and PIP 5-kinase are now also recognized as effectors of Rho-dependent Rho-kinase (Schmidt et al, 1999; Oude Weernink et al, 2000, 2004b; Kam and Exton, 2001; Cummings et al, 2002; Yamazaki et al, 2002). Vice versa, components of the actin regulatory machinery have been found to affect PLD. PLD activity, primarily PLD2, is negatively regulated by β-actin and the actin-binding protein α-actinin (Park et al, 2000; Lee et al 2001). Meanwhile, it has been reported that G-actin inhibits PLD1 as well, but that F-actin has the opposite effect, suggesting that specifically PLD1 may act as an actin dynamic responsive cellular element (Kusner et al, 2002). Here we report on a novel molecular link between the actin cytoskeleton and PLD1. We demonstrate that receptor-induced and Rho/Rho-kinase-dependent activation of PLD1 is under control of LIM-kinase1 and its substrate, cofilin. Cofilin directly interacts with and stimulates in its phosphorylated state PLD1. Our data, thus, indicate for the first time that phospho-cofilin, until now considered to be biologically inactive, is an active cellular component, which by its signaling to PLD1 can regulate essential cellular functions known to be under control of PLD.
Results
Involvement of cofilin in muscarinic receptor signaling to PLD
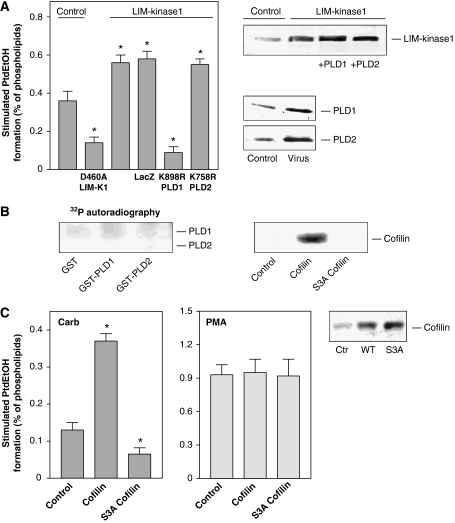
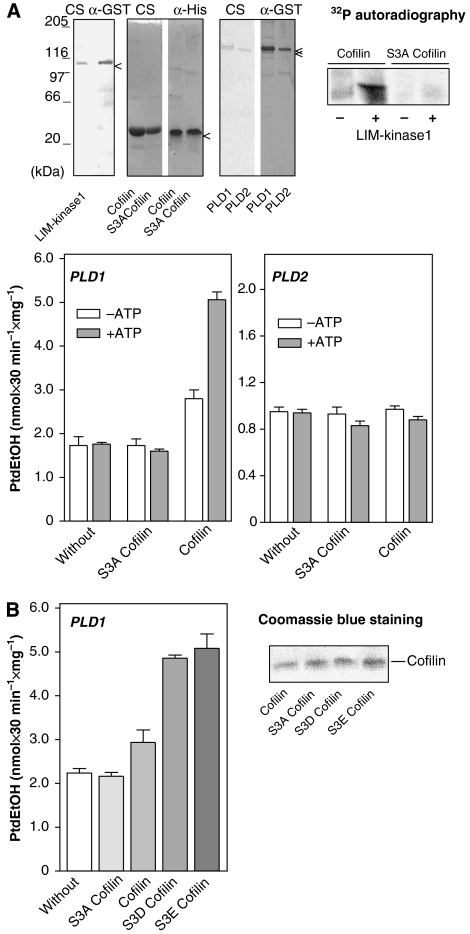
The Rho effector, Rho-kinase, mediates PLD stimulation by G-protein-coupled muscarinic acetylcholine receptors (mAChRs) in HEK-293 and N1E-115 neuroblastoma cells (Schmidt et al, 1999; Kam and Exton, 2001; Cummings et al, 2002). As PLD enzymes neither directly interact with, nor are phosphorylated by Rho-kinase (Schmidt et al, 1999), we considered the involvement of the Rho-kinase effector, LIM-kinase, in mAChR signaling to PLD. As shown in Figure 1A, overexpression of LIM-kinase1 in HEK-293 cells strongly increased PLD stimulation by the mAChR agonist, carbachol, whereas expression of kinase-deficient D460A LIM-kinase1 had the opposite effect. The carbachol-induced and LIM-kinase1-reinforced PLD stimulation was almost fully blunted by adenoviral expression of lipase-inactive K898R PLD1, whereas lipase-inactive K758R PLD2 had no effect. Similar data were obtained for mAChR regulation of PLD activity in N1E-115 neuroblastoma cells (data not shown). These data suggested that the receptor-induced PLD stimulation not only involves Rho-kinase, but probably also its substrate, LIM-kinase, and that the PLD isozyme responsible for the increased PLD activity is PLD1. Consequently, we studied whether LIM-kinase1 may phosphorylate PLD1. However, as shown in Figure 1B, purified recombinant LIM-kinase1 did neither phosphorylate purified GST-tagged PLD1 nor PLD2. LIM-kinase1 did also not bind to the PLD enzymes (data not shown). The purified LIM-kinase1 was active, as it phosphorylated, as expected, wild-type cofilin, but not its mutant, S3A cofilin (Figure 1B).
Figure 1.
Cofilin, the sole substrate of LIM-kinase, regulates stimulation of PLD1 by the M3 mAChR. HEK-293 cells were transfected with kinase-deficient LIM-kinase1 (D460A LIM-K1) or wild-type LIM-kinase1, either alone or with adenoviruses encoding LacZ, lipase-inactive K898R PLD1 or lipase-inactive K758R PLD2 (A), or transfected with empty vector (Control), wild-type cofilin and Unphosphorylatable S3A cofilin (C). After 48 h, stimulated [3H]PtdEtOH accumulation was determined in the presence of 1 mM carbachol (Carb) (A, C) or 100 nM PMA (C). Data shown are means±s.e. (n=3–4). The immunoblots demonstrate expression of LIM-kinase1, PLD enzymes and cofilin in cell lysates. (B) [γ-32P]ATP phosphorylation (32P autoradiography) of GST, GST-tagged PLD1, GST-tagged PLD2 (full-length each) and wild-type cofilin and S3A cofilin by LIM-kinase1. Data are representative of three to four similar experiments. *P<0.05.
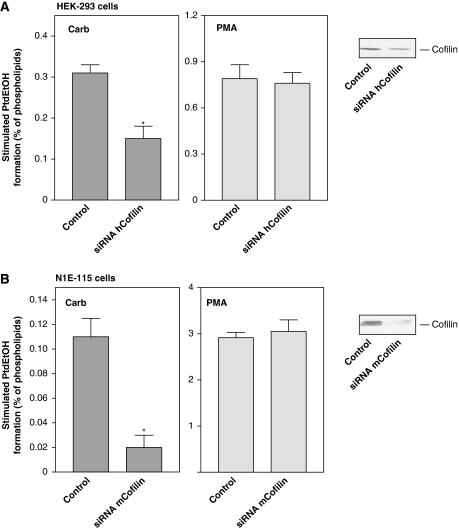
These data prompted us to study whether the LIM-kinase substrate, cofilin, may be involved in PLD stimulation. As illustrated in Figure 1C, overexpression of wild-type cofilin in HEK-293 cells greatly enhanced, by about 2.5-fold, stimulation of PLD by the mAChR agonist, carbachol, whereas expression of Unphosphorylatable S3A cofilin suppressed PLD stimulation by about 50%. In contrast, expression of wild-type and S3A cofilin did not alter PLD stimulation by the phorbol ester, phorbol 12-myristate 13-acetate (PMA), which is Rho- and Rho-kinase independent in HEK-293 cells (Voß et al, 1999), and which was also not affected by expression of LIM-kinase1 variants (data not shown). To confirm that cofilin mediates mAChR-induced PLD stimulation, human HEK-293 and mouse N1E-115 cells were transfected with siRNA pSUPER expression plasmids, which direct the synthesis of siRNAs targeting either human or mouse cofilin, respectively (Nishita et al, 2005; Kiuchi et al, 2007). As shown in Figure 2, these maneuvers greatly reduced the cellular content of cofilin in both cell types. Silencing of cellular cofilin in HEK-293 and N1E-115 neuroblastoma cells reduced the carbachol-induced PLD stimulation by about 60 and 80%, respectively (Figure 2). In contrast, knockdown of cofilin expression did not alter PLD stimulation by PMA in either cell type.
Figure 2.
Depletion of cellular cofilin reduces PLD stimulation by carbachol. HEK-293 cells were transfected with human cofilin siRNA pSUPER plasmid (siRNA hCofilin), or with the empty pSUPER vector (Control) (A). N1E-115 neuroblastoma cells were transfected with mouse cofilin siRNA pSUPER plasmid (siRNA mCofilin), or with the empty pSUPER vector (Control) (B). After 48 h, stimulated [3H]PtdEtOH accumulation was determined in the presence of 1 mM carbachol (Carb) or 100 nM PMA. Data shown are means±s.e. (n=3–4). The immunoblots demonstrate endogenous expression of cofilin in lysates of cells transfected with empty pSUPER vector (Control) or the indicated siRNA pSUPER plasmids. *P<0.05.
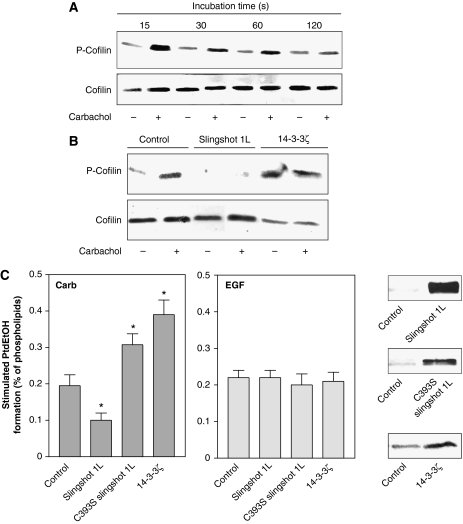
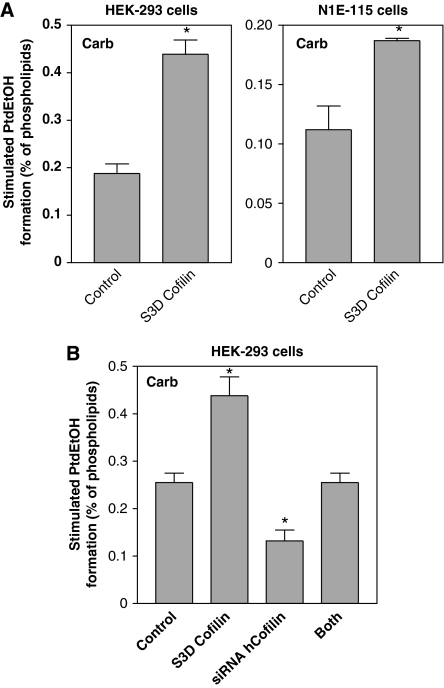
These findings together suggested that cofilin, possibly in its phosphorylated state, may control the receptor stimulation of PLD. To substantiate this assumption, we first studied whether receptor activation leads to cofilin phosphorylation, and whether this phosphorylation is under control of a cofilin-specific phosphatase and the scaffold protein, 14-3-3ζ. As illustrated in Figure 3A, agonist activation of the mAChR induced a strong, but rather transient phosphorylation of cofilin in HEK-293 cells. The stimulatory effect of carbachol reached its maximum at 15 s and rapidly declined thereafter. Expression of slingshot phosphatase 1L completely abolished phosphorylation of cofilin, both in the basal state and after stimulation by carbachol (Figure 3B). In contrast, expression of 14-3-3ζ and inactive slingshot phosphatase 1L (not shown) strongly increased phosphorylation of cofilin in unstimulated cells, and there was no additional increase by the receptor activation. We then examined the effects of the slingshot and 14-3-3ζ on mAChR stimulation of PLD activity. Expression of slingshot phosphatase 1L significantly decreased PLD stimulation by carbachol, whereas inactive slingshot phosphatase 1L and 14-3-3ζ enhanced PLD stimulation by carbachol (Figure 3C, left panel). In contrast, PLD stimulation by epidermal growth factor (EGF), which similar to PLD stimulation by PMA, is independent of Rho and Rho-kinase in these cells (Voß et al, 1999), was not altered by expression of these proteins controlling the phosphorylation state of cofilin (Figure 3C, right panel). In line with a role of cofilin phosphorylation in PLD stimulation, expression of the phosphorylation-mimic S3D cofilin mutant strongly increased the carbachol-induced PLD stimulation, both in HEK-293 and N1E-115 neuroblastoma cells (Figure 4A). We next studied whether the expression of phosphorylation-mimic S3D cofilin could counteract the inhibitory effect of cofilin silencing on the PLD response. As shown in Figure 4B, expression of phosphorylation-mimic S3D mouse cofilin in cofilin-silenced HEK-293 cells considerably rescued PLD stimulation by carbachol.
Figure 3.
Regulation of the cellular phosphorylation state of cofilin and its impact on the PLD response. HEK-293 cells were stimulated without (−) or with (+) 1 mM carbachol for the indicated periods of time (A), or transfected with empty vector (Control), c-myc-tagged wild-type slingshot 1L, inactive slingshot 1L (C393S slingshot 1L) or VSV-G-tagged 14-3-3ζ, and stimulated with carbachol for 15 s (B). Phosphorylation of cofilin (P-Cofilin) and the total cellular content of cofilin were detected in cell lysates with anti-phospho-cofilin and anti-cofilin antibodies, respectively. Data are representative of three experiments. (C) Stimulated [3H]PtdEtOH accumulation was determined in the presence of 1 mM carbachol (Carb) or 100 ng/ml EGF. Data shown are means±s.e. (n=3–4). The immunoblots show expression of c-myc-tagged slingshot 1L and myc-tagged inactive slingshot 1L (C393S slingshot 1L), or VSV-G-tagged 14-3-3ζ. *P<0.05.
Figure 4.
Phosphorylation-mimic S3D cofilin potentiates PLD1 stimulation by carbachol and rescues PLD1 stimulation in cofilin-depleted cells. HEK-293 cells or N1E-115 cells were transfected with phosphorylation-mimic S3D cofilin, or with empty vector (Control) (A), or transfected with either phosphorylation-mimic S3D mouse cofilin, human cofilin siRNA pSUPER plasmid (siRNA hCofilin), or both constructs together (B). After 48 h, stimulated [3H]PtdEtOH accumulation was determined in the presence of 1 mM carbachol (Carb). Data shown are means±s.e. (n=3) (A), or are representative of three experiments (B). *P<0.05.
Cofilin binds to and alters subcellular localization of PLD1
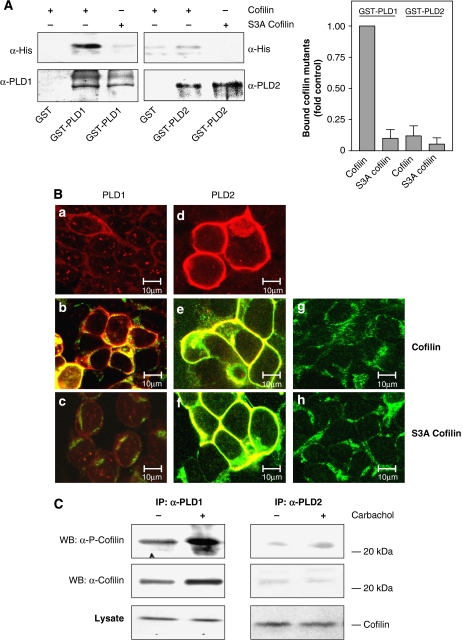
For analysis of cofilin–PLD interaction, studies were performed both in vitro with purified components and in intact cells. As illustrated in Figure 5A, purified wild-type His-tagged cofilin strongly bound to GST-tagged PLD1. In contrast, binding of Unphosphorylatable S3A cofilin to GST-tagged PLD1 was hardly detectable, similar as binding of wild-type or S3A cofilin to PLD2. Thus, cofilin can directly and specifically interact with PLD1 and this interaction apparently requires the phosphorylatable serine 3 of cofilin. To examine whether cofilin also interacts with PLD1 in intact cells, we transfected HEK-293 cells with PLD1 or PLD2 and cofilin mutants for immunofluorescence laser confocal microscopy analysis. As reported before in other cell types (Bamburg, 1999; Bamburg and Wiggan, 2002; Exton, 2002), we found that PLD1 localized to intracellular compartments and the plasma membrane, whereas PLD2 exclusively localized to the plasma membrane (Figure 5B, panels a and d); the cofilin mutants were found to be localized to the plasma membrane and intracellular compartments (Figure 5B, panels g and h). Coexpression of wild-type cofilin altered subcellular localization of PLD1 (Figure 5B, panel b). In cells coexpressing PLD1 and wild-type cofilin, PLD1 was found primarily at the plasma membrane. Coexpression of S3A cofilin and PLD1 caused only a minor subcellular redistribution of PLD1 (Figure 5B, panel c). In contrast to PLD1, the plasma membrane localization of PLD2 was not altered by coexpression of wild-type or S3A cofilin (Figure 5B, panels e and f). Thus, cofilin can specifically alter subcellular localization of PLD1.
Figure 5.
Direct cofilin–PLD1 interaction is reflected by cofilin-induced subcellular redistribution of PLD1, but not PLD2. (A) Immobilized GST, GST-tagged PLD1 and GST-tagged PLD2 were incubated with recombinant His6-tagged wild-type or unphosphorylatable S3A cofilin overnight at 4°C. Specifically bound proteins were separated by SDS–PAGE, transferred onto nitrocellulose membrane and detected by immunoblotting with anti-PLD and anti-His antibodies. Bar graph illustrates mean±s.e.m. (n=3), with the amount of wild-type cofilin bound to GST-PLD1 set to 1 (Control). (B) HEK-293 cells were transfected with PLD1 (a–c), PLD2 (d–f), HA-tagged wild-type cofilin (b, e, g) or with HA-tagged unphosphorylatable S3A cofilin (c, f, h), either alone (a, d, g, h) or with the indicated combinations. After 48 h, immunofluorescence laser confocal microscopy was performed as described in the Materials and methods section. Yellow color: merge of red (PLD) and green (cofilin) colors. Data are characteristic of three similar experiments. Scale bar, 10 μm. (C) HEK-293 cells were transfected with wild-type PLD1 or PLD2. After 48 h, the cells were treated for 15 s without (−) or with (+) 1 mM carbachol, followed by cell lysis and immunoprecipitation with anti-PLD antibodies. The PLD immunoprecipitates (IP) and total lysates were resolved by SDS–PAGE and probed with anti-phospho-cofilin (α-P-Cofilin) or anti-cofilin antibodies (α-cofilin) as indicated. The results shown are representative of 3–4 experiments. WB, Western blot.
To substantiate that cofilin interacts with PLD1 also in intact cells, co-immunoprecipitation studies were performed. As illustrated in Figure 5C, left panel, cofilin and phospho-cofilin were co-immunoprecipitated with PLD1 from lysates of HEK-293 cells coexpressing PLD1 and cofilin, demonstrating their in vivo interaction. Most important, stimulation of the mAChR with carbachol for 15 s strongly enhanced the amount of cofilin and phospho-cofilin co-immunoprecipitated with PLD1. In contrast, cofilin and phospho-cofilin poorly co-immunoprecipitated with PLD2, and there was no effect of carbachol (Figure 5C, right panel).
Phosphorylation of cofilin is essential for stimulation of PLD1
We then determined whether cofilin not only interacts with PLD1, but also controls its activity, and whether such regulation is dependent on the phosphorylation state of cofilin in vitro. For this, wild-type and Unphosphorylatable S3A cofilin were first treated with LIM-kinase1, in the absence and presence of MgATP, to allow for cofilin phosphorylation, and then the cofilin proteins were added to purified recombinant PLD enzymes for measurement of enzyme activity. Addition of purified recombinant wild-type or S3A cofilin pretreated with LIM-kinase1, in the absence or presence of MgATP had no effect on the activity of the PLD2 enzyme (Figure 6A, right bar graph). Furthermore, similar to buffer control, S3A cofilin was without effect on PLD1 activity. Wild-type cofilin pretreated with LIM-kinase1, but without MgATP, increased the activity of PLD1 only slightly, by about 50% (Figure 6A, left bar graph), whereas wild-type cofilin pretreated without LIM-kinase1 was without effect on PLD1 activity (data not shown). In contrast, wild-type cofilin phosphorylated by LIM-kinase1 (in the presence of MgATP; Figure 6A, 32P autoradiography) strongly enhanced PLD1 activity, by about three-fold (Figure 6A, left bar graph). To corroborate the hypothesis that it is the phosphorylated cofilin, which stimulates PLD1, purified recombinant phosphorylation-mimic cofilin mutants were added to purified recombinant PLD1. Similar to wild-type cofilin phosphorylated by LIM-kinase1, the phosphorylation-mimic S3D and S3E cofilin mutants strongly enhanced PLD1 activity, by about three-fold (Figure 6B). Thus, cofilin not only directly and specifically interacts with PLD1, but also strongly increases its activity, and this activation is dependent on the phosphorylation state of cofilin.
Figure 6.
Phospho-cofilin stimulates PLD1. (A) With LIM-kinase1, phosphorylated (+ATP) and unphosphorylated (−ATP) wild-type or unphosphorylatable S3A cofilin were incubated with GST-tagged PLD1 or GST-tagged PLD2 to measure PLD activity. The upper blots show purified GST-tagged LIM-kinase1, GST-tagged PLD1 and PLD2 and His6-tagged wild-type and S3A cofilin by Coomassie blue staining (CS), and immunoblotting with anti-GST and anti-His antibodies, respectively, as well as the specific phosphorylation of wild-type cofilin with [γ-32P]ATP (32P autoradiography) in the absence (−) and presence (+) of LIM-kinase1. (B) GST-tagged PLD1 was incubated without and with recombinant unphosphorylatable S3A cofilin, wild-type cofilin, phospho-mimetic S3D cofilin or phospho-mimetic S3E cofilin to measure PLD activity. Purified His6-tagged cofilin mutants are presented by Coomassie blue staining. The results shown are representative of 3–4 experiments.
Identification of the PLD1 region responsible for interaction with cofilin
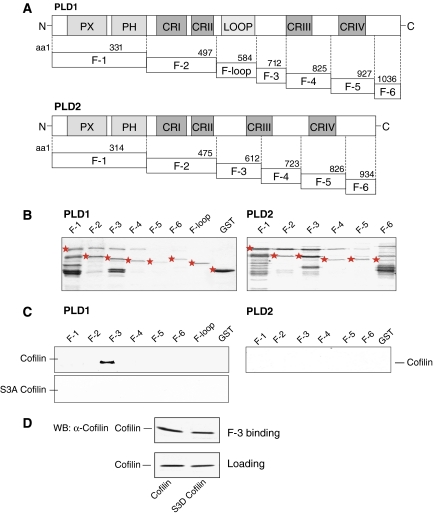
To identify the PLD1 region involved in cofilin binding, we constructed GST-tagged PLD1 fragments (Figure 7A and B) and analyzed their ability to bind purified cofilin. The F-3 fragment of PLD1, encompassing the amino acids 585–712, was found to exclusively bind wild-type cofilin (Figure 7C). None of the PLD1 fragments analyzed, bound Unphosphorylatable S3A cofilin. Furthermore, there was no binding of cofilin to the corresponding PLD2 fragments (Lee et al, 2001; Chae et al, 2005) (Figure 7C). These data identify the PLD1-specific region encoded by the amino acids 585–712 to be important for the direct interaction with cofilin. To study whether cofilin phosphorylation alters binding to PLD1, binding of the phosphorylation-mimic S3D cofilin to the F-3 fragment of PLD1 was studied. As illustrated in Figure 7D, S3D cofilin bound to the F-3 fragment, but somewhat less than wild-type cofilin. Cellular cofilin phosphorylated by treatment of HEK-293 cells for 15 s with carbachol, also bound to the PLD1 F-3 fragment and to full-length PLD1, but again somewhat less than cofilin (data not shown). Thus, phosphorylation of cofilin strongly increases PLD1 activity, but not binding of cofilin to PLD1.
Figure 7.
Interaction of cofilin with the F-3 fragment of PLD1. (A) A schematic representation of the highly conserved regions in PLD1 and PLD2. PX, phox; PH, pleckstrin homology; CRI–CRIV, conserved regions I to IV. (B) Purified GST and GST-tagged fragments of PLD1 and PLD2, visualized by Coomassie blue staining. (C) Binding of purified recombinant wild-type or unphosphorylatable S3A cofilin to GST-tagged fragments of PLD1 or PLD2 was determined as described under Materials and methods. (D) Binding of purified recombinant wild-type or phospho-mimetic S3D cofilin to GST-tagged F-3 fragment of PLD1. The lower blot shows the loading control by Western blot (WB). The results shown are representative of 2–4 experiments.
Inhibition of PLD stimulation and actin stress fiber formation by the cofilin-binding F-3 fragment
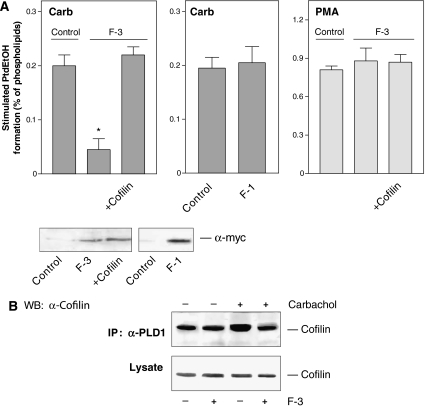
As cofilin directly interacts with the F-3 fragment of PLD1, we used this fragment to interfere with the mAChR-induced cofilin-PLD1 interaction, as well as PLD stimulation and actin stress fiber formation. Expression of the F-3 fragment in HEK-293 cells strongly and specifically reduced PLD stimulation by carbachol, without affecting PLD stimulation by PMA (Figure 8A). In contrast, expression of the F-1 fragment of PLD1, that does not interact with cofilin, did not alter PLD stimulation by carbachol. The inhibitory effect of the F-3 fragment could be completely rescued by coexpression of wild-type cofilin (Figure 8A). These data indicate that the PLD1-specific F-3 fragment specifically interferes with the activation of PLD1, most probably by attenuating the mAChR-induced association of cofilin with PLD1. Indeed, the enhanced co-immunoprecipitation of cofilin with PLD1 induced by activation of the mAChR by carbachol was completely abolished by expression of the F-3 fragment (Figure 8B).
Figure 8.
The F-3 fragment of PLD1 specifically reduces PLD stimulation by carbachol and interaction of PLD1 with cofilin. (A) HEK-293 cells were transfected with empty vector (Control), F-1 or F-3, either alone or together with wild-type cofilin. After 48 h, stimulated [3H]PtdEtOH accumulation was determined in the presence of 1 mM carbachol (Carb) or 100 nM PMA. Data shown are means±s.e. (n=3–4). The immunoblots show expression of c-myc-tagged F-3 and F-1 fragments in cell lysates. (B) HEK-293 cells were transfected with wild-type PLD1 alone or together with the F-3 fragment. After 48 h, the cells were treated for 15 s without (−) or with (+) 1 mM carbachol, followed by cell lysis and immunoprecipitation with the anti-PLD1 antibody. The PLD1 immunoprecipitates (IP) and total lysates were resolved by SDS–PAGE and probed with anti-cofilin antibodies (α-cofilin) as indicated. The results shown are representative of 3–4 experiments. WB, Western blot.
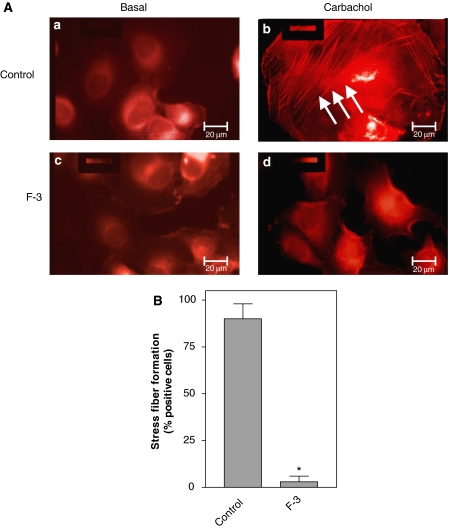
Cofilin is known to profoundly affect actin dynamics (Bamburg, 1999; DesMarais et al, 2005), and also PLD1 has been reported to alter the organization of the actin cytoskeleton (Ha and Exton, 1993; Cross et al, 1996; Kam and Exton, 2001; Porcelli et al, 2002; Komati et al, 2005). To determine whether interaction of cofilin with PLD1 may affect Rho-dependent actin dynamics, we studied stress fiber formation in human bladder carcinoma (J82) cells coexpressing the M3 mAChR and the PLD1-specific F-3 fragment. As expected, activation of the M3 mAChR by carbachol induced the formation of stress fibers (Figure 9, panel b). Coexpression of the F-3 fragment completely suppressed stress fiber formation (Figure 9B, panels b and d), suggesting that interaction of cofilin with PLD1 is involved in the regulation of the actin cytoskeleton architecture.
Figure 9.
The F-3 fragment of PLD1 abolishes stress fiber formation in J82 cells. (A) J82 cells were transfected with the M3 mAChR alone or together with the PLD1-specific F-3 fragment. Forty-eight hours after transfection, cells stimulated without (Basal) or with 1 mM carbachol for 5 min were examined for the actin cytoskeleton by TRITC–phalloidin staining. Arrows indicate carbachol-induced stress fiber formation. Scale bar, 20 μm. (B) Carbachol-induced stress fiber formation in M3 mAChR-positive cells in the absence (Control) or presence of the F-3 fragment (F3). Data shown are means±s.e. (n=3). *P<0.05.
Discussion
Cofilin plays a prominent role in the regulation of actin dynamics and is involved in the initiation of actin-driven processes, including cell motility, neuronal pathfinding, assembly of cell polarity, cell division and apoptosis (Bamburg, 1999; Chua et al, 2003; Pollard and Borisy, 2003; DesMarais et al, 2005). Cofilin can actually exert two opposing effects on the actin filament assembly/disassembly machinery: (1) barbed-end formation and subsequent actin polymerization, as well as (2) actin depolymerization. At present, the precise contribution of these cofilin-mediated mechanisms to actin reorganization is rather undefined and may be even different in various cell types (for recent review see DesMarais et al, 2005). Cellular functions of cofilin are tightly controlled by a unique phosphorylation–dephosphorylation cycle, involving specific kinases (LIM- and TES-kinases) and cofilin-specific phosphatases (slingshot phosphatases and chronophin), as well as the scaffold protein 14-3-3ζ. Overall, phosphorylation of cofilin at serine 3 results in its inactivation, and reactivation is catalyzed by its dephosphorylation (reviewed in Huang et al, 2006).
PLD enzymes generating PA from phosphatidylcholine, in particular PLD1, have also been found to regulate actin organization and to induce stress fiber formation in a cell-dependent manner (Ha and Exton, 1993; Cross et al, 1996; Kam and Exton, 2001; Porcelli et al, 2002; Komati et al, 2005). PLD-driven regulation of the actin cytoskeleton architecture likely occurs in concert with PA-responsive PIP 5-kinase (for recent review see Oude Weernink et al, 2004a). As it has become evident that various components of the actin regulatory network conversely affect PLD activity (Park et al, 2000; Lee et al 2001; Kusner et al, 2002), this reciprocal modulation of PLD activity and actin cytoskeleton dynamics, together with the observation that both PLD and actin dynamics are regulated by Rho GTPases and their effectors (Edwards and Gill, 1999; Kaibuchi et al, 1999; Exton, 2002), points to concerted mechanisms in cellular actions, involving acute, localized alterations in actin architecture.
Here we report that the pivotal actin regulatory protein cofilin, specifically interacts with and controls in its phosphorylated state the activity of PLD1. By this, we define a novel cellular function for the phosphorylation–dephosphorylation cycle of cofilin. While non-phosphorylated cofilin controls actin dynamics, the phosphorylated cofilin is inactive in this regard (Bamburg, 1999; DesMarais et al, 2005; Huang et al, 2006). In contrast, it is apparently the phosphorylated cofilin, which preferentially stimulates PLD1 activity. Furthermore, stimulation of PLD1 by cofilin is under control of G-protein-coupled muscarinic receptors. Several lines of evidence support this notion. First, by expression in intact cells, we found that factors known to increase cofilin phosphorylation, such as LIM-kinase1, inactive slingshot phosphatase 1L and 14-3-3ζ, enhanced receptor-mediated stimulation of PLD1, whereas expression of wild-type slingshot phosphatase 1L, which abolished cofilin phosphorylation, and the Unphosphorylatable S3A cofilin mutant had the opposite effect on PLD stimulation. This regulation of PLD activity was specific for the G-protein-coupled receptors, known to be controlled by Rho and the LIM-kinase activator, Rho-kinase (Schmidt et al, 1999), whereas PLD stimulation by PMA and EGF, which is independent of Rho and Rho-kinase in HEK-293 and N1E-115 neuroblastoma cells (Voß et al, 1999), was not altered. Most important, silencing of cellular cofilin largely prevented PLD stimulation by carbachol in both cell types, whereas expression of phosphorylation-mimic S3D cofilin had the opposite effect. Second, mAChR activation induced a very rapid association of cofilin specifically with PLD1 and a phosphorylation of cofilin. Third, in vitro studies with purified components showed that cofilin specifically bound to PLD1. Evidence for such a specific cofilin–PLD1 interaction was also observed by expression of these proteins in intact cells, where cofilin induced a specific subcellular redistribution of PLD1. Finally, cofilin phosphorylated by LIM-kinase1 as well as two phosphorylation-mimic cofilin mutants (S3D and S3E) strongly and specifically increased enzyme activity of PLD1.
Phosphorylation of cofilin as studied in detail for the M3 mAChR in HEK-293 cells, was a very rapid and transient response, indicating that cellular phosphocycling of cofilin is indeed tightly controlled by kinases and phosphatases (Huang et al, 2006). Several previous studies designed to analyze the regulation of cofilin dephosphorylation may not have been aware of such rapid induction of cofilin phosphorylation (Zhan et al, 2003; Nagata-Ohashi et al, 2004; Nebl et al, 2004; Nishita et al, 2004; Wang et al, 2005). The activity of the slingshot phosphatases is controlled by cellular Ca2+, in part mediated by the Ca2+/calmodulin-dependent protein phosphatase calcineurin, cyclic AMP and the Ras effector, phosphatidylinositol 3-kinase (Zhan et al, 2003; Nagata-Ohashi et al, 2004; Nebl et al, 2004; Nishita et al, 2004; Wang et al, 2005). As the M3 mAChR not only induces PLD stimulation in HEK-293 cells, but also increases intracellular Ca2+ concentration and cyclic AMP, and induces activation of Ras GTPases (Evellin et al, 2002; López de Jesús et al, 2006), such mechanisms may contribute to the rapid dephosphorylation of cofilin. The rapid and transient phosphorylation of cofilin perfectly reflects its well-established pivotal role in the regulation of actin filament dynamics (Bamburg, 1999; Bamburg and Wiggan, 2002; Pollard and Borisy, 2003; DesMarais et al, 2005). Similarly interesting, this receptor-induced cofilin phosphorylation profile is in line with the rapid and very transient PLD stimulation by G-protein-coupled receptors, including muscarinic receptors (Schmidt et al, 1995; reviewed in Exton, 2002). Recently, it has been reported that tubulin specifically interacts with the F-3 fragment of PLD2, and that inhibition of receptor-induced PLD2 stimulation, probably by direct interaction with tubulin, may contribute to the rapid decline of the PLD2 response (Chae et al, 2005). Thus, different PLD isozymes may use and associate with different cytoskeletal proteins for rapid deactivation.
Finally, we analyzed the region in PLD1 responsible for cofilin binding. For this, PLD1 fragments were generated essentially as the corresponding PLD2 fragments described before (Lee et al, 2001; Chae et al, 2005). Cofilin and phosphorylated cofilin cofilin were found to specifically bind to the F-3 fragment of PLD1, encompassing the amino acids 585–712. Although phosphorylation-mimic cofilin strongly increased PLD1 activity, binding to PLD1 was not enhanced. The somewhat reduced affinity of phospho-cofilin may contribute to the rapid termination of PLD1 stimulation by muscarinic receptors. In line with the binding on the full-length PLD enzymes, there was no binding of Unphosphorylatable S3A cofilin to any of the PLD1 fragments, and also no binding of cofilin to PLD2 fragments. These data indicate that this region in PLD1 is necessary and sufficient for cofilin binding. This cofilin-binding F-3 fragment was then used to further probe the role of cofilin–PLD1 interaction for receptor signaling to PLD and actin stress fiber formation. Expression of the F-3 fragment profoundly and specifically suppressed PLD stimulation by the M3 AChR, an effect fully reversed by expression of cofilin. This was accompanied by a similar inhibition of receptor-induced association of cofilin with PLD1, indicating that indeed this PLD1 fragment binds cofilin also in intact cells. Expression of the cofilin-binding F-3 fragment also abrogated M3 mAChR-induced stress fiber formation. These findings together indicate that cofilin-induced actin rearrangements (Chan et al, 2000; Endo et al, 2003; Ghosh et al, 2004; Marcoux and Vuori, 2005) may be partly mediated by PLD1 and its product PA, which have been shown to participate in receptor-induced stress fiber formation in various cell types (Ha and Exton, 1993; Cross et al, 1996; Kam and Exton, 2001; Porcelli et al, 2002; Komati et al, 2005).
In conclusion, we report here on a novel molecular link between the actin cytoskeleton and PLD1, and provide evidence that inactive phospho-cofilin exerts an unexpected biological function. By its stimulation of PLD1, known to regulate many early and late cellular functions, from calcium mobilization, glucose transport, mitogenesis to apoptosis (Exton, 2002), phospho-cofilin is most likely an active signaling component, and may control essential cellular functions.
Materials and methods
Cell culture, transfection, plasmid construction and adenoviruses
HEK-293 and N1E-115 cells grown to near confluence on 145-mm culture dishes were transfected as reported (Schmidt et al, 2001; López de Jesús et al, 2006). PLD1 and PLD2 (each subcloned into pCGN), wild-type LIM-kinase1 and kinase-deficient D460A LIM-kinase1 (each subcloned into pUCD2), wild-type cofilin, Unphosphorylatable S3A cofilin and phosphorylation-mimic S3D cofilin (each subcloned into pcDL-SRα), slingshot phosphatase 1L, inactive C393S slingshot phosphatase 1L, VSV-G (vesicular stomatitis virus glycoprotein)-tagged 14-3-3ζ, as well as the M3 mAChR (each subcloned into pCDNA3.1/myc-His) were kindly provided by Drs MA Frohman, A Morris, K Mizuno, T Uemura, H Betz and CJ van Koppen, respectively. Typically 50–100 μg of DNA for cell transfection were used, except for Unphosphorylatable S3A cofilin amounting to 150 μg of DNA. For transfection of the cells with siRNA pSUPER plasmids (Brummelkamp et al, 2002), cells on 35-mm dishes were transfected with 3 μl Lipofectamine 2000 in 1 ml Opti-MEM containing 6–9 μg of human cofilin siRNA or mouse cofilin siRNA. If phosphorylation-mimic S3D mouse cofilin was cotransfected with human cofilin siRNA, 6 μg of the vector DNA was used in a total amount of 12 μg. Human cofilin siRNA pSUPER plasmid (target sequence GGAGGATCTGGTGTTTATC) and mouse cofilin siRNA pSUPER plasmid (target sequence GGAGGACCTGGTGTTCATC) were constructed as described previously (Nishita et al, 2005; Kiuchi et al, 2007). Expression of the encoded proteins was verified by immunoblotting of cell lysates with specific antibodies. Assays were performed 48 h after transfection. To generate the c-myc-tagged F-1 and F-3 fragments of hPLD1b, HindIII to XbaI 842 and 442-bp fragments from pGEX4T1 were subcloned into pCDNA3.1, respectively (Invitrogen). Adenoviruses encoding lipase-inactive PLD mutants were generated essentially as described before (He et al, 1998; Rümenapp et al, 2001). Briefly, K898R human PLD1b or K758R mouse PLD2 XbaI–SmaI fragments from pCGN were subcloned into pAdTrack-CMV XbaI–EcoRV, followed by homologous recombination with the pAdEasy vector (both vectors kindly provided by Dr B Vogelstein), the constructs were linearized and transfected into HEK-293 cells using LipofectAMINE (Invitrogen). The vector constructs were verified by sequencing. After several cycles of amplification, the adenoviruses were purified using a CsCl2 gradient and titration with the Adeno-X Rapid titer kit, according to the manufacturer's instructions (BD Biosciences). HEK-293 cells were infected with adenoviral bacterial β-galacosidase LacZ serving as a control (a kind gift from Dr T Eschenhagen), or adenoviral-encoded PLD mutants, 48 h before PLD assays, at a multiplicity of infection of 50. Before the assay, infected cells were visualized by fluorescence microscopy (Zeiss Axiovert S100).
Purification of proteins and protein binding assays
To obtain GST-tagged PLD1, PLD2 (cDNAs kindly provided by Drs A Morris and MA Frohman) and His6-tagged phosphorylation-mimic S3E cofilin, Spodoptera frugiperda cells were infected with appropriate recombinant baculoviruses. GST/flag-tagged LIM-kinase1 (kindly provided by Dr K Mizuno) was expressed in HEK-TsA201 cells. The recombinant proteins were purified with glutathione Sepharose (López de Jesús et al, 2006). GST-tagged PLD2 fragments F-1–F-6, His6-tagged wild-type cofilin, unphosphorylatable S3 cofilin and phosphorylation-mimic S3D cofilin (cDNAs kindly provided by Drs SH Ryu and K Mizuno) were expressed in Escherichia coli, and purified with glutathione Sepharose or Ni-NTA Superflow Agarose beads (López de Jesús et al, 2006). To obtain the soluble and untagged proteins, the immobilized cofilin proteins were eluted with 400 mM imidazole, and for protease digestion of GST-tagged proteins, the washed beads were resuspended in thrombin buffer (50 mM Tris/HCl, pH 8.6, 150 mM NaCl, 2.5 mM CaCl2, 0.1% 2-mercaptoethanol), and incubated for 2 h at room temperature with thrombin (followed by overnight incubation at 4°C). GST-tagged PLD1 fragments were generated essentially as the corresponding PLD2 fragments described before (Lee et al, 2001; Chae et al, 2005). Briefly, fragment 1 (F-1, aa 1–331, 64 kDa) encompassed the PX- and the PH-domains, fragment 2 (F-2, aa 332–497, 45 kDa) the conserved regions CRI and CRII, which are followed by the PLD1-specific loop (F-loop, aa 498–585, 37 kDa). Fragment 3 (F-3, aa 585–712, 41 kDa) comprised the region between the PLD1-specific loop and the conserved region CRIII, fragment 4 (F-4, aa 713–825, 40 kDa) the CRIII region, fragment 5 (F-5, aa 826–927, 38 kDa) the CRIV region, and fragment 6 (F-6, aa 928–1036, 39 kDa) encompassed the entire COOH-terminus of human PLD1b (Ensembl Protein Report; www.ensembl.org). The fragments were amplified with Pfu polymerase (Fermentas) from a human PLD1b vector, using modified PCR primers harboring an artificial SalI- (sense) or NotI-site (antisense). The PCR fragments were ligated in the SalI/NotI-restricted multiple cloning site of the bacterial expression vector pGEX4T1 (Amersham). The vector clones were verified by sequencing. The resulting GST-tagged hPLD1b fragments were expressed in E. coli, and purified with glutathione Sepharose (López de Jesús et al, 2006). Purification of the proteins was analyzed by SDS–PAGE. GST, GST-tagged PLD1, PLD2 (full length each) and their corresponding fragments were incubated with purified wild-type, unphosphorylatable S3 cofilin or phosphorylation-mimic S3D cofilin overnight at 4°C, and the ratio of beads to cofilin (approximately 5–30 μg each) was assessed by calibration on the basis of Coomassie-stained gels. After three washes, the beads were resolved in Laemmli buffer, subjected to SDS–PAGE and transferred to nitrocellulose for Western blotting to visualize bound recombinant cofilin proteins using anti-His (Santa Cruz) or anti-cofilin antibodies (Cell Signaling Technology).
Confocal laser scanning microscopy and fluorescence microscopy
HEK-293 cells transfected with PLD1 or PLD2 either alone or together with HA-tagged wild-type or Unphosphorylatable S3A cofilin were grown on coverslips (Falcon) pretreated with 0.1 mg/ml poly-D-lysine (Stope et al, 2004). After 48 h, the cells were rinsed with Moscona, containing 13.6 mM NaCl, 4 mM KCl, 12 mM NaHCO3, 10 mM D-glucose, 0.36 mM NaH2PO4 and 0.18 mM KH2PO4, pH 7.4, fixed and permeabilized with ethanol/acetone (1/1, v/v) for 10 min at RT, followed by two washes with Moscona. Unspecific binding was blocked for 15 min by incubation with Moscona/BSA (Moscona supplemented with 0.5% BSA). The cells were rinsed and incubated for 1 h at RT with anti-PLD antibodies (dilution 1:50; kindly provided by Dr S Bourgoin) or with an anti-HA antibody (12CA5, dilution 1:20; Roche Molecular Biochemicals). Afterwards, the cells were rinsed three times with Moscona/BSA, incubated for 45 min in darkness with the corresponding fluorescence-conjugated secondary antibodies (Alexa-633 red for PLD, Alexa-488 green for cofilin; dilution 1:200; MoBiTec) at RT. The secondary antibodies were washed out in darkness with Moscona/BSA, and the cell specimens were mounted in Moscona supplemented with 90% (v/v) glycerol and 1.0% (v/v) p-phenylenediamine. Confocal immunofluorescence imaging was performed using Zeiss LSM 510 Axiovert 100 M confocal laser scanning microscopy system (Plan-Neofluor × 40/1.3 oil objective, 488/633 nm excitation). Confocal laser microscopy images were processed with the ImageProPlus software (Version 4.5; Media cybernatics) equipped with a Gaussian filter module (Stope et al, 2004). J82 bladder carcinoma cells transfected with M3 mAChR alone or together with F-3 fragment of PLD1 (50 μg DNA each) were stained for actin filaments with tetramethyl-rhodamine isothiocyanate (TRITC)-conjugated phalloidin (Sigma-Aldrich) (Rümenapp et al, 1999). Fluorescent images were obtained using Zeiss Axiovert S100 microscope.
Phosphorylation of recombinant proteins and phosphorylation of cellular cofilin
To study phosphorylation of recombinant proteins by LIM-kinase1, immobilized LIM-kinase1 (1.5 μM) was equilibrated with phosphorylation buffer (25 mM Tris/HCl, pH 7.4, 5 mM MgCl2, 1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol) and mixed with 10–50 μg recombinant GST, GST-tagged PLD1, GST-tagged PLD2, wild-type or Unphosphorylatable S3A cofilin. After preincubation for 5 min at 37°C (or at 25°C to measure cofilin phosphorylation), the reaction was started by adding 100 mM Tris/HCl, pH 7.5, 25 mM MgCl2, 1 mM CaCl2, 250 μM [γ-32P]ATP (0.75 MBq per reaction tube) at a dilution of 1:5. After 15 min (or 45 min for the cofilin phosphorylation), the reaction was stopped by addition of Laemmli sample buffer and heating for 5 min at 95°C. The proteins were separated by SDS–PAGE, and phosphorylated proteins detected by autoradiography using Kodak X-Omat AR films. To measure phosphorylation of cellular cofilin, transfected cells were incubated for the indicated periods of time at 37°C with 1 mM carbachol, followed by cell lysis in a buffer containing 1% SDS and 10 mM Tris/HCl, pH 7.4, and five passages through a 25-gauge needle (Keiper et al, 2004). The lysates were clarified by centrifugation, followed by determination of protein concentration and incubation in Laemmli buffer for 10 min at 95°C. After SDS–PAGE and transfer to nitrocellulose membranes, phosphorylated cofilin and the total cellular content of cofilin were detected with anti-phospho-cofilin (anti-P-Cofilin) and anti-cofilin antibodies, respectively (Cell Signaling Technology).
Immunoprecipitation and immunoblotting
Cells were transfected with the expression plasmids indicated in the figure legends and grown on 60-mm culture dishes in serum-free medium. Cells were rinsed in Hank's balanced salt solution and incubated without and with carbachol for 15 s at 37°C. Then, the cells were washed twice with ice-cold phosphate-buffered saline, containing 137 mM NaCl, 2.7 mM KCl, 6.5 mM NaH2PO4, 1.5 mM KH2PO4, 0.9 mM CaCl2, 0.5 mM MgCl2 and 100 μM orthovanadate, pH 7.2. Meanwhile, the anti-PLD antibodies (7 μg each) were gently mixed with protein A–Sepharose (20 μl per reaction tube) in immunoprecipitation buffer containing 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 10 mM sodium pyrophosphate, 50 mM NaF, 1 mM EGTA, 1% Nonidet P-40, 0.5% sodium deoxycholate and 0.1% sodium dodecylsulfate, for 1 h at 4°C. Cells were scraped in 500 μl ice-cold immunoprecipitation buffer, supplemented with 1 mM orthovanadate, 1 mM phenylmethylsulfonylfluoride, 10 μg/ml leupeptin and 25 μg/ml aprotinin, transferred to precooled reaction tubes, vortexed for 10 s and incubated on ice for at least 10 min. Lysates were clarified by centrifugation, the supernatants (4 mg of protein/reaction tube) were gently mixed overnight at 4°C and precipitates were washed four times with immunoprecipitation buffer. Precipitated proteins were incubated in Laemmli buffer for 10 min at 95°C and separated by SDS–PAGE. Bound phosphorylated cofilin and cofilin were detected by immunoblotting with specific cofilin antibodies. Analysis of the total cellular cofilin content in the lysates served as control. For detection of c-myc, LIM-kinase1, VSV-G, PLD1 and PLD2, equal amounts of protein from cell lysates were separated by SDS–PAGE on 5 or 15% acrylamide gels. After a transfer to nitrocellulose membranes and a 1-h incubation with the appropriate antibodies, the proteins were visualized by enhanced chemiluminescence.
Measurement of PLD activity
PLD activity was measured in transfected cells labeled with [3H]oleic acid (5 Ci/mmol, PerkinElmer Life Sciences) as formation of the specific PLD product, [3H]phosphatidylethanol ([3H]PtdEtOH), for 30 min at 37°C in the presence of ethanol. Basal PLD activity amounted to 0.08±0.03 [3H]PtdEtOH formation expressed as the precentage of total labeled phospholipids in HEK-293, cells and to 0.01±0.002 [3H]PtdEtOH formation expressed as the precentage of total labeled phospholipids in N1E-115 cells, respectively. As basal PLD activity was not altered by any maneuver, agonist-induced PLD stimulation is expressed as stimulated [3H]PtdEtOH formation (Rümenapp et al, 2001). To study the role of phosphorylated cofilin on recombinant PLD activity, GST-tagged LIM-kinase1 bound to glutathione Sepharose beads (30 μg) and His6-tagged wild-type or S3A cofilin (1 mg each) were incubated together in the presence or absence of 1 mM MgATP for 45 min at 25°C. LIM-kinase1 was removed by centrifugation for 3 min at 2400 r.p.m. at 4°C. The supernatant containing phosphorylated or unphosphorylated cofilin was incubated with GST-tagged PLD1 or PLD2 immobilized on glutathione Sepharose beads (20–30 μg per reaction tube), [3H]PtdCho/PIP2 vesicles (1-palmitoyl-2-[9,10-3H]palmitoyl-glycerophosphocholine (89 Ci/mmol, PerkinElmer Life Sciences) mixed with PIP2 (Roche Applied Science) in a molar ratio of 8:1), 2% (v/v) ethanol and stimulatory agents for 30 min at 30°C (Schmidt et al, 1999; Wilde et al, 2002). Under identical experimental conditions, the activity of GST-tagged PLD1 immobilized on glutathione Sepharose beads (20–30 μg per reaction tube) was measured in the presence or absence of recombinant wild-type cofilin, Unphosphorylatable S3A cofilin, phospho-mimetic S3D cofilin or phospho-mimetic S3E cofilin (2.5 μg per reaction tube). Data shown in the figures are either representative experiments or means±s.e.m. of n independent experiments, each performed in triplicate. Comparisons between means were either with the Student's paired t-test or one-way analysis of variance test, and a difference was regarded significant at P<0.05.
Acknowledgments
We thank Y Mahlke and S Schimanski for exceptional technical assistance, supported by assistance from K Baden, M Hagedorn, H Geldermann, D Petermeyer and A Kötting-Dorsch. C Heneweer and M Thie supported the microscopic studies. The gifts of various plasmids and antisera from R Agami, H Betz, S Bourgoin, T Eschenhagen, MA Frohman, A Morris, CJ van Koppen, SH Ryu, B Vogelstein and T Uemura are greatly appreciated. This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, the Interne Forschungsförderung Essen and the Fakultät für Klinische Medizin Mannheim. M López de Jesús is recipient of a Basque Government Fellowship. M Schmidt is recipient of a Rosalind Franklin Fellowship from the University of Groningen.
References
- Acevedo K, Moussi N, Li R, Soo P, Bernard O (2006) LIM kinase 2 is widely expressed in all tissues. J Histochem 54: 487–501 [DOI] [PubMed] [Google Scholar]
- Bamburg JR (1999) Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol 15: 185–230 [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Wiggan OP (2002) ADF/cofilin and actin dynamics in disease. Trends Cell Biol 12: 598–605 [DOI] [PubMed] [Google Scholar]
- Birkenfeld J, Betz H, Roth D (2003) Identification of cofilin and LIM-domain-containing protein kinase 1 as novel interaction partners of 14-3-3ζ. Biochem J 369: 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalina cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Chae YC, Lee S, Lee HY, Heo K, Kim JH, Kim JH, Suh P-G, Ryu SH (2005) Inhibition of muscarinic receptor-linked phospholipase D activation by association with tubulin. J Biol Chem 274: 3723–3730 [DOI] [PubMed] [Google Scholar]
- Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS (2000) Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J Cell Biol 148: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua TB, Volbracht C, Tan KO, Li R, Yu VC, Li P (2003) Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat Cell Biol 5: 1083–1089 [DOI] [PubMed] [Google Scholar]
- Condeelis J (2001) How is actin polymerization nucleated in vivo? Trends Cell Biol 11: 288–293 [DOI] [PubMed] [Google Scholar]
- Cross MJ, Roberts S, Ridley AJ, Hodgkin MN, Stewart A, Claesson-Welsh L, Wakelam MJO (1996) Stimulation of actin stress fibre formation mediated by activation of phospholipase D. Curr Biol 6: 588–597 [DOI] [PubMed] [Google Scholar]
- Cummings RJ, Parinandi NL, Zaiman A, Wang LX, Usatyuk PV, Garcia JG, Natarajan V (2002) Phospholipase D activation by sphingosine 1-phosphate regulates interleukin-8 secretion in human bronchial epithelial cells. J Biol Chem 277: 30227–30235 [DOI] [PubMed] [Google Scholar]
- DesMarais V, Ghosh M, Eddy R, Condeelis J (2005) Cofilin takes the lead. J Cell Sci 118: 19–26 [DOI] [PubMed] [Google Scholar]
- Endo M, Ohashi K, Sasaki Y, Goshima Y, Niwa R, Uemura T, Mizuno K (2003) Control of growth cone motility and morphology by LIM kinase and slingshot via phosphorylation and dephosphorylation of cofilin. J Neurosci 23: 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DC, Gill GN (1999) Structural features of Lim-kinase that control effects on the actin cytoskeleton. J Biol Chem 274: 11352–11361 [DOI] [PubMed] [Google Scholar]
- Evellin S, Nolte J, Tysack K, vom Dorp F, Thiel M, Oude Weernink PA, Jakobs KH, Webb E, Lomasney JW, Schmidt M (2002) Stimulation of phospholipase C-ɛ by the M3 muscarinic acetycholine receptor mediated by cyclic AMP and the GTPase Rap2B. J Biol Chem 277: 16805–16813 [DOI] [PubMed] [Google Scholar]
- Exton JH (2002) Phospholipase D—structure, regulation and function. Rev Physiol Biochem Pharmacol 144: 1–94 [DOI] [PubMed] [Google Scholar]
- Foletta VC, Moussi N, Sarmieri PD, Bamburg JR, Bernard O (2004) LIM kinase1, a key regulator of actin dynamics, is widely expressed in embryonic and adult tissues. Exp Cell Res 294: 392–405 [DOI] [PubMed] [Google Scholar]
- Gohla A, Bokoch GM (2002) 14-3-3 regulates actin dynamics by stabilizing phosphorylated cofilin. Curr Biol 12: 1704–1710 [DOI] [PubMed] [Google Scholar]
- Gohla A, Birkenfeld J, Bokoch GM (2005) Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat Cell Biol 7: 21–29 [DOI] [PubMed] [Google Scholar]
- Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS (2004) Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304: 743–746 [DOI] [PubMed] [Google Scholar]
- Ha KS, Exton JH (1993) Activation of actin polymerization by phosphatidic acid derived from phosphatidylcholine in IIC9 fibroblasts. J Cell Biol 123: 1789–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Zhou S, Da Costa LT, Yu J, Kinzler KW, Vogelstein B (1998) A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA 95: 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilpela P, Vartiainen MK, Lappalainen P (2004) Regulation of the actin cytoskeleton by PI(4,5)P2 and PI(3,4,5)P3. Curr Top Microbiol Immunol 282: 117–163 [DOI] [PubMed] [Google Scholar]
- Huang TY, DerMardirossian C, Bokoch GM (2006) Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol 18: 26–31 [DOI] [PubMed] [Google Scholar]
- Kam Y, Exton JH (2001) Phospholipase D activity is required for actin stress fiber formation in fibroblasts. Mol Cell Biol 21: 4055–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaibuchi K, Kuroda S, Amano M (1999) Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem 68: 459–486 [DOI] [PubMed] [Google Scholar]
- Kaji N, Ohashi K, Shuin M, Niwa R, Uemura T, Mizuno K (2003) Cell cycle-associated changes in slingshot phosphatase activity and roles in cytokinesis in animal cells. J Biol Chem 278: 33450–33455 [DOI] [PubMed] [Google Scholar]
- Keiper M, Stope MB, Szatkowski D, Böhm A, Tysack K, vom Dorp F, Saur O, Oude Weernink PA, Evellin S, Jakobs KH, Schmidt M (2004) Epac- and Ca2+-controlled activation of Ras and extracellular signal-regulated kinases by Gs-coupled receptors. J Biol Chem 279: 46497–46508 [DOI] [PubMed] [Google Scholar]
- Kiuchi T, Ohashi K, Kurita S, Mizuno K (2007) Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J Cell Biol 177: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komati H, Naro F, Mebarek S, De Arcangelis V, Adamo S, Lagarde M, Prigent A-F, Némoz G (2005) Phospholipase D is involved in myogenic differentiation through remodeling of actin cytoskeleton. Mol Biol Cell 16: 1232–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusner DJ, Barton JA, Wen K-K, Wang X, Rubenstein PA, Iyer SS (2002) Regulation of phospholipase D activity by actin. Actin exerts bidirectional modulation of mammalian phospholipase D activity in a polymerization-dependent, isoform-specific manner. J Biol Chem 277: 50683–50692 [DOI] [PubMed] [Google Scholar]
- Lee S, Park JB, Kim JH, Kim Y, Kim JH, Shin KJ, Lee JS, Ha SH, Suh PG, Ryu SH (2001) Actin directly interacts with phospholipase D, inhibiting its activity. J Biol Chem 276: 28252–28260 [DOI] [PubMed] [Google Scholar]
- López de Jesús M, Stope MB, Oude Weernink PA, Börgermann C, Ananaba VN, Michel MC, Jakobs KH, Schmidt M (2006) Cyclic AMP-dependent and Epac-mediated activation of R-Ras by G proteins-coupled receptors leads to phospholipase D stimulation. J Biol Chem 281: 21837–21847 [DOI] [PubMed] [Google Scholar]
- Marcoux N, Vuori K (2005) EGF receptor activity is essential for adhesion-induced stress fibre formation and cofilin phosphorylation. Cell Signal 17: 1449–1455 [DOI] [PubMed] [Google Scholar]
- Nagata-Ohashi K, Ohta Y, Goto K, Chiba S, Mori R, Nishita M, Ohashi K, Kousaka K, Iwamatsu A, Niwa R, Uemura T, Mizuno K (2004) A pathway of neuregulin-induced activation of cofilin-phosphatase slingshot and cofilin in lamellipodia. J Cell Biol 165: 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebl G, Fischer S, Penzel R, Samstag Y (2004) Dephosphorylation of cofilin is regulated through Ras and requires the combined activities of the Ras-effectors MEK and PI3K. Cell Signal 16: 235–243 [DOI] [PubMed] [Google Scholar]
- Nishita M, Wang Y, Tomizawa C, Suzuki A, Niwa R, Uemura T, Mizuno K (2004) Phosphoinositide 3-kinase-mediated activation of cofilin phosphatase slingshot and its role for insulin-induced membrane protrusion. J Biol Chem 279: 7193–7198 [DOI] [PubMed] [Google Scholar]
- Nishita M, Tomizawa C, Yamamoto M, Horita Y, Ohashi K, Mizuno K (2005) Spatial and temporal regulation of cofilin activity by LIM kinase and slingshot is critical for directional cell migration. J Cell Biol 171: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T (2002) Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 108: 233–246 [DOI] [PubMed] [Google Scholar]
- Ohta Y, Kousaka K, Nagata-Ohashi K, Ohashi K, Muramoto A, Shima Y, Niwa R, Uemura T, Mizuno K (2003) Differential activities, subcellular distribution and tissue expression patterns of three members of the slingshot family phosphatases that dephosphorylate cofilin. Genes Cell 8: 811–824 [DOI] [PubMed] [Google Scholar]
- Oude Weernink PA, Schulte P, Guo YJ, Wetzel M, Amano M, Kaibuchi K, Haverland S, Voß M, Schmidt M, Mayr GW, Jakobs KH (2000) Stimulation of phosphatidylinositol-4-phosphate 5-kinase by Rho-kinase. J Biol Chem 275: 10168–10174 [DOI] [PubMed] [Google Scholar]
- Oude Weernink PA, Schmidt M, Jakobs KH (2004a) Regulation and cellular roles of phosphoinositide 5-kinases. Eur J Pharmacol 500: 87–99 [DOI] [PubMed] [Google Scholar]
- Oude Weernink PA, Meletiadis K, Hommeltenberg S, Hinz M, Ishihara H, Schmidt M, Jakobs KH (2004b) Activation of type I phosphatidylinositol-4-phosphate 5-kinase isoforms by the Rho GTPases, RhoA, Rac1, and Cdc42. J Biol Chem 279: 7840–7849 [DOI] [PubMed] [Google Scholar]
- Park JB, Kim JH, Kim Y, Ha SH, Kim JH, Yoo JS, Du G, Frohman MA, Suh PG, Ryu SH (2000) Cardiac phospholipase D2 localizes to sarcolemmal membranes and is inhibited by α-actinin in an ADP-ribosylation factor-reversible manner. J Biol Chem 275: 21295–21301 [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG (2003) Cellular motility by assembly and disassembly of actin filaments. Cell 112: 453–465 [DOI] [PubMed] [Google Scholar]
- Porcelli AM, Ghelli A, Hrelia S, Rugolo M (2002) Phospholipase D stimulation is required for sphingosine-1-phosphate activation of actin stress fibre assembly in human airway epithelial cells. Cell Signal 14: 75–81 [DOI] [PubMed] [Google Scholar]
- Rümenapp U, Asmus M, Schablowski H, Wozniki M, Han L, Jakobs KH, Fahimi-Vahid M, Michalek C, Wieland T, Schmidt M (2001) The M3 muscarinic acetylcholine receptor expressed in HEK-293 cells signals to phospholipase D via G12 but not Gq-type G-proteins. Regulators of G-proteins as tools to dissect pertussis toxin-resistant G-proteins in receptor–effector coupling. J Biol Chem 276: 2474–2479 [DOI] [PubMed] [Google Scholar]
- Rümenapp U, Blomquist A, Schwörer G, Schablowski H, Psoma A, Jakobs KH (1999) Rho-specific binding and guanine nucleotide exchange catalysis by KIAA0380, a Dbl family member. FEBS Lett 459: 313–318 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Evellin S, Oude Weernink PA, vom Dorp F, Rehmann H, Lomasney JW, Jakobs KH (2001) A new phospholipase-C-calcium signalling pathway mediated by cyclic AMP and a Rap GTPase. Nat Cell Biol 3: 1020–1024 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Fasselt B, Rümenapp U, Bienek C, Wieland T, van Koppen CJ, Jakobs KH (1995) Rapid and persistent desensitization of m3 muscarinic acetylcholine receptor-stimulated phospholipase D. Concomitant sensitization of phospholipase C. J Biol Chem 270: 19949–19956 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Voß M, Oude Weernink PA, Wetzel J, Amano M, Kaibuchi K, Jakobs KH (1999) A role for Rho-kinase in Rho-controlled phospholipase D stimulation by the m3 muscarinic acetylcholine receptor. J Biol Chem 274: 14648–14654 [DOI] [PubMed] [Google Scholar]
- Stope MB, vom Dorp F, Szatkowski D, Böhm A, Keiper M, Nolte J, Oude Weernink PA, Rosskopf D, Evellin S, Jakobs KH, Schmidt M (2004) Rap2B-dependent stimulation of phospholipase C-ɛ by epidermal growth factor receptor mediated by c-Src phosphorylation of RasGRP3. Mol Cell Biol 24: 4664–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soosairajah J, Maiti S, Wiggan O, Sarmieri P, Moussi N, Sarcevic B, Sampath R, Bamburg JR, Bernard O (2005) Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. EMBO J 24: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voß M, Oude Weernink PA, Haupenthal S, Möller U, Cool RH, Bauer B, Camonis JH, Jakobs KH, Schmidt M (1999) Phospholipase D stimulation by receptor tyrosine kinases mediated by protein kinase C and a Ras/Ral signaling cascade. J Biol Chem 274: 34691–34698 [DOI] [PubMed] [Google Scholar]
- Wang Y, Shibasaki F, Mizuno K (2005) Calcium signal-induced cofilin dephosphorylation is mediated by slingshot via calcineurin. J Biol Chem 280: 12683–12689 [DOI] [PubMed] [Google Scholar]
- Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS (2006) The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol 173: 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde C, Barth H, Sehr P, Han L, Schmidt M, Just I, Aktories K (2002) Interaction of the Rho-ADP-ribosylating C3 exoenzyme with RalA. J Biol Chem 277: 14771–14776 [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Miyazaki H, Watanabe H, Sasaki T, Maehama T, Frohman MA, Kanaho Y (2002) Phosphatidylinositol 4-phosphate 5-kinase is essential for ROCK-mediated neurite remodeling. J Biol Chem 277: 17226–17230 [DOI] [PubMed] [Google Scholar]
- Yin HL, Janmey PA (2003) Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol 65: 761–789 [DOI] [PubMed] [Google Scholar]
- Zhan Q, Bamburg JR, Badwey JA (2003) Products of phosphoinositide specific phospholipase C trigger dephosphorylation of cofilin in chemoattractant stimulated neutrophils. Cell Motil Cytoskeleton 54: 1–15 [DOI] [PubMed] [Google Scholar]