Abstract
The mammalian Per1 gene is expressed in the suprachiasmatic nucleus of the hypothalamus, where it is thought to play a critical role in the generation of circadian rhythms. Per1 mRNA also is expressed in other tissues. Its expression in the pars tuberalis (PT) of the pituitary is noteworthy because, like the suprachiasmatic nucleus, it is a known site of action of melatonin. The duration of the nocturnal melatonin signal encodes photoperiodic time, and many species use this to coordinate physiological adaptations with the yearly climatic cycle. This study reveals how the duration of photoperiodic time, conveyed through melatonin, is decoded as amplitude of Per1 and ICER (inducible cAMP early repressor) gene expression in the PT. Syrian hamsters display a robust and transient peak of Per1 and ICER gene expression 3 h after lights-on (Zeitgeber time 3) in the PT, under both long (16 h light/8 h dark) and short (8 h light/16 h dark) photoperiods. However, the amplitude of these peaks is greatly attenuated under a short photoperiod. The data show how amplitude of these genes may be important to the long-term measurement of photoperiodic time intervals.
The selective advantage conferred by the ability to anticipate periodic phenomena has led to the evolution of endogenous, self-sustaining rhythmicity in disparate groups of organisms. In mammals, it generally is accepted that circadian rhythms are generated by the suprachiasmatic nucleus (SCN) of the hypothalamus, and this is considered to be the master biological clock (1). The expression of circadian output from the SCN determines the circadian activities of the organism. Thus, the basis of biological timing depends not only on the generation of circadian rhythms, but also on the measurement of time intervals. One of the best-studied examples of interval timing is the photoperiodic regulation of seasonal changes in reproduction, body weight, and pelage. This phenomenon is based on the duration-dependent interpretation of the melatonin signal, generated by the pineal gland, which is under circadian control by the SCN.
Over the last 2 years, there have been some major advances in our understanding of the molecular basis of the mammalian circadian clock, following on from earlier studies in Drosophila. This was realized after the identification of the first component gene of the mammalian clock, Per1, which was shown to be expressed with a self-sustaining rhythm in the SCN (2, 3). Despite this progress, difficulties exist in explaining what makes the biological clock unique. This is because clock genes are expressed in a range of nonoscillatory tissues outside the SCN (2–4). Among these nonoscillatory sites, the pars tuberalis (PT) of the pituitary has been studied extensively to examine how melatonin-dependent interval timing takes place (5). Melatonin provides the humoral interval signal for photoperiod (6), and considerable advances in knowledge of its mode of action have been made by using the PT, a tissue that expresses high-affinity receptors for melatonin (5). However, the way by which the melatonin message is decoded at the cellular level is still largely unresolved.
To investigate how the photoperiodic signal is decoded at the cellular level, we have studied, by quantitative in situ hybridization, the changes in the temporal expression of Per1 and ICER (inducible cAMP early repressor) in the PT and the SCN of a highly photoperiodic mammal, the Syrian hamster, placed under either long days (LD) (16 h light/8 h dark) or short days (SD) (8 h light/16 h dark).
Per1 is known to be expressed with a circadian rhythm in the SCN, and this rhythm persists in constant darkness and constant light (2, 3, 7). Per1 expression also can be activated by light during the dark phase (7–9). In the mouse, Per1 transcripts also are detected with a circadian rhythm in the PT [which also persist in constant darkness (DD) (2)]. In this gland, Per1 expression seems to be driven by melatonin, since it is not detected in the PT of mice bearing a genetic defect in melatonin synthesis (2). In the sheep, Per1 is expressed with a circadian and a seasonal rhythm in the PT, with higher levels at daytime than at night and higher levels in LD than in SD (10). Moreover, Per1 is an early-response gene whose expression is increased after 2 h of stimulation by forskolin in ovine PT cells (10).
ICER is a transcription factor repressor that was described first in the rat pineal gland (11). ICER represses cAMP-induced transcription and exerts an autonegative feedback on its own transcription (11). In the pineal gland, temporal expression of ICER varies depending on the photoperiod, with a shorter peak of expression and higher inducibility in LD than in SD (12). This pattern of expression is responsible for the correlation between the duration of melatonin synthesis and the length of the night (12), because ICER inhibits the synthesis of the rate-limiting enzyme of melatonin synthesis, the N-acetyltransferase (12). ICER is also constitutively expressed in the SCN of the rat (13), where it is inducible by light (14), and in other tissues, including adrenal gland, testis, and liver (11). Recently, we have demonstrated that ICER is expressed in the ovine PT (15), where its level of expression depends on photoperiod (unpublished data).
These two genes were chosen for study because they have been shown to play an important role in the activity of tissues implicated in seasonal functions. Also, as early-response genes activated through the cAMP pathway, they constitute good indicators of cellular activity that are likely to be regulated by melatonin.
MATERIALS AND METHODS
Animals.
All animal experiments were carried out in accordance with the Animals (Scientific Procedures) Act of 1986.
Male Syrian hamsters weighing 76–100 g were purchased from Charles River Breeding Laboratories and housed under an LD photoperiod of 16 h light/8 h darkness, with free access to food and water. Constant red, dim light (<1 lux) allowed manipulation of the animals during the dark phase.
Effect of photoperiod on Per1 and ICER expression in the PT and the SCN.
Hamsters were kept in LD for 2 weeks. Half of the animals then were sacrificed every 2 h during 24 h. The remaining hamsters were kept for 2 further weeks under an SD photoperiod of 8 h light/16 h darkness, with an extension of the dark period of 8 h toward lights-off (Fig. 1D). Animals were sacrificed by decapitation. The brains were removed rapidly, frozen on dry ice, and stored at −80°C. Serial coronal sections (20 μm) of the hypothalamic areas containing the SCN and including the PT were cut on a cryostat, thaw-mounted onto poly-l-lysine-coated slides, and kept at −80°C until in situ hybridization or autoradiography.
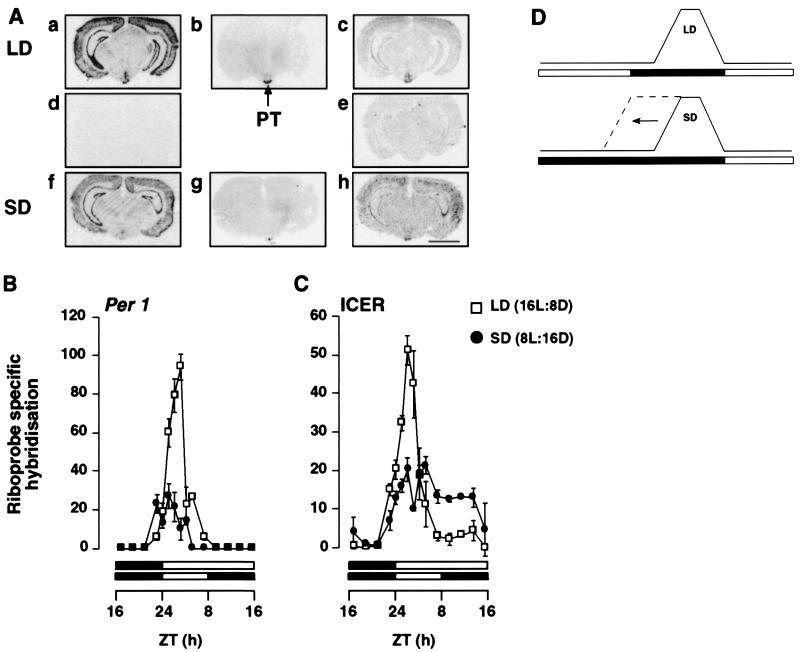
Figure 1.
Effect of photoperiod on the temporal pattern of expression of Per1 and ICER mRNA in the PT of the Syrian hamster. (A) Autoradiographs showing the expression of Per1 and ICER mRNA and 2-[125I]iodomelatonin binding in the PT and adjacent brain areas. Coronal sections (20 μm) show gene expression or radioligand binding at ZT3 under LD and SD. Sections a–c are consecutive sections showing antisense labeling for Per1 (a) and ICER (c) and 2-[125I]iodomelatonin binding (b) under LD. Sections f–h are consecutive sections showing antisense labeling for Per1 (f) and ICER (h) and 2-[125I]iodomelatonin binding (g) under SD. Sections d and e show sense labeling for Per1 (d) and ICER (e) under LD. (Bar = 4 mm.) (B and C) Diurnal pattern of expression of Per1 (B) and ICER (C) riboprobe-specific hybridization in the pars tuberalis in LD (□) and SD (●). Each value is the mean ± SEM of two to six animals per time point. The horizontal solid bars represent the dark period for LD and SD. (D) A schematic showing the effect of changing from LD to SD on the endogenous melatonin rhythm when the time of lights-on is kept the same for both photoperiods, as used in this experiment. The melatonin rhythm decompresses slowly over several cycles toward lights-off (20).
Effect of melatonin injection on Per1 and ICER expression.
Hamsters were kept in LD for 2 weeks. Melatonin (25 μg in 100 μl of a 0.1% ethanolic saline solution) then was injected s.c. into animals 1 h before lights-on. Another group of animals received an injection of 100 μl of ethanolic saline (vehicle), and the remaining animals were used as untreated controls. All animals were sacrificed 4 h after the time of melatonin injection [i.e., Zeitgeber time 3 (ZT3)].
Effect of transfer from LD to SD on Per1 and ICER expression.
Animals were kept in LD for 2 weeks. Half of the animals then were transferred to SD conditions, with an extension of the dark period by 8 h toward lights-on time relative to the LD schedule. One, 2, 5, and 14 days after the transfer, animals (LD and SD) were sacrificed each day at a time point corresponding to 3 h after lights-on in LD (the same time point was used to sacrifice the SD animals, which thus were sacrificed during the dark period).
Cloning Studies.
Cloning of Per1 and ICER was performed as described previously (10, 15). Total RNA was extracted from ovine PT, and reverse transcription—PCR was used to generate a fragment of the ovine Per1 (GenBank accession no. AF044911) gene corresponding to bases 287–690 of the human Per1 coding sequence (GenBank accession no. AB002107) and a fragment of the ovine ICER gene corresponding to bases 16–149 of the mouse ICER coding sequence (GenBank accession no. S66024). Per1 and ICER genes fragments were ligated into pGEM-T and PCRscript, respectively, and then cloned. The cloned inserts then were sequenced to verify that the amplified products corresponded to a bone fide fragment of ovine Per1 and ICER.
In situ Hybridization and Autoradiography.
In situ hybridization and autoradiography were performed as described previously (10, 15, 16). Slides were apposed to Hyperfilm β-max (Amersham) together with 14C- or 125I microscale standards (Amersham) for 5 days (Per1), 12 days (ICER), or 7 days (iodomelatonin). OD values were measured by computing densitometry (Image-Pro plus; Media Cybernetics, Silver Spring, MD) and converted into the relative OD produced by microscale standards. The riboprobe-specific hybridization or melatonin-specific binding was calculated by measuring the OD of the PT and the SCN and subtracting a background OD of equal area.
Statistical Analysis.
All statistical comparisons were made by using the computing software sigmastat. Multiple comparisons were made by using one-way ANOVA followed by the Tukey test or ANOVA on ranks by Dunn’s method as appropriate.
RESULTS
Localization of Per1 and ICER Expression in the Hamster Brain.
In situ hybridization revealed a range of structures that express Per1 and ICER genes in the Syrian hamster brain (Figs. 1A and 3A). High levels of Per1 mRNA expression were observed in the PT and the SCN at specific time points (Figs. 1A and 3A), similar to the mouse (2, 3). Strong labeling by 2-[125I]iodomelatonin, which is consistent with the localization of melatonin receptors, was observed in the PT and the SCN, but with a lower level of labeling in the SCN (Figs. 1A and 3A), as described previously (17, 18).
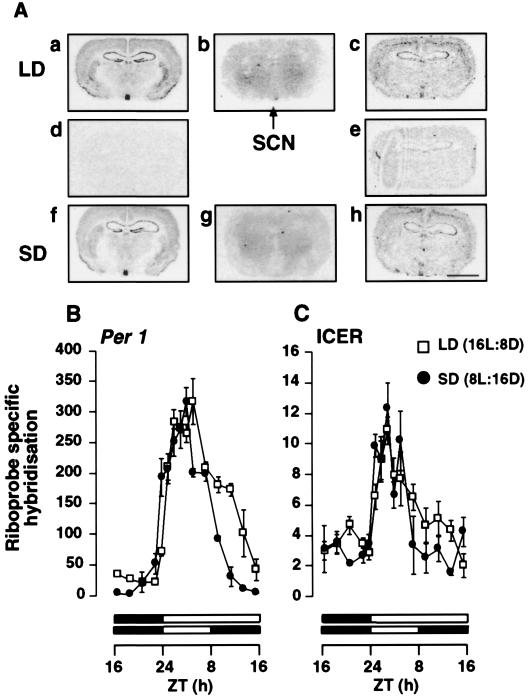
Figure 3.
Effect of photoperiod on the temporal pattern of expression of Per1 and ICER mRNA in the SCN of the Syrian hamster. (A) Autoradiographs showing the expression of Per1 and ICER mRNA and 2-[125I]iodomelatonin binding in the SCN and adjacent brain areas. Coronal sections (20 μm) show gene expression or radioligand binding at ZT3 under LD and SD. Sections a–c are consecutive sections showing antisense labeling for Per1 (a) and ICER (c) and 2-[125I]iodomelatonin binding (b) under LD. Sections f–h are consecutive sections showing antisense labeling of Per1 (f) and ICER (h) and 2-[125I]iodomelatonin binding (g) under SD. Sections d and e show sense labeling for Per1 (d) and ICER (e) under LD. (Bar = 4 mm.) (B and C) Diurnal pattern of expression of Per1 (B) and ICER (C) riboprobe-specific hybridization in the SCN in LD (□) and SD (●). Each value is the mean ± SEM of two to six animals per time point. The horizontal solid bars represent the dark period for LD and SD. The time of lights-on was held constant between LD and SD as described in Fig. 1.
Effect of Photoperiod on Per1 and ICER Expression in the PT.
Quantification of Per1 mRNA levels in the PT taken from animals on LD revealed a significant diurnal rhythm of expression (P < 0.001). A rapid increase in mRNA levels occurred immediately after lights-on, with a peak of expression at ZT3. During the dark phase, levels of expression were weak or undetectable (Fig. 1B). Animals held in SD for 2 weeks showed a significant difference in the diurnal pattern of Per1 expression relative to LD animals. The amplitude of the Per1 mRNA peak at ZT3 under SD was one-third of that under LD (P < 0.005) (Fig. 1B). (The lights-on time was common to each photoperiod; see Fig. 1D.) Similarly, ICER mRNA in the PT showed a diurnal rhythm of expression under LD and SD (P < 0.001) (Fig. 1C). Peak mRNA levels were observed at ZT3 under both LD and SD, but as for Per1, the amplitude of the peak was influenced by photoperiod, being 2.5-fold higher under LD than under SD. Following the peak, levels of ICER mRNA expression declined but did not return to baseline for several hours, remaining higher under SD than at corresponding time points under LD (P < 0.05). The difference in the levels of Per1 and ICER gene expression at ZT3 between SD and LD animals was maintained even after 8 weeks in their respective photoperiods (data not shown).
Effect of Melatonin Injection on Per1 and ICER Expression in the PT.
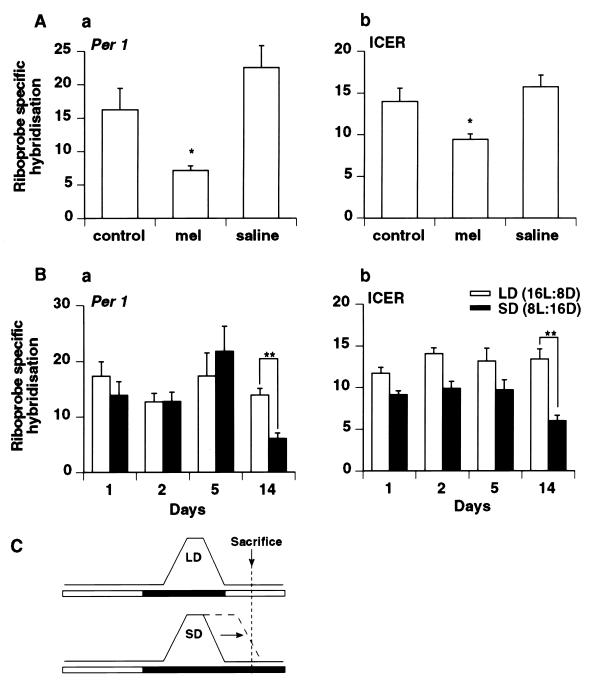
For both Per1 and ICER, the rise in mRNA expression in the PT begins before lights-on. These data could be explained if it is the fall in nocturnal levels of melatonin that permits and synchronizes the induction of Per1 and ICER mRNA. To examine this hypothesis, Syrian hamsters were injected with 25 μg of melatonin 1 h before lights-on to extend the nocturnal melatonin signal, thus delaying the fall in melatonin levels. In agreement with the hypothesis, animals sacrificed at ZT3 (4 h after injection) had lower levels of Per1 and ICER mRNA in the PT relative to control animals injected with saline (Fig. 2A), although the levels of each gene after melatonin injection were not as low as nighttime values. Melatonin is cleared rapidly after injection so that normal daytime levels are reached by 4 h after injection (19). This suggests, therefore, that melatonin present during the early-light phase either delays or prevents the onset of the Per1 and ICER peaks.
Figure 2.
(A) Effect of exogenous melatonin on the expression of Per1 (a) and ICER (b) mRNA at ZT3 in the PT of Syrian hamsters held under LD. Animals received no treatment (control), were injected with 25 μg of melatonin 1 h before lights-on (mel), or were injected with vehicle (saline). Each value is the mean ± SEM from three animals (∗, P < 0.05). (B) Expression of Per1 (a) and ICER (b) mRNA in the PT 1, 2, 5, and 14 days after a switch from LD to SD. Each value is the mean ± SEM of three animals (∗∗, P < 0.001). (C) Schematic showing the effect of changing from LD to SD on the endogenous melatonin rhythm when the timing of lights-off is held constant as in this experiment. The melatonin rhythm decompresses slowly over many cycles toward lights-on (20).
Effect of Transfer from LD to SD on Per1 and ICER Expression in the PT.
In further support of the hypothesis, transfer of animals from LD to SD showed a time-dependent effect on Per1 and ICER mRNA levels when sacrificed 3 h after lights-on relative to the LD group. (In this experiment lights-off time was common to each photoperiod, and, thus, the lights-on time changed for the SD animals; see Fig. 2C.) After 1, 2, and 5 days in SD, no significant differences in the levels of Per1 or ICER mRNA could be detected between these and the LD animals. However, after 14 days in SD, the animals showed a lower level of Per1 and ICER expression (Fig. 2B), consistent with a delay in the onset of gene expression. These data are compatible with the time course of decompression of the melatonin rhythm, which alters the timing of the fall of nocturnal melatonin.
Effect of Photoperiod on Per1 and ICER Expression in the SCN.
Next, we examined the expression of Per1 and ICER mRNA in the SCN. Both genes displayed robust diurnal patterns of expression (P < 0.01), where the peaks in mRNA level were detected during the day at ZT3–5. As for the PT, Per1 mRNA expression was found to rise before lights-on. The duration of elevated Per1 mRNA expression was longer in the SCN than in the PT under each photoperiod. In contrast to the PT, photoperiod had no influence on the amplitude of gene expression. However, photoperiod did influence the duration of elevated expression of Per1 mRNA, being extended under a longer photoperiod (Fig. 3B). Although similarly rhythmic to Per1, the level of ICER mRNA expression in the SCN was considerably lower than in the PT (Figs. 1C and 3C). Notably, unlike Per1, ICER did not rise before lights-on, but immediately after the light–dark transition. There also was no effect of photoperiod on either the duration or amplitude of the peak of ICER mRNA expression in the SCN. However, there did appear to be a trend for a longer duration of ICER expression under LD than under SD, but this difference was not statistically significant.
Effect of Melatonin Injection on Per1 and ICER Expression in the SCN.
In contrast to the PT, melatonin injection, 1 h before lights-on under LD, did not affect Per1 or ICER mRNA expression in the SCN relative to vehicle or control animals (data not shown).
Effect of Transfer from LD to SD on Per1 and ICER Expression in the SCN.
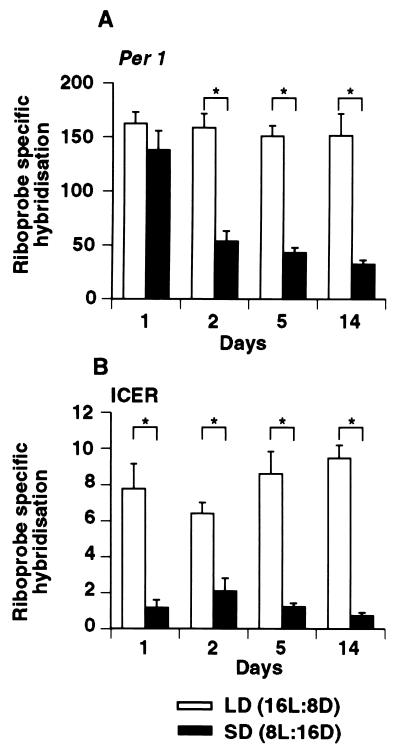
The level of expression of both genes was influenced by the transfer from LD to SD. Per1 mRNA expression measured at 3 h after lights-on relative to the LD group decreased in a time-dependent manner, reaching values equivalent to nighttime levels by day 14 (Fig. 4A). In contrast, ICER levels decreased to nighttime levels within the first day (Fig. 4B).
Figure 4.
Expression of Per1 (A) and ICER (B) mRNA at ZT3 in the SCN 1, 2, 5, and 14 days after a switch from LD to SD. The time of lights-off was held constant between LD and SD (see Fig. 2C). Each value is the mean ± SEM of three animals. (∗, P < 0.01).
DISCUSSION
Here we show how photoperiod regulates gene expression in the PT and the SCN. Namely, photoperiod is translated into gene expression amplitude in the PT, with higher expression under a long photoperiod than under a short one.
Because melatonin translates photoperiod into a neuroendocrine signal, it is likely that this hormone is responsible for the amplitude difference in Per1 and ICER expression between LD and SD in the PT. Indeed, the inhibitory effect of melatonin injection on Per1 and ICER is in favor of this hypothesis. Similarly, the transfer of Syrian hamsters from LD to SD leads to a decrease in gene expression only after 14 days. In Syrian hamsters, after a transfer from LD to SD, the duration of melatonin synthesis slowly increases; this phenomenon is known as decompression and can take up to 8 weeks to complete (20). However, after only 2 weeks, the duration of melatonin synthesis under SD is already double that of LD (20), and this could allow the melatonin signal to inhibit the expression of the Per1 and ICER peaks.
This study improves our understanding of the intracellular effects of melatonin in two ways. First, the need for melatonin levels to fall before gene expression can be induced is consistent with the known inhibitory function of melatonin receptors in the PT, which prevents activation of the gland through cAMP and other signaling pathways (5). Thus, the findings implicate an unidentified factor in the stimulation of the PT, after the fall in melatonin, to account for the induction of Per1 and ICER, because in primary cultures no independent effect of melatonin on signal transduction has been observed (21, 22). Second, because photoperiod determines the amplitude of Per1 and ICER gene expression, it seems likely that it is the duration of melatonin that defines the magnitude of gene expression in the PT. In support of this, Per1 is not detected in the PT of mice bearing a genetic defect in melatonin synthesis (2).
In the SCN, no effect of photoperiod was observed on amplitude of expression of both genes, but Per1 is expressed longer under LD relative to SD. Light induces Per1 mRNA expression in SCN during the subjective night (7–9), and, notably, at the beginning or at the end of the subjective night (7). In keeping with this, the extended period of Per1 mRNA expression observed in the SCN under LD appears directly related to the duration of light exposure. In Drosophila, phosphorylated Per protein dimerizes with phosphorylated TIM (Timeless protein) to negatively regulate their own transcription. A similar autoregulatory feedback mechanism is thought to occur in mammals, although this may involve Per–Per homodimers (23). The data presented in this study suggest that photoperiod (light) may influence the temporal dynamics of such a putative feedback mechanism.
Our observations provide an interesting contrast between the SCN and the PT, for which there are at least two potential explanations. First, the density of melatonin receptors in the SCN of the Syrian hamster is known to decrease with age (18). It has been suggested that this may account for the inability of melatonin injections to entrain circadian activity rhythms in this species (24, 25). In this study, and as described previously (17, 18), the level of 2-[125I]iodomelatonin binding in the SCN is much lower than in the PT (see Figs. 1A and 3A), and, therefore, the difference in the Per1 and ICER responses to photoperiod may reflect differences in the sensitivity to melatonin. Second, because Per1 is part of the mechanism of the biological clock, it may be important that the amplitude of the Per1 oscillation is unaffected so that intrinsic clock function is maintained. The difference between the responses of Per1 and ICER to transfer from LD to SD in the SCN may reflect differences in the regulation and function of the two transcripts. Whereas both Per1 and ICER are inducible by light in the SCN (7–9, 14), only Per1 is expressed as a self-sustaining rhythm (2, 3, 13). Therefore, the residual levels of Per1 mRNA expression on day 1 after transfer from LD to SD (Fig. 4A) may reflect spontaneous expression of Per1 occurring independently of light. In contrast, no ICER mRNA induction occurs in the absence of light.
The PT already has been implicated in the regulation of seasonal endocrine changes (5, 26). However, the new results of this study, showing the influence of photoperiod on the amplitude of two early-response genes in this tissue, indicate how the duration of the light–dark cycle may be decoded at a cellular level. The data also show how the fall in the nocturnal melatonin signal synchronizes the timing of the peak of gene expression. The expression of other early-response genes may be affected similarly by the ambient photoperiod. Thus, there is likely to be a wave of early-response gene expression and associated protein synthesis at the beginning of the light period, which sets in train further transcriptional activation. The amplitude of this wave of gene expression at the beginning of the light period may define the seasonal, physiological response of the PT tissue. Downstream consequences of either Per1 or ICER mRNA expression in the PT are not yet understood, but it is known that ICER protein acts as a repressor of cAMP-dependent gene expression (11). ICER protein can be induced in sheep PT glands (unpublished observations), and, thus, it may contribute to terminating cAMP-dependent gene expression in the PT. In addition to gene induction, we also have observed an increase in melatonin receptor expression after lights-on under long days relative to short days (Fig. 1A; data not shown), consistent with previous findings (27). Overall, these observations suggest a state of enhanced stimulation of the PT under LD relative to SD that is apparent at the level of mRNA and protein synthesis. Potentially, this may provide a general mechanism through which melatonin conveys its photoperiodic information to target sites involved in seasonal responses. Clock-related genes in addition to Per1 are expressed in the PT, such as TIM (23), BMAL1, and CLOCK (unpublished data). It will be interesting to study how these genes and their protein products interact to contribute to the interval-timing function of the PT as opposed to their role as intrinsic clock genes in the SCN. Similarly, it will be interesting to address whether the changes in gene expression as observed under entrained conditions also occur after transfer to free-running conditions.
Acknowledgments
We acknowledge the financial support of the Scottish Office for Agriculture, Environment, and Food Department (A.W.R., P.B., and P.J.M.) and of the Fondation Singer-Polignac, France (S.M.).
ABBREVIATIONS
- SCN
suprachiasmatic nucleus
- PT
pars tuberalis
- LD
long day
- SD
short day
- ICER
inducible cAMP early repressor
- DD
constant darkness
- ZT
Zeitgeber time
References
- 1.Klein D C, Moore R Y, Reppert S M. Suprachiasmatic Nucleus: The Mind’s Clock. Oxford: Oxford Univ. Press; 1991. [Google Scholar]
- 2.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 3.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Nature (London) 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre A, Damiola F, Schibler U. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 5.Morgan P J, Williams L M. Rev Reprod. 1996;1:153–161. doi: 10.1530/ror.0.0010153. [DOI] [PubMed] [Google Scholar]
- 6.Bartness T J, Powers J B, Hastings M H, Bittman E L, Goldman B D. J Pineal Res. 1993;15:161–190. doi: 10.1111/j.1600-079x.1993.tb00903.x. [DOI] [PubMed] [Google Scholar]
- 7.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros J, Dunlap J C, Okamura H. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 8.Shearman L, Zylka M J, Weaver D R, Kolakowski L F, Reppert S M. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 9.Albrecht U, Sun Z S, Eichele G, Lee C C. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 10.Morgan P J, Ross A W, Graham E S, Adam C, Messager S, Barrett P. J Neuroendocrinol. 1998;10:319–323. doi: 10.1046/j.1365-2826.1998.00232.x. [DOI] [PubMed] [Google Scholar]
- 11.Stehle J H, Foulkes N S, Molina C A, Simmoneaux V, Pévet P, Sassone-corsi P. Nature (London) 1993;365:314–320. doi: 10.1038/365314a0. [DOI] [PubMed] [Google Scholar]
- 12.Foulkes N S, Duval G, Sassone-Corsi P. Nature (London) 1996;381:83–85. doi: 10.1038/381083a0. [DOI] [PubMed] [Google Scholar]
- 13.Stehle J H, Foulkes N S, Pévet P, Sassone-Corsi P. Mol Endocrinol. 1995;9:706–716. doi: 10.1210/mend.9.6.8592516. [DOI] [PubMed] [Google Scholar]
- 14.Stehle J H, Pfeffer M, Kuhn R, Korf H W. Neurosci Lett. 1996;217:169–172. [PubMed] [Google Scholar]
- 15.Fleming J V, Barrett P, Coon S L, Klein D C, Morgan P J. Endocrinology. 1999;140:972–978. doi: 10.1210/endo.140.2.6496. [DOI] [PubMed] [Google Scholar]
- 16.Messager S, Caillol M, George D, Martinet L. J Neuroendocrinol. 1997;9:523–528. doi: 10.1046/j.1365-2826.1997.d01-1122.x. [DOI] [PubMed] [Google Scholar]
- 17.Williams L M, Morgan P J, Hastings M H, Lawson W, Davidson G, Howell H E. J Neuroendocrinol. 1989;1:315–320. doi: 10.1111/j.1365-2826.1989.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 18.Gauer F, Schuster C, Poirel V J, Pévet P, Masson-Pévet M. Mol Brain Res. 1998;60:193–202. doi: 10.1016/s0169-328x(98)00177-6. [DOI] [PubMed] [Google Scholar]
- 19.Maywood E S, Hastings M H, Max M, Ampleford E, Menaker M, Loudon A S I. J Endocrinol. 1993;136:65–73. doi: 10.1677/joe.0.1360065. [DOI] [PubMed] [Google Scholar]
- 20.Hastings M H, Walker A P, Herbert J. J Endocrinol. 1987;114:221–229. doi: 10.1677/joe.0.1140221. [DOI] [PubMed] [Google Scholar]
- 21.Morgan P J, Lawson W, Davidson G, Howell H E. J Mol Endocrinol. 1989;3:R5–R8. [Google Scholar]
- 22.Morgan P J, Barrett P, Howell H E, Helliwell R. Neurochem Int. 1994;24:101–146. doi: 10.1016/0197-0186(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 23.Zylka M J, Shearman L M, Levine J D, Jin X, Weaver D R, Reppert S M. Neuron. 1998;21:1115–1122. doi: 10.1016/s0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]
- 24.Hastings M H, Mead S M, Vindlacheruvu F J P, Ebling E S, Maywood E S, Grosse J. Brain Res. 1992;591:20–26. doi: 10.1016/0006-8993(92)90973-d. [DOI] [PubMed] [Google Scholar]
- 25.Grosse J, Davis F C. J Neurosci. 1998;18:8032–8037. doi: 10.1523/JNEUROSCI.18-19-08032.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lincoln G A, Clarke I J. J Neuroendocrinol. 1994;6:251–260. doi: 10.1111/j.1365-2826.1994.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 27.Recio J, Gauer F, Schuster C, Pévet P, Masson-Pévet M. J Pineal Res. 1998;21:162–167. doi: 10.1111/j.1600-079x.1998.tb00529.x. [DOI] [PubMed] [Google Scholar]