Abstract
The Notch and Epidermal Growth Factor Receptor (EGFR) signaling pathways interact cooperatively and antagonistically to regulate many aspects of Drosophila development, including the eye. How output from these two signaling networks is fine-tuned to achieve the precise balance needed for specific inductive interactions and patterning events remains an open and important question. Previously, we reported that the gene split ends (spen) functions within or parallel to the EGFR pathway during midline glial cell development in the embryonic central nervous system. Here, we report that the cellular defects caused by loss of spen function in the developing eye imaginal disc place spen as both an antagonist of the Notch pathway and a positive contributor to EGFR signaling during retinal cell differentiation. Specifically, loss of spen results in broadened expression of Scabrous, ectopic activation of Notch signaling, and a corresponding reduction in Atonal expression at the morphogenetic furrow. Consistent with Spen’s role in antagonizing Notch signaling, reduction of spen levels is sufficient to suppress Notch-dependent phenotypes. At least in part due to loss of Spen-dependent down-regulation of Notch signaling, loss of spen also dampens EGFR signaling as evidenced by reduced activity of MAP kinase (MAPK). This reduced MAPK activity in turn leads to a failure to limit expression of the EGFR pathway antagonist and the ETS-domain transcriptional repressor Yan and to a corresponding loss of cell fate specification in spen mutant ommatidia. We propose that Spen plays a role in modulating output from the Notch and EGFR pathways to ensure appropriate patterning during eye development.
Keywords: Spen, Ras signal transduction, Notch, Yan, morphogenetic furrow
1. Introduction
Signaling pathways are responsible for a host of cellular functions during development, including cell fate specification, morphogenesis, proliferation, differentiation, polarity establishment, programmed cell death, and motility. These signaling pathways do not operate in isolation, but integrate with other networks to elicit specific activities (reviewed in Doroquez and Rebay, 2006; Hurlbut et al., 2007; Sundaram and Han, 1996; Sundaram, 2005; Tan and Kim, 1999; Voas and Rebay, 2003). Thus, a major challenge in molecular developmental genetics is to identify those genes that facilitate such coordination.
The compound eye of the fruitfly Drosophila melanogaster is a paradigm for gaining insight into mechanisms that govern combinatorial signaling (Freeman, 1997; Ready et al., 1976; Voas and Rebay, 2003). The adult eye is composed of ~750 regularly repeated, light-sensing ommatidia that each contains eight photoreceptors (R cells) and 12 accessory cells (Ready et al., 1976). The eye develops from an imaginal disc primordium that originates embryonically and proliferates until the third larval instar, when cells arrest at the G1 stage of the cell cycle and apically constrict to form the morphogenetic furrow (MF) at the disc’s posterior edge (Wolff and Ready, 1991). As the MF travels anteriorly, Notch signaling establishes and then refines an initially broad expression pattern of the proneural bHLH transcription factor Atonal (Ato) for specification of evenly-spaced R8 photoreceptors, the founder cells of developing ommatidia, in processes known as proneural enhancement and lateral inhibition, respectively (Baker et al., 1996; Baker and Yu, 1997; Baonza and Freeman, 2001; Dokucu et al., 1996; Jarman et al., 1994; Li and Baker, 2001).
Reiterative signaling by the Epidermal Growth Factor Receptor (EGFR)/Ras/MAP Kinase (MAPK) pathway (hereafter termed EGFR pathway) is then required for progressive and sequential recruitment of cell types, with the remaining neuronal photoreceptor cells added in a stereotypical pair-wise manner (R2/5, R3/4, R1/6, and finally R7), followed by non-neuronal cone and pigment cells (Freeman, 1996). Mechanistically, in response to EGFR activation, dual-phosphorylated ERK (dpERK/activated MAPK) translocates into the nucleus to phosphorylate proteins including two ETS-domain transcription factors, Pointed-P2 (Pnt-P2) and Yan. Phosphorylation of Pnt-P2 potentiates its activator function, whereas phosphorylation abrogates Yan-mediated repression (Brunner et al., 1994; O’Neill et al., 1994; Rebay and Rubin, 1995). This results in genes formerly repressed by Yan becoming activated by Pnt-P2, thereby initiating a cellular response appropriate to the original stimulus.
The Notch and EGFR pathways interact intimately through genetic cooperation or antagonism; however, the molecular mechanisms that underlie these relationships remain largely unclear (reviewed in Doroquez and Rebay, 2006; Sundaram, 2005). One way by which the Notch pathway interfaces with the EGFR pathway is through regulation of Ato at the MF (Baonza and Freeman, 2001; Chen and Chien, 1999). Here, Ato is required for the expression of rhomboid (rho), which processes TGF-α-like ligands that activate EGFR (Baonza and Freeman, 2001; Lee et al., 2001; Urban et al., 2001; Urban et al., 2002), and for the activity of dpERK (Chen and Chien, 1999). Therefore, Notch-mediated lateral inhibition represses ato and limits EGFR signaling output. Since both Notch and EGFR networks orchestrate virtually all aspects of eye development, numerous mechanisms will undoubtedly be required for precise and context-appropriate coordination of pathway outputs.
We initially identified split ends (spen) in a screen directed to uncover novel downstream effectors and regulators of the EGFR pathway (Rebay et al., 2000). Spen belongs to a family of nuclear proteins defined by two domains: (1) three RNA-recognition motifs (RRMs) at the N-terminus and (2) a C-terminal domain termed the Spen Paralog and Ortholog C-terminus (SPOC) (Kuang et al., 2000; Rebay et al., 2000; Wiellette et al., 1999). Consistent with the conservation of signaling pathways with which Spen has been implicated, Spen orthologs have been reported from worms to humans (Ludewig et al., 2004; Ma et al., 2001; Mercher et al., 2001; Newberry et al., 1999; Shi et al., 2001).
Our genetic analysis showed that Spen functions in a positive manner with respect to EGFR signaling during Drosophila embryonic central nervous system development (Chen and Rebay, 2000; Rebay et al., 2000) whereas a parallel study suggested spen antagonizes EGFR output during embryonic neural development (Kuang et al., 2000). Additional studies have suggested that Spen may regulate a variety of other processes including the cell cycle, planar cell polarity, and cell death (Dickson et al., 1996; Firth et al., 2000; Kuang et al., 2000; Lane et al., 2000; Lin et al., 2003; Mace and Tugores, 2004; Mace et al., 2005; Sanchez-Pulido et al., 2004; Staehling-Hampton et al., 1999). This pleiotropy of developmental requirements positions Spen as an appealing candidate for integrating information from multiple signaling networks and the Drosophila eye provides an ideal context in which to explore such questions.
Spen family proteins have been best characterized as transcriptional co-repressors (Ariyoshi and Schwabe, 2003; Kuroda et al., 2003; Li et al., 2005; Ludewig et al., 2004; Ma et al., 2007; Oswald et al., 2002; Oswald et al., 2005; Shi, 2002; Shi et al., 2001; Tsuji et al., 2007; Vadlamudi et al., 2005; Yang et al., 2005). Notably, the mammalian orthologs human SHARP and mouse MINT interact with the Notch pathway transcriptional effector RBP-J(κ), an ortholog of Drosophila Suppressor of Hairless [Su(H)], and recruit a repressor complex (Ariyoshi and Schwabe, 2003; Kuroda et al., 2003; Li et al., 2005; Ma et al., 2007; Oswald et al., 2002; Shi et al., 2001; Tsuji et al., 2007; Yang et al., 2005). When Notch signaling is initiated, the repressor complex is competed away from RBP-J(κ) by Notchintra, allowing for activators to promote expression from target genes (Kuroda et al., 2003; Oswald et al., 2002; Oswald et al., 2005). The developmental significance of spen function with respect to Notch signaling remains poorly understood in Drosophila.
In this study, we explore the requirement for spen function during Drosophila eye development, toward the goal of elucidating how Spen influences output from the Notch and the EGFR pathways. Using somatic mosaic analysis to induce patches of spen mutant tissue in the developing eye imaginal disc, we examined expression of a collection of molecular markers that provide sensitive readouts of both Notch and EGFR pathway activity. Our results indicate that spen functions as a negative regulator of Notch signaling at the MF. Consistent with the elevated Notch signaling that occurs in spen mutant tissue, we observe a concomitant loss of Ato and dpERK at the MF, and thus reduced output from the EGFR pathway as evidenced further by cell fate specification defects. Together, our results reveal a role for Spen as an antagonist of Notch and a promoter of EGFR signaling during Drosophila retinal development.
2. Results
2.1. spen is required for proper proneural gene expression at the morphogenetic furrow
To explore a coordinate role for spen in Notch and EGFR signaling, we chose to study its function in the developing compound eye, a tissue whose proper patterning requires reiterative inputs from both pathways. To circumvent the embryonic lethality of loss-of-function spen alleles (Chen and Rebay, 2000; Gellon et al., 1997; Kuang et al., 2000; Wiellette et al., 1999), we generated and analyzed somatic mosaic spen clones (see EXPERIMENTAL PROCEDURES). Because we did not observe proliferative or other defects suggestive of a critical role early in eye development (data not shown), we focused our investigation on the requirement for spen during the specification and differentiation events that occur in and posterior to the MF.
We began by asking whether loss of spen altered the expression pattern of the proneural protein Ato. The pattern of proneural Ato in the eye occurs in four stages in wild-type imaginal discs (reviewed in Doroquez and Rebay, 2006; Frankfort and Mardon, 2002). First, Ato is expressed in a broad D-V stripe in the progressing MF during a process termed proneural enhancement (Stage 1) (Jarman et al., 1995). Ato is then restricted posteriorly via lateral inhibition into the intermediate groups (IGs), the earliest stage of ommatidial cluster formation (Stage 2), of which two or three cells become equipotent in their ability to become the founding photoreceptor R8 (Stage 3) (Fig. 1A–C/Ato, GFP-positive tissue). A single cell within this R8 equivalence group becomes the R8 and maintains Ato expression (Stage 4) (Dokucu et al., 1996).
Fig. 1.
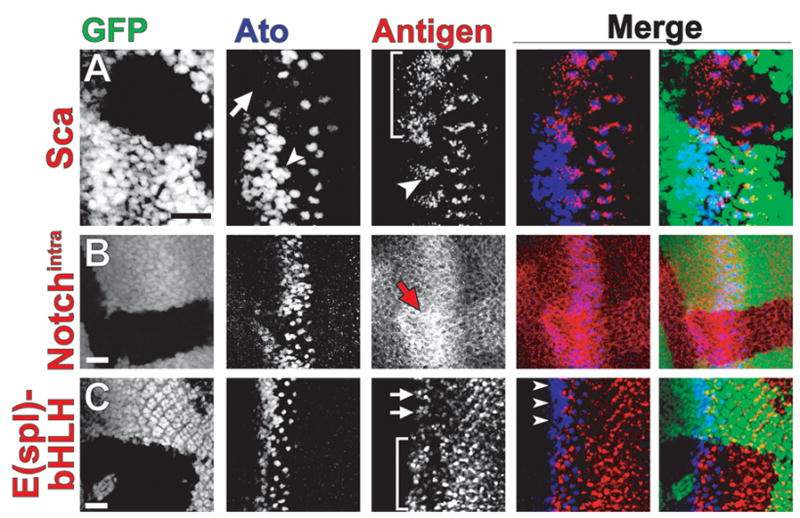
Up-regulation of Notch pathway components in spen clones affects Atonal expression. Confocal micrograph of spen eye disc clones stained (in red) for (A) Scabrous, (B) Notchintra, (C) E(spl)-bHLH (mAb323). Discs were also co-stained for Ato (blue). spen mutant tissue lacks GFP (green). Ato staining is reduced in spen clones (A, arrow; B). Sca expression is broadened in spen clones (A, bracket) as compared to its restricted expression in IG cells (A, arrowhead). Notch expression is elevated in clones (B, arrow). E(spl)-bHLH expression is broadened in spen clones (C, bracket) and elevated posterior to the MF. E(spl)-bHLH is normally restricted to alternating groups of cells (C, arrows) complementary to Ato-positive IG cell expression in the MF. Discs are oriented anterior to the left. Scale bar = 20 μm.
In the majority of spen clones that cross the MF, Ato is significantly reduced during Stages 1–3 (Fig. 1A–C, GFP-negative tissue). However, in all cases, Ato expression ‘recovers’ and becomes refined to single cells—the presumptive R8s—that are spaced abnormally as compared to wild-type tissue (Fig. 1A–C). These data suggest that Spen is required for the maintenance of Ato proneural expression at the MF.
2.2. Loss of spen leads to elevated levels of Sca protein and transcript
Like Ato, the fibrinogen-related protein Scabrous (Sca) is expressed in the IGs, but is secreted to nearby cells for the restriction of Ato expression during lateral inhibition (Baker et al., 1990; Baker and Zitron, 1995; Lee et al., 1996; Mlodzik et al., 1990). Since we observed down-regulation of Ato in spen eye clones, we hypothesized that spen loss could promote Ato inhibition via broadening or elevating Sca expression. Indeed, in spen clones, the expression domain of Sca is dramatically broadened at the MF, especially in the IG zone where Ato expression is down-regulated (Fig. 1A, GFP-negative tissue; Fig. S1A,B [Supplementary Data]).
To test whether the increase in Sca expression reflected increased transcription, we performed qRT-PCR to compare levels of sca transcript levels in spen mutant discs to those in wild-type discs. We find that sca mRNA levels are significantly elevated (1.6-fold; p = 0.009) in spen mutant discs as compared to wild-type (Fig. 2A). This suggests that in wild-type tissue, Spen directly or indirectly represses sca transcription or mRNA stability.
Fig. 2.
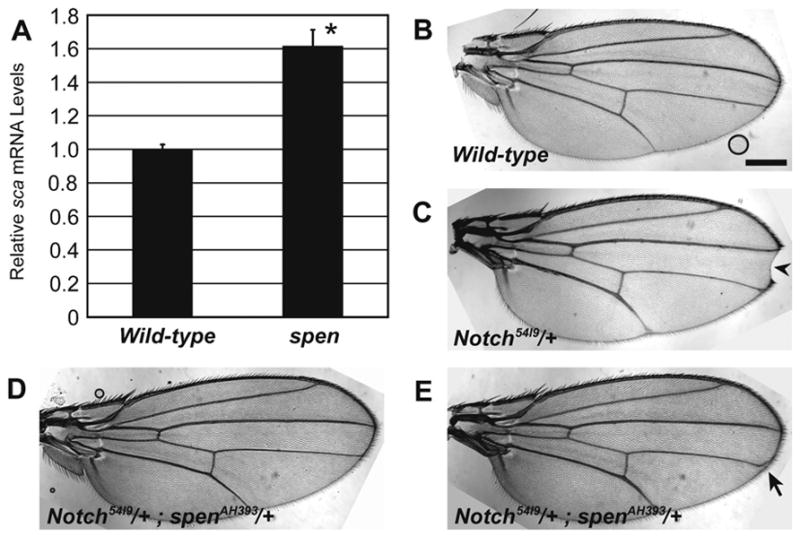
Antagonism of Notch by Spen: loss of spen leads to elevated sca transcript levels and heterozygous reduction of spen suppresses the Notch wing phenotype. (A) qRT-PCR was performed to measure relative sca mRNA levels in wild-type and spen/cell lethal eye discs (three samples, mean + st. dev.). Elevated levels are significant (*), p = 0.009. (B–E) Adult wing phenotypes of (B) wild-type, (C) Notch54l9/+, (D,E) Notch54l9/+; spenAH393/+ flies. Notch heterozygotes have loss of distal wing margin tissue (C, arrowhead). This is suppressed in Notch/+;spen/+ double-heterozygotes (D,E), where 16.7% of flies (n = 48) have aberrant LIII wing veins targeting to the margin (E, arrow). Scalebar = 0.3 mm.
2.3. Loss of spen results in elevated Notch signaling at the morphogenetic furrow
Because Sca facilitates Notch-mediated lateral inhibition (Baker and Zitron, 1995; Lee et al., 1996), broadly expressed Sca in spen clones would be predicted to activate the Notch pathway ectopically. To test this, markers for activation of Notch signaling were measured to determine the status of this pathway. First, we examined Notch expression with a monoclonal antibody specific to the intracellular domain of Notch that detects the cleaved, activated form of Notch (Notchintra) as well as full-length membrane-bound Notch (Fehon et al., 1990). In wild-type eye disc tissue, Notch is expressed predominantly apically throughout the eye imaginal disc tissue with a peak of expression at the MF and tapering anteriorly (Fig. 1B, GFP-positive tissue) (Baker and Yu, 1998; Baker and Zitron, 1995; Kidd et al., 1989). We found Notch protein levels elevated in spen clones with significant up-regulation at the MF (Fig. 1B, GFP-negative tissue). We also detected elevated levels of Notch using an antibody specific for the extracellular domain of Notch (Diederich et al., 1994) but did not detect significant change in Notch transcripts in spen mutants (Fig. S1C,D and data not shown), consistent with the previous demonstration that elevated Sca expression stabilizes Notch protein levels (Powell et al., 2001).
Because elevated receptor levels need not imply increased signaling output, we asked whether the Notch pathway was activated in spen clones. First, we examined the expression of the E(spl)-bHLH transcription factors, whose expression provides a direct read-out of Notch signal transduction (Bailey and Posakony, 1995; Baker et al., 1996; Dokucu et al., 1996; Jennings et al., 1994; Lecourtois and Schweisguth, 1995). With an antibody that interacts with four of the seven E(spl) bHLH proteins [E(spl)-mδ, -mγ, -mβ, and -m3], these proteins are detected at the MF in an alternating pattern with Ato-positive IG cells and in cells posterior to the MF (Fig. 1C, GFP-positive tissue; Fig. S1E,F). The MF expression of E(spl)-bHLHs detected in wild-type tissue is consistent with a role for E(spl)-bHLHs in the repression of ato during Notch-mediated lateral inhibition (Baker et al., 1996; Dokucu et al., 1996). In spen clones, however, the alternating E(spl)-bHLH/Ato expression is disrupted such that E(spl)-bHLH is elevated and broadened at the MF (Fig. 1C, GFP-negative tissue; Fig. S1E,F). Consistent with the activation of Notch signaling in this tissue, we also detected elevated levels of E(spl)-bHLH transcripts in spen mutant discs (Fig S2A,B). The expanded E(spl)-bHLH pattern is consistent with the repression of Ato observed in spen clones. In addition to broadening of E(spl)-bHLH expression at the MF, we observe modestly elevated levels of E(spl)-bHLH posterior to the MF in spen clones as compared to wild-type tissue (Fig. 1C; Fig. S1E,F), indicating that loss of spen leads to prolonged hyperactivation of the Notch pathway.
2.4. Reduction of spen suppresses Notch haploinsufficiency phenotypes in the wing
The results just described are all consistent with Spen antagonizing the Notch pathway. To test this model further, we looked for dose-sensitive genetic interactions between spen and Notch. Although Notch null mutations are homozygous lethal, heterozygosity for Notch results in a dominant loss of tissue at the distal wing margin ([N54l9/+] 88% notched wing, n = 256) as compared to wild-type adult wings (0% notched wing, n = 100) (Fig. 2B,C) (Mohler, 1956), providing a sensitized genetic background in which to test for interactions. Consistent with our model that predicts a reduction in spen should elevate Notch levels, we found that adult flies doubly heterozygous for Notch and spen showed complete suppression of the wing notching phenotype ([N54l9/+; spenAH393/+] 0% notched wing, n = 48) (Fig. 2D,E).
2.5. Loss of spen reduces dpERK levels in the developing eye
What might be the functional consequences of reduced Ato expression and up-regulated Notch signaling in spen mutant eye tissue? Given that the Notch and EGFR pathways often operate antagonistically, we asked whether output from the EGFR pathway was perturbed. Specifically we asked whether dpERK (the dually-phosphorylated and activated form of MAPK) levels are affected. In wild-type discs, dpERK is most prominent in the IGs at the MF, and its expression pattern depends on Ato (Baonza et al., 2001; Chen and Chien, 1999). Posterior to the MF, dpERK is expressed at lower levels where it is predominantly cytoplasmic but also transits to the nucleus to phosphorylate target proteins (Fig. 3, GFP-positive tissue; Fig. S3A,B) (Chen and Chien, 1999; Gabay et al., 1997; Kumar et al., 2003; Kumar et al., 1998; Spencer et al., 1998; Yang and Baker, 2003). Consistent with the loss of Ato expression in the IGs, we find that the dpERK expression in the IGs is drastically reduced in spen mutant clones (Fig. 3, arrow). dpERK levels posterior to the MF appear unchanged (Fig. 3, arrowhead). The reduced dpERK levels that result from loss of spen suggest Spen may potentiate EGFR signaling via antagonism of the Notch pathway (see DISCUSSION).
Fig. 3.
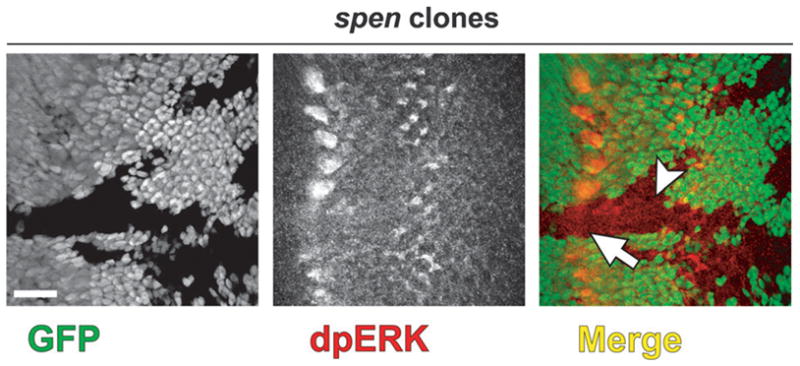
dpERK expression is lost at the MF in spen clones. Confocal micrograph of eye discs oriented with anterior to the left in spen clones stained for dpERK (red). spen mutant tissue lacks GFP (green). dpERK is lost at the MF in spen clones (B, arrow) but appears unchanged posterior to the MF (B, arrowhead). Scalebar = 20 μm.
2.6. Transcriptional output from the EGFR pathway is perturbed in the absence of spen
Do the reduced dpERK levels in spen mutant tissue dampen output from the EGFR signaling pathway? In wild-type animals, upon stimulation of EGFR signaling, phosphorylation of the transcriptional repressor Yan by dpERK leads to Yan nuclear export and degradation (Rebay and Rubin, 1995; Tootle et al., 2003). Therefore, we asked if Yan protein levels in wild-type versus spen mutant imaginal disc clones were different. In wild-type eye discs, Yan is expressed at the MF and in the basally located nuclei of undifferentiated cells posterior to the MF (Fig. 4A), but is down-regulated in the apically localized nuclei of differentiating cells (Fig. 4C) (Lai and Rubin, 1992). Thus, in developing wild-type eye discs, Yan expression is complementary to that of the pan-neuronal nuclear marker Elav, consistent with Yan’s function to block neuronal differentiation (Fig. 4A,C) (O’Neill et al., 1994; Rebay and Rubin, 1995). In spen clones, Yan expression is elevated in cells both at and posterior to the MF (Fig. 4B,D; Fig. S3C,D). Western blot analysis of eye discs mutant for spen confirms that Yan protein levels are elevated relative to wild-type (data not shown). Posterior to the MF, Yan up-regulation is evident throughout the clone in basally-localized nuclei (Fig. 4B) and in a subset of apical nuclei (Fig. 4D). As in wild-type, Yan and Elav expression in spen clones is largely complementary despite Yan’s encroachment into the apical plane, consistent with up-regulated Yan blocking these cells from adopting a neuronal fate (Fig. 4B,D).
Fig. 4.
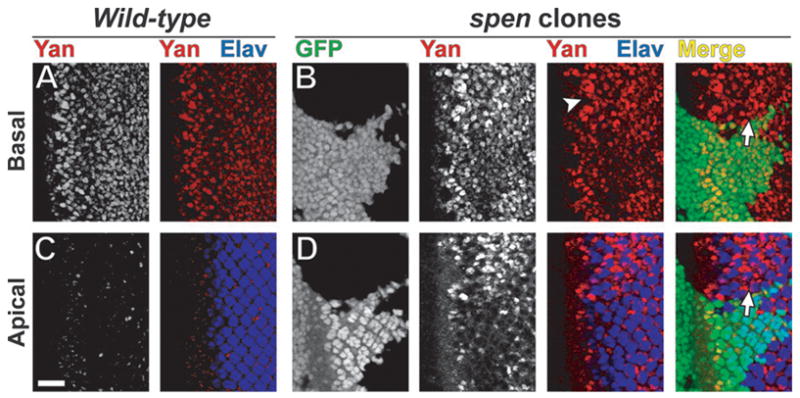
Yan is up-regulated in spen clones. Confocal micrographs of third instar eye imaginal discs oriented anterior to the left in wild-type (A,C) and spen clones (B,D) at basal (A,B) and apical (C,D) levels. Discs were stained for Yan (red) and Elav (blue), which are complementary in expression. spen mutant tissue is marked by lack of GFP (B,D; green). Yan is up-regulated in spen clones both in (B, arrowhead) and posterior (D, arrow) to the MF. Scalebar = 20 μm.
Mechanistically, elevated Yan protein levels should inappropriately repress expression of EGFR pathway target genes. For example, expression of argos (aos), which encodes a feedback inhibitor of EGFR signaling, is normally repressed by Yan in the absence of pathway activation and activated by Pnt upon pathway stimulation (Freeman, 1994; Gabay et al., 1996; Golembo et al., 1996; Sawamoto et al., 1996; Sawamoto et al., 1994; Schweitzer et al., 1995). Using quantitative real-time RT-PCR (qRT-PCR), we find that aos mRNA levels are significantly reduced approximately two-fold (p = 0.001) in spen mutant eye discs relative to wild-type, indicating that transcriptional output downstream of the EGFR signaling pathway is compromised (Fig. 5A).
Fig. 5.
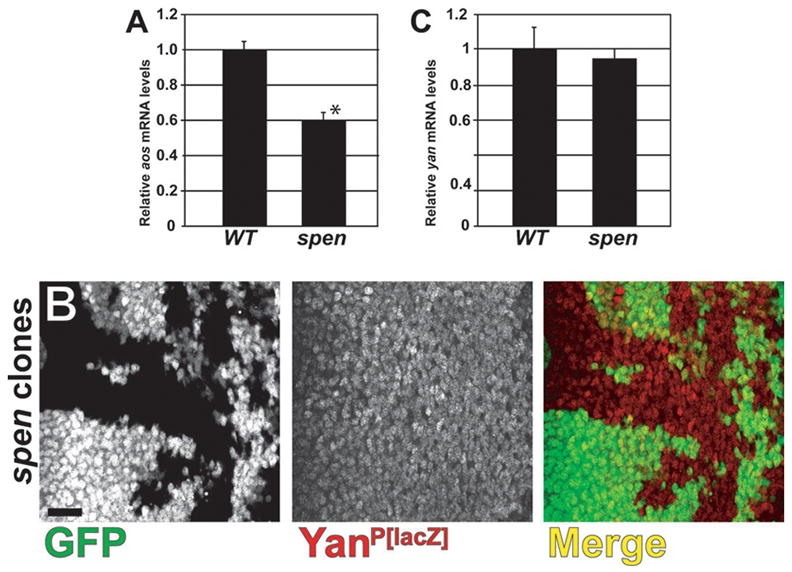
Loss of spen leads to reduced EGFR pathway target expression, but does not alter yan transcript levels. (A,C) qRT-PCR was performed to measure relative (A) argos and (C) yan mRNA levels in wild-type (w1118 and Oregon R, respectively) and spen/cell lethal eye discs (three samples, mean + st. dev.). The reduced aos transcript levels in spen discs is significant (*), p = 0.001, but yan transcript levels do not change, p = 0.602. (B) Confocal micrograph of spen clones generated in eye discs carrying the yanP[lacZ] enhancer-trap. β-Galactosidase levels are indistinguishable between control (GFP-positive) and spen mutant (GFP-negative) tissue. Discs are oriented anterior to the left. Scalebar = 20 μm.
2.7. yan is not transcriptionally regulated by Spen
Our results suggest that Spen regulates Yan levels post-translationally via regulation of dpERK levels, but it is alternatively possible that yan is regulated transcriptionally by Spen. This seems reasonable for two reasons: first, Notch signaling has been previously shown to transcriptionally activate yan expression directly (Rohrbaugh et al., 2002); and second, Spen family proteins have been implicated as transcriptional repressors of Notch pathway target genes (Kuroda et al., 2003; Oswald et al., 2002; Oswald et al., 2005).
To address this question, we asked whether loss of spen changes the expression of an enhancer-trap reporter, yanP[lacZ], that recapitulates the eye-specific yan mRNA expression (Lai and Rubin, 1992; Rohrbaugh et al., 2002). If Spen negatively regulates yan transcription, β-Gal reporter signal should be elevated in spen mutant clones. Analysis of spen clones generated in the yanP[lacZ] background detected no change in β-GAL levels relative to wild-type control tissue (Fig. 5B). In situ hybridization analysis similarly showed no change in yan expression in spen clones (data not shown).
To confirm the above finding and to further analyze whether Spen might negatively regulate Yan expression by affecting the production or stability of yan mRNA, we examined yan mRNA levels via qRT-PCR analysis of spen mutant eye discs. We find that yan transcript levels in spen mutants are not significantly different than those measured in wild-type eye discs (Fig. 5C). Together, these data indicate that Spen does not inhibit yan mRNA production or stability.
Having ruled out transcriptional regulation of yan by Spen, we next asked if Spen regulates Yan post-transcriptionally. To investigate this, we compared the levels of Yan expressed from a cDNA transgene lacking all introns, the entire 5′UTR and the majority (all but 249 bp) of the 3′UTR in the presence or absence of spen. Because epitope tags significantly compromise Yan subcellular localization and function (T.L. Tootle and I. Rebay, unpublished results), we performed these experiments with untagged transgene constructs and detected expression with anti-Yan antibody (Rebay and Rubin, 1995). This precluded us from performing the experiment in the developing eye, as it would be impossible to distinguish endogenous from ectopically-expressed Yan. Therefore, we expressed wild-type Yan (YanWT) in the developing wing imaginal discs where, like the eye, Spen is ubiquitously expressed (Lin et al., 2003) and the EGFR signaling pathway is active (Gabay et al., 1997). However, Yan is not expressed in the wing (data not shown), allowing us to follow specifically the protein expressed from the cDNA transgene.
To circumvent the larval lethality that results from ubiquitous expression of YanWT, we expressed UAS-yan transgenes under control of the dpp-GAL4 driver, which targets expression to a stripe of cells along the anterior-posterior (A-P) margin of the wing imaginal disc (Fig. 6A) (Morimura et al., 1996). As a control, we co-expressed YanWT and GFP and compared the two expression patterns. Interestingly, YanWT protein expression is limited to only a subset of GFP-positive cells; specifically, YanWT is not detected at the lateral edges of the dpp stripe (Figure 6A; Fig. S4A,B). Based on prior work showing that endogenous EGFR signaling induces robust down-regulation of Yan expressed from this transgene in a variety of developmental contexts (Rebay and Rubin, 1995), we reasoned that a similar phenomenon was occurring in the wing. Supporting the idea, dpERK is elevated at the lateral sides of the A-P border of the wing pouch (Gabay et al., 1997), precisely the region where ectopic dpp>YanWT is lost. Therefore, to test our model, we drove expression of the dpERK-unresponsive YanACT transgene (Rebay and Rubin, 1995) and found a close correlation between Yan-positive and GFP-positive nuclei (Fig. 6B; Fig. S4C,D). These data show that accumulation of ectopic Yan protein in the wing is regulated by EGFR signaling as it is in the eye, suggesting that the wing provides a relevant context in which to investigate Yan post-translational regulation by Spen.
Fig. 6.
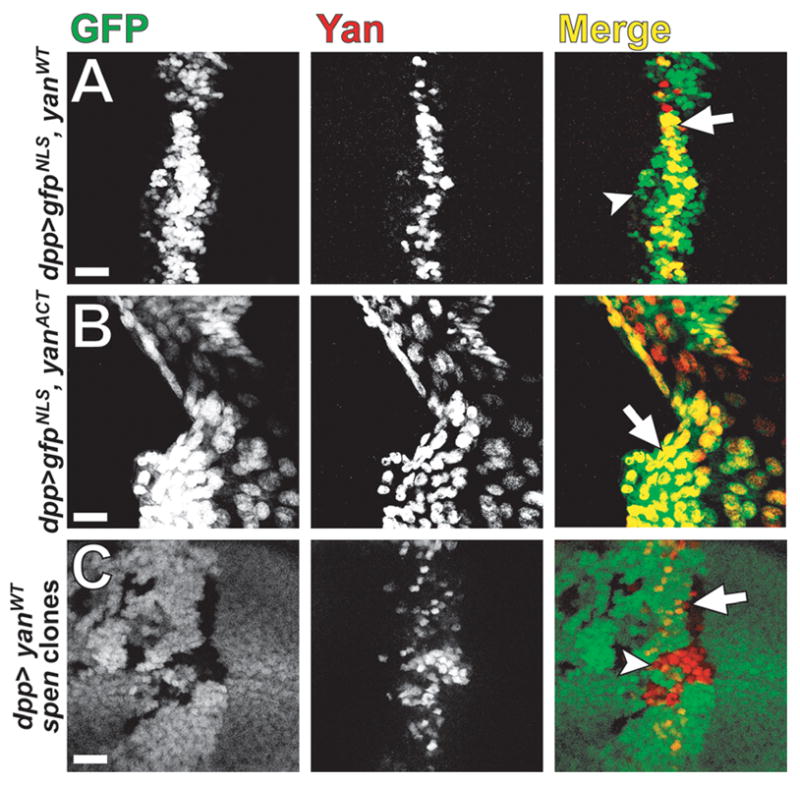
Yan is up-regulated in spen wing clones that express ectopic Yan from a cDNA trangene. (A,B) Confocal micrographs of third instar wing imaginal discs that co-express NLS-tagged GFP with (A) YanWT or (B) YanACT under the control of the dpp-GAL4 driver. dpp>YanWT (A, arrow) is expressed in a subset of cells where dpp>GFPNLS is driven (B, arrowhead). However, dpp>YanACT co-localizes well (B, arrow) with dpp>GFPNLS. (C) dpp>YanWT is expressed in a spen clones background. spen clones lack GFP (green). More red Yan-positive cells are seen in spen mutant tissue (C, arrowhead) than in control tissue (C, arrow). Discs are oriented dorsal up. Scalebar = 20 μm.
To ask if loss of spen function affects Yan protein levels when expressed under an exogenous promoter, we next generated spen clones in wing imaginal discs expressing dpp>YanWT. We found that in spen mutant wing clones, ectopic Yan is more broadly expressed compared to wild-type cells (Fig. 6C, Fig. S4E,F). Importantly, generation of spen clones in an otherwise wild-type wing disc is not sufficient to induce expression of yan from the endogenous locus (data not shown). The observation that ectopic expression of Yan is required to observe up-regulation upon loss of spen and that the wing experiments are performed with a cDNA transgene, argue that Spen-mediated regulation of Yan occurs post-translationally, most likely via activation of ERK.
2.8. spen is required for patterning of neuronal and non-neuronal cell types during eye development
Because we observed elevated Notch and reduced EGFR pathway signaling activities in spen mutant clones, we were interested in how this disrupted signaling might affect recruitment and differentiation of the different cell types that comprise each ommatidium of the eye. To begin, we analyzed the phenotype of spen mutant clones in adult eyes. In wild-type adult eyes, tangential sections reveal regularly-spaced ommatidia, each containing eight trapezoidally-arrayed photoreceptor R cells (only seven of which are observed in any given plane) separated by a non-neuronal pigment cell lattice (Fig. 7A). Disorganization of ommatidia in spen mutant tissue reflects loss of R cells and likely disruption of the supporting lattice of non-neuronal accessory cells (Fig. 7B). The spen phenotype is highly penetrant between ommatidia. Sixty-percent of spen mutant ommatidia have reduced number of rhabdomeres. Four percent of spen mutant ommatidia have more than seven rhabdomeres, likely due to the failure of R7 and/or R8 positioning or the splitting of rhabdomeres within an ommatidium (data not shown). Of the 29 percent that showed seven rhabdomeres within tangential sections, 91 percent had abnormal rhabdomere morphology, orientation, and/or positioning of rhabdomeres in a trapezoidal pattern within an ommatidium. Thus, spen is required for the proper patterning and complement of ommatidial cell types, consistent with earlier descriptions of spen mutant eyes (Dickson et al., 1996).
Fig. 7.
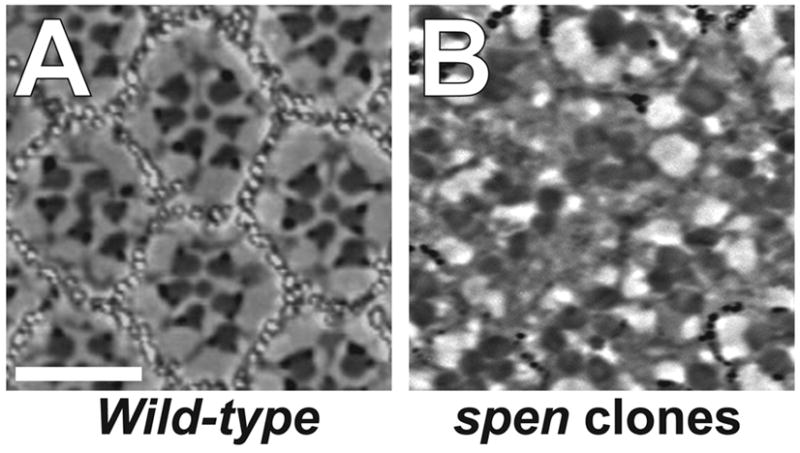
Loss of spen disrupts morphology of the adult eye. Tangential sections of adult compound eyes from (A) wild-type (Oregon R) and (B) spen clones. Photoreceptor loss and ommatidial disorganization is evident in spen mutant tissue, which is marked by lack of the white gene product. Scalebar = 10 μm.
Considering the prominent role EGFR signaling plays in specifying cell fates in the developing eye (Freeman, 1997; Schweitzer and Shilo, 1997; Shilo and Raz, 1991), we investigated whether differentiation of specific cell types was compromised by loss of spen by examining expression of markers for neuronal and non-neuronal cell specification in third instar eye imaginal discs. In wild-type tissue, R cells, positive for the pan-neuronal nuclear marker Elav (Fig. 8A) (Robinow and White, 1991), are recruited sequentially to each ommatidium posterior to the MF, followed by addition of non-neuronal Cut-positive cone cells (Fig. 8K) (Blochlinger et al., 1990). In spen mutant discs, fewer Elav-positive cells are present relative to wild-type, suggesting that R cell specification is perturbed (Fig. 8B, arrows). spen’s effect on R cell development is quite penetrant, as demonstrated by analysis of later pupal stages in which the morphology of the tissue makes the reduced numbers of Elav-positive cells easier to view (Fig. S5A–D).
Fig. 8.
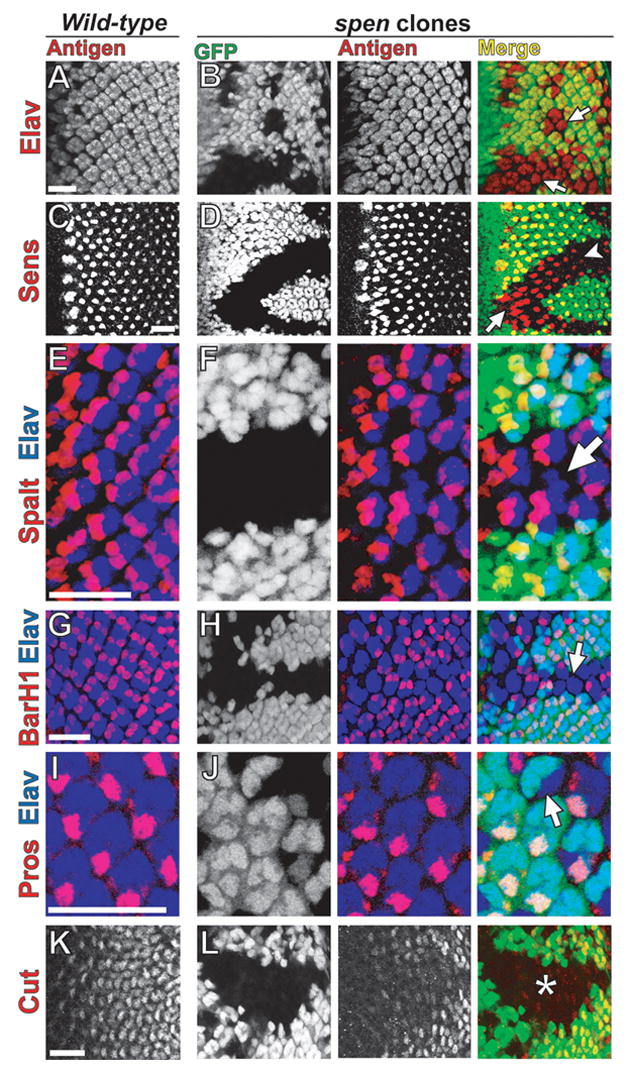
Loss of neuronal and non-neuronal ommatidial cell types in spen mutant tissue. Confocal micrographs of third instar eye imaginal discs in wild-type (A,C,E,G,I,K) and spen clones (B,D,F,H,J,L). Discs, oriented anterior to the left, were stained (red) for Elav (neuronal nuclei; A,B), Senseless (R8; C,D), Spalt (R3/4; E,F), BarH1 (R1/6; G,H), Prospero (R7; I,J), or Cut (cone; K,L). Elav (blue) was used to co-stain for ommatidial clusters (E–J). spen mutant tissue is marked by lack of GFP (green). In spen clones, there are fewer Elav-positive R cells per ommatidial cluster (B, arrows). The initial R8 spacing defect in spen clones (D, arrow) is later resolved (D, arrowhead). spen clones have ommatidial clusters that lack specific R cells [R3/4 (F, arrow), R1/6 (H, arrow), R7 (J, arrow)] and also have reduced expression of cone cell marker (L, asterisk). Scalebar = 20 μm.
To determine which photoreceptors failed to develop within spen mutant ommatidia, cell-type specific R cell markers were examined. R8 is the first photoreceptor to differentiate in each ommatidium as marked by Senseless (Sens) expression (Fig. 8C) (Nolo et al., 2000). In spen clones, R8 spacing appears irregular (Fig. 8D, arrow) near the MF as compared to WT; however, this defect is resolved more posterior to the MF and R8 differentiation does not seem compromised in spen mutant tissue (Fig. 8D, arrowhead). The R8 spacing defect is consistent with Spen’s role in affecting Notch-mediated lateral inhibition of Ato at the MF (Fig. 1A). Furthermore, the R8 spacing defect seen here is also consistent with reduced EGFR signaling in spen tissue, as it has been previously shown that loss of dpERK activity in EGFR mutant clones at the MF leads to abnormal R8 spacing (Baonza and Freeman, 2001).
We also analyzed the differentiation of R3/4, R1/6, and R7 as determined by Spalt (de Celis and Barrio, 2000), BarH1 (Higashijima et al., 1992), and Prospero (Pros) (Spana and Doe, 1995) expression, respectively (Fig. 8E–J). In spen mutants, loss of R3 and R4 occurs, but at fairly low penetrance (Fig. 8F, arrow). Loss of R1 and R6 in spen clones is more striking, although not fully penetrant between ommatidia (Fig. 8H, arrow). Like R3/4 and R1/6, R7 differentiation is also compromised in spen mutants (Fig. 8J, arrow). These results are consistent with the terminal phenotype of penetrant but irregularly patterned photoreceptor loss observed in adult eye sections (Fig. 7B). In contrast to variable loss of Elav-positive cells, loss or severe reduction in Cut expression is completely penetrant (Fig. 8L, asterisk), suggesting a role for spen in cone cell differentiation. As expected, during pupal development, fewer than the normal complement of four cone cells are present within most ommatidia (Fig. S5E,F). Together these data indicate a requirement for spen in specification and differentiation of neuronal and non-neuronal cell types during eye development, a function consistent with Spen playing a positive role within the EGFR pathway.
3. Discussion
In this study, we demonstrate that loss of spen perturbs the normal balance between the EGFR and Notch pathways as evidenced by the patterning disruptions and aberrant expression of multiple pathway components. These findings raise the question of whether Spen functions primarily in the Notch pathway, primarily in the EGFR pathway, or as a critical component of both. Although definite resolution is difficult given the extensive and intricate feedback regulation within and between these two signaling networks, we propose a model in which Spen-mediated antagonism of the Notch pathway regulates the signaling flow through the EGFR pathway to achieve proper retinal cell fate specification.
3.1. Spen antagonizes Notch signaling
Loss of spen results in hyperactivation of the Notch pathway as evidenced by elevated levels of both Notch and its transcriptional targets, the E(spl)-bHLHs. Therefore, a normal function of Spen in the developing eye is to limit the activity of Notch. Consistent with this model, we observed that heterozygous reduction of spen is sufficient to suppress the heterozygous Notch wing margin phenotype. However, loss of spen does not lead to the anti-neurogenic phenotypes typically associated with overexpression/overactivation of canonical members of the Notch pathway (reviewed in Artavanis-Tsakonas et al., 1999), suggesting that although Notch signaling output is elevated, the increase is below the threshold needed to achieve such phenotypes. Consistent with this interpretation, recruitment of the initial R8 photoreceptor neuron, a process influenced at multiple stages by Notch signaling, occurs normally in the absence of spen.
Where might Spen interface with the Notch signaling pathway? The striking increase in Sca expression in spen mutant clones at the MF is consistent with Spen regulating Notch activation by limiting the expression of sca either through transcriptional repression or by destabilizing the transcript. This suggests that in the Drosophila eye Spen may have an upstream role in the Notch pathway in contrast to the downstream role described for Spen mammalian orthologs (Kuroda et al., 2003; Oswald et al., 2002; Oswald et al., 2005) On the other hand, because of extensive feedback regulation in Notch signaling, it is plausible that Spen interfaces with the network at a more downstream point. For example, ectopic expression of Notchintra was shown to promote Sca expression (Baker and Yu, 1997), which in turn activates Notch signaling. Additionally, although we did not detect such a role with respect to yan, it is possible that Spen limits Notchintra/Su(H)-mediated transactivation at the level of transcriptional repression of other Notch pathway targets, including the E(spl)-bHLHs, as is the case for the mammalian Spen orthologs (Kuroda et al., 2003; Oswald et al., 2002; Oswald et al., 2005). This latter mechanism might also be relevant posterior the MF, where Notch signaling remains elevated as judged by increased levels of both Notch and the E(spl)-bHLHs in spen mutant clones, but where Sca is no longer expressed
Although pinpointing where Spen interfaces with the Notch signaling pathway remains a challenge, the simplest interpretation of our data is that at the MF, Spen either directly or indirectly regulates Sca expression to restrict Notch pathway output. Posterior to the MF, as discussed below, mutual antagonism between the Notch and EGFR pathways may stabilize the initial signaling imbalance independent of Sca, leading to sustained up-regulation of Notch and down-regulation of EGFR output in spen mutant tissue.
3.2. Elevated Notch pathway output leads to reduced EGFR signaling in spen mutant tissue
What might the consequences of a moderate increase in Notch pathway output be? Given the extensive functional antagonism that has been previously reported between the Notch and EGFR pathways in the eye (reviewed in Doroquez and Rebay, 2006; Sundaram, 2005), a likely outcome is that the increased Notch signaling in spen mutant tissue would dampen EGFR pathway output. Supporting the idea that spen plays a positive role with respect to EGFR signaling, the cell fate specification defects observed in spen mutant clones are highly reminiscent of phenotypes associated with hypomorphic mutants in positive components of the EGFR pathway (Dickson et al., 1992; Dominguez and de Celis, 1998; O’Neill et al., 1994; Silver et al., 2004; Simon et al., 1991). Thus, the defective specification of neuronal and non-neuronal cell types and the perturbed R8 spacing adjacent to the MF all suggest reduced, but not ablated, EGFR pathway function in spen clones.
Lending further support to a model in which elevated Notch signaling in spen clones dampens EGFR pathway output, both Ato and dpERK expression at the MF are reduced. Because previous work has shown that Ato is required for activation of the EGFR pathway at the MF (Baonza et al., 2001; Chen and Chien, 1999), one possibility is that Spen stabilizes dpERK levels at the MF by antagonizing Notch-mediated lateral inhibition to ensure appropriate Ato expression. Another plausible mechanism for Spen-mediated regulation of dpERK activity would be downstream of or in parallel to Ras as we had proposed previously (Chen and Rebay, 2000). In this scenario, Spen might mediate transcriptional repression of an inhibitor such as a MAPK phosphatase (reviewed in Farooq and Zhou, 2004). However, qRT-PCR analysis in imaginal discs predominantly mutant for spen do not indicate a role for Spen in regulating the expression of two characterized Drosophila MAPK phosphatases—dMKP3 and PTP-ER (D.B. Doroquez and I. Rebay, unpublished observations; (Karim and Rubin, 1999; Kim et al., 2004; Kim et al., 2002; Rintelen et al., 2003). Thus, validation of such a mechanism will require identification of other MAPK phosphatases or pathway inhibitors that might be regulated by Spen
It should be noted that the results of our analysis of spen function in the eye appear contradictory to those from a prior study that suggested spen antagonizes EGFR output and promotes Notch signaling during embryonic neural development (Kuang et al., 2000). Specifically, the authors reported elevated EGFR signaling in spen maternal/zygotic null embryos, as evidenced by increased numbers of midline glial cells and loss of Yan expression. However, we have been unable to reproduce these results (F. Chen and I. Rebay, unpublished). On the contrary, our analysis of spen function during midline glial cell development in the embryonic central nervous system was entirely consistent with a role for spen as a positive contributor to EGFR signaling (Chen and Rebay, 2000). Thus, at least with respect to EGFR signaling, we believe Spen serves an analogous role in multiple developing tissues.
With respect to Notch signaling, Kuang and colleagues report a strong reduction in E(spl)-bHLH expression throughout the embryo but no change in Notch levels, exactly opposite to our findings in the eye. Additional work will be needed to determine whether and how spen interfaces with the Notch pathway during embryogenesis, and whether distinct or identical mechanisms operate in retinal versus embryonic neural development.
3.3. Spen modulates EGFR and Notch signaling posterior to the morphogenetic furrow
It is not yet clear whether spen’s role in Notch-EGFR interactions posterior to the MF is identical to its role in events occurring at the MF. The failure to down-regulate Yan and the resulting cell fate specification defects show that EGFR signaling posterior to the MF is compromised in spen mutant tissue. Given that Yan up-regulation in spen clones does not result from loss of Spen-mediated transcriptional repression, but rather reflects loss of post-translational control, two models for Spen function seem likely. First, if our inability to detect changes in dpERK protein levels posterior to the MF in situ accurately indicates unaltered dpERK levels, then the ability of dpERK to phosphorylate Yan must be compromised in spen mutants. Alternatively, dpERK levels may be sufficiently reduced to increase Yan stability, but the change may be below our immunohistochemical detection threshold.
In terms of the signals that impinge on dpERK, whereas Notch signaling and Ato expression are critical for proper dpERK expression in the MF, reiterative EGFR signaling takes over posterior to the MF to maintain dpERK activity (Freeman, 1996). Thus, it is possible that a Spen-dependent, Notch-independent mechanism may regulate EGFR output posterior to the MF. Alternatively, because Notch, E(spl) and Yan expression are all elevated in spen mutant tissue both in and posterior to the MF, Spen-mediated antagonism of Notch signaling may be relevant to EGFR regulation in both contexts. An extension of this idea that results in perhaps the most appealing model is that the initial increase in Notch output at the MF dampens EGFR signaling, which in turn leads to elevated Notch signaling in more posterior regions resulting in reduced EGFR output. In this way, the initial signaling imbalance created by loss of spen at the MF could be maintained over the entire eye disc through mutual antagonism and feedback regulation between the Notch and EGFR pathways.
3.4. Conclusions
In summary, we have analyzed the requirement for spen in regulating the EGFR and Notch pathways during Drosophila eye development and propose that increased Notch pathway activity upon loss of spen may be sufficient to dampen EGFR signaling, but not to disrupt other downstream effects of Notch signaling. Therefore, because the effects of spen loss appear to be at a threshold below the production of bona fide Notch-related phenotypes, we suggest that Spen plays a subtle role in the regulation of the Notch pathway or functions redundantly alongside other components. An equally likely hypothesis is that Spen regulates the Notch and EGFR pathways separately and that the phenotypes we have reported reflect a composite of independent disruptions to both signaling networks.
Although much of the literature focuses on a primary role for Spen family proteins as co-repressors, recent findings suggest members of this family may also regulate non-coding RNA sequestration, mRNA export, RNA splicing, and proteolysis (Hiriart et al., 2005; Li et al., 2006; Lindtner et al., 2006; Shi et al., 2001). Therefore, future identification of the precise molecular mechanisms by which Spen interfaces with the EGFR and Notch pathways may reveal novel modes of interaction between these two critical and conserved signaling networks.
4. Experimental Procedures
4.1. Fly Strains and Genetics
Fly stocks were maintained at 25°C and obtained from the Bloomington Stock Center. w1118 was used as the control strain, unless otherwise indicated. For the generation of mitotic clones in the eye imaginal disc, the following genotype was analyzed: P[ey-FLP/+; spenAH393 P[FRT]40A/P[w+, ubi-GFPnls] P[FRT]40A. For tangential sections, adult eyes were fixed, embedded in plastic, sectioned, and mounted as described (Wolff, 2000). Mutant tissue was recognizable by the absence of red eye pigment (Xu and Rubin, 1993). To generate eye discs predominantly mutant for spen, the EGUF/hid cell lethal technique (Stowers and Schwarz, 1999) was employed to produce the genotype: spenAH393 P[FRT]40 P[GMR-hid] l(2)cl-L31 P[FRT]40A; P[ey-Gal4] P[UAS-FLP]. yan post-transcriptional regulation by Spen was analyzed in the following genotype: spenAH393 P[FRT]40A/P[ubi-GFPnls] P[FRT]40A; P[dpp3-GAL4] P[UAS-FLP]/P[UAS-YanWT].
To analyze adult wing phenotypes, N54l9/+ and N54l9/+; spenAH393/+ flies were generated. Wings were fixed in 70% ethanol and mounted in Aquamount (BDH). Microscopy and image acquisition was performed on a Zeiss AxioPhot using a Spot digital imaging system (Diagnostic Instruments).
4.2. Immunohistochemistry and Immunofluorescence
Eye and wing imaginal discs were dissected from wandering third instar larvae, fixed with 4% paraformaldehyde in PBT (0.1% Triton X-100 in Phosphate-Buffered Saline [PBS]) for 15 min, washed three times in PBT (5 min each), incubated in primary antibody overnight in PBT with 5% normal goat serum (PNT) at 4°C, and then washed three times in PBT (5 min each). Discs were incubated in secondary antibody for 2 hr in PNT at room temperature, washed three times in PBT (5 min each), and mounted in Vectashield mountant (Vector Laboratories). Microscopy and image acquisition was performed on a Zeiss LSM510 confocal microsope.
For immunofluorescence, the following primary antibodies were used: 1:5,000 rabbit anti-Atonal (Jarman et al., 1993), 1:30 mouse anti-Scabrous (Sca1, DSHB), 1:20 mouse anti-Notchintra (C17.9C6, DSHB), 1:10 mouse anti-E(spl)-bHLH mAb323 (Jennings et al., 1994), 1:200 mouse anti-dpERK (M8159, Sigma), 1:200 mouse anti-Yan (8B12H9, DSHB), 1:50 rat anti-Elav (7E8A10, Developmental Studies Hybridoma Bank [DSHB]), 1:20,000 rabbit anti-β-Galactosidase (Cappel Laboratories), 1:1000 guinea pig anti-Senseless (Nolo et al., 2000), 1:300 rabbit anti-Spalt, 1:500 rabbit anti-BarH1 (Higashijima et al., 1992), 1:10 mouse anti-Prospero (MR1A, DSHB), and 1:100 mouse anti-Cut (2B10, DSHB). Secondary antibodies were Alexa Fluor 568-conjugated goat anti-mouse IgG (Molecular Probes), or appropriate Rhodamine Red-X- and Cy5-conjugated antibodies (Jackson ImmunoResearch).
4.3. Quantitative RT-PCR
Forty pairs of eye imaginal discs were dissected from control and spen/cell lethal clone third instar larvae. Total RNA was isolated using Trizol Reagent (Invitrogen) according to the manufacturer’s protocol, treated with DNase I (Invitrogen), and used as template to generate cDNA using the random or oligo-dT primers provided in the Reverse Transcription System (Promega). Quantitative/real-time PCR from 1 μL cDNA template was performed using 2× ABI SYBR Green Master Mix (Applied Biosystems), 80 nM forward primer, and 80 nM reverse primer in 25 μL reactions for 35 thermocycling reactions. Each experimental reaction was performed in triplicate using the relative quantitation method (ABI) or alongside four ten-fold dilutions of standard [wild-type eye disc cDNA] and no-template control reactions (in triplicate) as previously described in (Claycomb et al., 2002). For each sample, relative fluorescence was measured in comparison to standard curves. Mean and standard deviations of the triplicate reactions were calculated (ABI Prism 7300 software). Fold-expression of the experimental transcript was normalized to RpS17 expression for each sample. Analysis to determine statistical significance was performed with paired, two-tailed Student’s t-tests.
PCR primers are as follows:
| sca forward | 5′-AACCGATTTCCCTAAACCAACC-3′ |
| sca reverse | 5′-CTTGATCTCTTTGGCATGCGACT-3′ |
| aos forward | 5′-CTTCCGTGACTACACTTGGACTT-3′ |
| aos reverse | 5′-CTATCTGCTCCGTCACATTCAAC-3′ |
| RpS17 forward | 5′-GTACGAACCAAGACGGTGAAGA-3′, |
| RpS17 reverse | 5′-GGCGAGTGTAGTACTTCTCGATG-3′, |
| yan forward | 5′-GGTATTAGCAGTGCCAGCAGTAA-3′, |
| yan reverse | 5′-ACTCCACCACTGGTCTCAGAGTA-3′ |
Supplementary Material
Fig. S1. Up-regulation of Notch pathway components in spen clones. Confocal micrograph of spen eye discs clones stained (in red) for (A,B) Scabrous, (C,D) extracellular Notch (NotchEC, 1:200 C458.2H, DSHB) and (E,F) E(spl)-bHLH (mAb323). spen mutant tissue lacks GFP (green). Sca expression is broadened in spen clones (A,B, brackets) as compared to its restricted expression in IG cells. Notch expression is elevated in clones (C,D, arrows). E(spl)-bHLH expression is broadened in spen clones (E,F, brackets) and elevated posterior to the MF. E(spl)-bHLH is normally restricted to alternating groups of cells (E, arrowheads). Discs are oriented anterior to the left. Scale bars = 20 μm.
Fig. S2. E(spl)-bHLH transcripts are elevated in spen mutant discs. qRT-PCR was performed to measure relative (A) E(spl)-m3 and (B) E(spl)-mβ mRNA levels in wild-type and spen/cell lethal eye discs (three samples, mean + st. dev.). Elevated levels are significant (*), p = 0.0026 and 0.018, respectively.
Fig. S3. spen loss affects EGFR signaling pathway components. (A,B) Confocal micrographs of third instar eye imaginal discs oriented with anterior to the left in spen clones stained (in red) for (A,B) dpERK or (C,D) Yan. spen mutant tissue lacks GFP (green). dpERK is lost at the MF in spen clones (A,B, asterisks *) but appears unchanged posterior to the MF. Yan is up-regulated in spen clones both in and posterior to the MF (B,D, arrows). Scale bars = 20 μm.
Fig. S4. Yan is up-regulated in spen wing clones that express ectopic Yan from a cDNA transgene. (A,B) confocal migraphs of third instar wing imaginal discs that co-expression NLS-tagged GFP with (A,B) YanWT or (B) YanACT under the control of the dpp-GAL4 driver. dpp>YanWT (A,B, arrows) is expressed in a subset of cells where dpp>GFPNLS is driven (A,B, arrowheads). However, dpp>YanACT co-localizes well (C,D, arrows) with dpp>GFPNLS. (E,F) dpp>YanWT is expressed in a spen clones background. spen clones lack GFP (green). More red Yan-positive cells are seen in spen mutant tissue (E,F, arrowheads) than in control tissue (E,F, arrow). Discs are oriented dorsal up. Scale bars = 20 μm.
Fig. S5. Loss of organization and cell types in spen mutant pupal tissue. Confocal micrographs of eye imaginal discs 42 hr APF (after puparium formation) in wild-type (A,C,E) and spen clones (B,D,F). Discs were stained for (A,B) DE-cadherin (DE-cad, 1:50 DCAD2, DSHB), (C,D) Elav, and (E,F) Cut. spen mutant tissue is marked by lack of GFP (green). C,E and D,F correspond to the same sample, respectively. DE-cad (A,B) is a cell boundary marker that especially marks cone, pigment, and bristle cells. spen mutant tissue (B) shows loss of these cell types and loss of the regular ommatidial organization found in wild-type (A). In spen clones, there are fewer Elav-positive R cells (D) and Cut-positive cone cells (F). Scale bars = 10 μm.
Acknowledgments
We would like to thank R.G. Fehon, P.A. Garrity, J.C. Jemc, P. Vivekanand, S. Maitra for comments on the manuscript; J.A. Lees and the Fehon, Orr-Weaver, and Rebay labs for stimulating discussion. We are grateful to A. Mukherjee, G. Hurlbut, and the Artavanis-Tsakonas and Garrity labs for reagents, and to N. Watson (WIBR/Keck Microscopy Facility) for microscopy assistance. D.B.D. was supported by a National Science Foundation Graduate Fellowship. This work was supported in part by National Institutes of Health grant R01 EY12549 to I.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ariyoshi M, Schwabe JW. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 2003;17:1909–20. doi: 10.1101/gad.266203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–22. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Baker NE, Mlodzik M, Rubin GM. Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science. 1990;250:1370–7. doi: 10.1126/science.2175046. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu S, Han D. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr Biol. 1996;6:1290–301. doi: 10.1016/s0960-9822(02)70715-x. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. Proneural function of neurogenic genes in the developing Drosophila eye. Curr Biol. 1997;7:122–32. doi: 10.1016/s0960-9822(06)00056-x. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. The R8-photoreceptor equivalence group in Drosophila: fate choice precedes regulated Delta transcription and is independent of Notch gene dose. Mech Dev. 1998;74:3–14. doi: 10.1016/s0925-4773(98)00054-9. [DOI] [PubMed] [Google Scholar]
- Baker NE, Zitron AE. Drosophila eye development: Notch and Delta amplify a neurogenic pattern conferred on the morphogenetic furrow by scabrous. Mech Dev. 1995;49:173–89. doi: 10.1016/0925-4773(94)00314-d. [DOI] [PubMed] [Google Scholar]
- Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol. 2001;11:396–404. doi: 10.1016/s0960-9822(01)00125-7. [DOI] [PubMed] [Google Scholar]
- Baonza A, Freeman M. Notch signalling and the initiation of neural development in the Drosophila eye. Development. 2001;128:3889–98. doi: 10.1242/dev.128.20.3889. [DOI] [PubMed] [Google Scholar]
- Blochlinger K, Bodmer R, Jan LY, Jan YN. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 1990;4:1322–31. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–9. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- Chen CK, Chien CT. Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc Natl Acad Sci U S A. 1999;96:5055–60. doi: 10.1073/pnas.96.9.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Rebay I. split ends, a new component of the Drosophila EGF receptor pathway, regulates development of midline glial cells. Curr Biol. 2000;10:943–6. doi: 10.1016/s0960-9822(00)00625-4. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, MacAlpine DM, Evans JG, Bell SP, Orr-Weaver TL. Visualization of replication initiation and elongation in Drosophila. J Cell Biol. 2002;159:225–36. doi: 10.1083/jcb.200207046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Barrio R. Function of the spalt/spalt-related gene complex in positioning the veins in the Drosophila wing. Mech Dev. 2000;91:31–41. doi: 10.1016/s0925-4773(99)00261-0. [DOI] [PubMed] [Google Scholar]
- Dickson B, Sprenger F, Morrison D, Hafen E. Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature. 1992;360:600–3. doi: 10.1038/360600a0. [DOI] [PubMed] [Google Scholar]
- Dickson BJ, van der Straten A, Dominguez M, Hafen E. Mutations Modulating Raf signaling in Drosophila eye development. Genetics. 1996;142:163–71. doi: 10.1093/genetics/142.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederich RJ, Matsuno K, Hing H, Artavanis-Tsakonas S. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development. 1994;120:473–81. doi: 10.1242/dev.120.3.473. [DOI] [PubMed] [Google Scholar]
- Dokucu ME, Zipursky SL, Cagan RL. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development. 1996;122:4139–47. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- Dominguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–8. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- Doroquez DB, Rebay I. Signal Integration During Development: Mechanisms of EGFR and Notch Pathway Function and Cross-Talk. Crit Rev Biochem Mol Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- Farooq A, Zhou MM. Structure and regulation of MAPK phosphatases. Cell Signal. 2004;16:769–79. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–34. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- Firth L, Manchester J, Lorenzen JA, Baron M, Perkins LA. Identification of genomic regions that interact with a viable allele of the Drosophila protein tyrosine phosphatase corkscrew. Genetics. 2000;156:733–48. doi: 10.1093/genetics/156.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankfort BJ, Mardon G. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development. 2002;129:1295–306. doi: 10.1242/dev.129.6.1295. [DOI] [PubMed] [Google Scholar]
- Freeman M. Misexpression of the Drosophila argos gene, a secreted regulator of cell determination. Development. 1994;120:2297–304. doi: 10.1242/dev.120.8.2297. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–60. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Freeman M. Cell determination strategies in the Drosophila eye. Development. 1997;124:261–70. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- Gabay L, Scholz H, Golembo M, Klaes A, Shilo BZ, Klambt C. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development. 1996;122:3355–62. doi: 10.1242/dev.122.11.3355. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–6. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Gellon G, Harding KW, McGinnis N, Martin MM, McGinnis W. A genetic screen for modifiers of Deformed homeotic function identifies novel genes required for head development. Development. 1997;124:3321–31. doi: 10.1242/dev.124.17.3321. [DOI] [PubMed] [Google Scholar]
- Golembo M, Schweitzer R, Freeman M, Shilo BZ. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development. 1996;122:223–30. doi: 10.1242/dev.122.1.223. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Kojima T, Michiue T, Ishimaru S, Emori Y, Saigo K. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 1992;6:50–60. doi: 10.1101/gad.6.1.50. [DOI] [PubMed] [Google Scholar]
- Hiriart E, Gruffat H, Buisson M, Mikaelian I, Keppler S, Meresse P, Mercher T, Bernard OA, Sergeant A, Manet E. Interaction of the Epstein Barr Virus (EBV) mRNA export factor EB2 with human Spen proteins SHARP, OTT1 and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J Biol Chem. 2005 doi: 10.1074/jbc.M501725200. [DOI] [PubMed] [Google Scholar]
- Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–21. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. Atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–30. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Jennings B, Preiss A, Delidakis C, Bray S. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120:3537–48. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- Karim FD, Rubin GM. PTP-ER, a novel tyrosine phosphatase, functions downstream of Ras1 to downregulate MAP kinase during Drosophila eye development. Mol Cell. 1999;3:741–50. doi: 10.1016/s1097-2765(01)80006-x. [DOI] [PubMed] [Google Scholar]
- Kidd S, Baylies MK, Gasic GP, Young MW. Structure and distribution of the Notch protein in developing Drosophila. Genes Dev. 1989;3:1113–29. doi: 10.1101/gad.3.8.1113. [DOI] [PubMed] [Google Scholar]
- Kim M, Cha GH, Kim S, Lee JH, Park J, Koh H, Choi KY, Chung J. MKP-3 has essential roles as a negative regulator of the Ras/mitogen-activated protein kinase pathway during Drosophila development. Mol Cell Biol. 2004;24:573–83. doi: 10.1128/MCB.24.2.573-583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kwon HB, Kim YS, Ryu JH, Kim KS, Ahn Y, Lee WJ, Choi KY. Isolation and characterization of a Drosophila homologue of mitogen-activated protein kinase phosphatase-3 which has a high substrate specificity towards extracellular-signal-regulated kinase. Biochem J. 2002;361:143–51. doi: 10.1042/bj3610143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang B, Wu SC, Shin Y, Luo L, Kolodziej P. split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development. 2000;127:1517–29. doi: 10.1242/dev.127.7.1517. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Hsiung F, Powers MA, Moses K. Nuclear translocation of activated MAP kinase is developmentally regulated in the developing Drosophila eye. Development. 2003;130:3703–14. doi: 10.1242/dev.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo BZ, Moses K. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–85. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T, Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S, Honjo T. Regulation of Marginal Zone B Cell Development by MINT, a Suppressor of Notch/RBP-J Signaling Pathway. Immunity. 2003;18:301–12. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Rubin GM. Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell. 1992;70:609–20. doi: 10.1016/0092-8674(92)90430-k. [DOI] [PubMed] [Google Scholar]
- Lane ME, Elend M, Heidmann D, Herr A, Marzodko S, Herzig A, Lehner CF. A screen for modifiers of cyclin E function in Drosophila melanogaster identifies Cdk2 mutations, revealing the insignificance of putative phosphorylation sites in Cdk2. Genetics. 2000;155:233–44. doi: 10.1093/genetics/155.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Lee EC, Hu X, Yu SY, Baker NE. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol Cell Biol. 1996;16:1179–88. doi: 10.1128/mcb.16.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–71. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Yang X, Qin H, Dong X, Zhu Y, Liang L, Liang Y, Han H. The Spen Homolog Msx2-Interacting Nuclear Target Protein Interacts with the E2 Ubiquitin-Conjugating Enzyme UbcH8. Mol Cell Biochem. 2006;288:151–7. doi: 10.1007/s11010-006-9131-9. [DOI] [PubMed] [Google Scholar]
- Li J, Yang X, Qin H, Zhou P, Liang Y, Han H. The C terminus of MINT forms homodimers and abrogates MINT-mediated transcriptional repression. Biochim Biophys Acta. 2005;1729:50–6. doi: 10.1016/j.bbaexp.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Li Y, Baker NE. Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol. 2001;11:330–8. doi: 10.1016/s0960-9822(01)00093-8. [DOI] [PubMed] [Google Scholar]
- Lin HV, Doroquez DB, Cho S, Chen F, Rebay I, Cadigan KM. Splits ends is a tissue/promoter specific regulator of Wingless signaling. Development. 2003;130:3125–35. doi: 10.1242/dev.00527. [DOI] [PubMed] [Google Scholar]
- Lindtner S, Zolotukhin AS, Uranishi H, Bear J, Kulkarni V, Smulevitch S, Samiotaki M, Panayotou G, Felber BK, Pavlakis GN. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J Biol Chem. 2006;281:36915–28. doi: 10.1074/jbc.M608745200. [DOI] [PubMed] [Google Scholar]
- Ludewig AH, Kober-Eisermann C, Weitzel C, Bethke A, Neubert K, Gerisch B, Hutter H, Antebi A. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 2004;18:2120–33. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Renda MJ, Wang L, Cheng EC, Niu C, Morris SW, Chi AS, Krause DS. Rbm15 Modulates Notch-induced Transcriptional Activation and Affects Myeloid Differentiation. Mol Cell Biol. 2007 doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Morris SW, Valentine V, Li M, Herbrick JA, Cui X, Bouman D, Li Y, Mehta PK, Nizetic D, Kaneko Y, Chan GC, Chan LC, Squire J, Scherer SW, Hitzler JK. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28:220–1. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- Mace K, Tugores A. The product of the split ends gene is required for the maintenance of positional information during Drosophila development. BMC Dev Biol. 2004;4:15. doi: 10.1186/1471-213X-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace KA, Pearson JC, McGinnis W. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science. 2005;308:381–5. doi: 10.1126/science.1107573. [DOI] [PubMed] [Google Scholar]
- Mercher T, Coniat MB, Monni R, Mauchauffe M, Khac FN, Gressin L, Mugneret F, Leblanc T, Dastugue N, Berger R, Bernard OA. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci U S A. 2001;98:5776–9. doi: 10.1073/pnas.101001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M, Baker NE, Rubin GM. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 1990;4:1848–61. doi: 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- Mohler JD. New mutants report. Drosophila Information Service. 1956;30:78–79. [Google Scholar]
- Morimura S, Maves L, Chen Y, Hoffmann FM. decapentaplegic overexpression affects Drosophila wing and leg imaginal disc development and wingless expression. Dev Biol. 1996;177:136–51. doi: 10.1006/dbio.1996.0151. [DOI] [PubMed] [Google Scholar]
- Newberry EP, Latifi T, Towler DA. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry. 1999;38:10678–90. doi: 10.1021/bi990967j. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–62. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–47. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knochel W, Liptay S, Schmid RM. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–90. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell PA, Wesley C, Spencer S, Cagan RL. Scabrous complexes with Notch to mediate boundary formation. Nature. 2001;409:626–30. doi: 10.1038/35054566. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–40. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rebay I, Chen F, Hsiao F, Kolodziej PA, Kuang BH, Laverty T, Suh C, Voas M, Williams A, Rubin GM. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif- containing protein. Genetics. 2000;154:695–712. doi: 10.1093/genetics/154.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–66. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Rintelen F, Hafen E, Nairz K. The Drosophila dual-specificity ERK phosphatase DMKP3 cooperates with the ERK tyrosine phosphatase PTP-ER. Development. 2003;130:3479–90. doi: 10.1242/dev.00568. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–61. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- Rohrbaugh M, Ramos E, Nguyen D, Price M, Wen Y, Lai ZC. Notch activation of yan expression is antagonized by RTK/pointed signaling in the Drosophila eye. Curr Biol. 2002;12:576–81. doi: 10.1016/s0960-9822(02)00743-1. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Rojas AM, van Wely KH, Martinez AC, Valencia A. SPOC: a widely distributed domain associated with cancer, apoptosis and transcription. BMC Bioinformatics. 2004;5:91. doi: 10.1186/1471-2105-5-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K, Okabe M, Tanimura T, Mikoshiba K, Nishida Y, Okano H. The Drosophila secreted protein Argos regulates signal transduction in the Ras/MAPK pathway. Dev Biol. 1996;178:13–22. doi: 10.1006/dbio.1996.0194. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Okano H, Kobayakawa Y, Hayashi S, Mikoshiba K, Tanimura T. The function of argos in regulating cell fate decisions during Drosophila eye and wing vein development. Dev Biol. 1994;164:267–76. doi: 10.1006/dbio.1994.1197. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Howes R, Smith R, Shilo BZ, Freeman M. Inhibition of Drosophila EGF receptor activation by the secreted protein Argos. Nature. 1995;376:699–702. doi: 10.1038/376699a0. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Shilo BZ. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 1997;13:191–6. doi: 10.1016/s0168-9525(97)01091-3. [DOI] [PubMed] [Google Scholar]
- Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9:459–70. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–51. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo BZ, Raz E. Developmental control by the Drosophila EGF receptor homolog DER. Trends Genet. 1991;7:388–92. doi: 10.1016/0168-9525(91)90261-n. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Chen F, Doyon L, Zink AW, Rebay I. New class of Son-of-sevenless (Sos) alleles highlights the complexities of Sos function. Genesis. 2004;39:263–72. doi: 10.1002/gene.20054. [DOI] [PubMed] [Google Scholar]
- Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–16. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–95. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Spencer SA, Powell PA, Miller DT, Cagan RL. Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development. 1998;125:4777–90. doi: 10.1242/dev.125.23.4777. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Ciampa PJ, Brook A, Dyson N. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics. 1999;153:275–87. doi: 10.1093/genetics/153.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–9. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Han M. Control and integration of cell signaling pathways during C. elegans vulval development. Bioessays. 1996;18:473–80. doi: 10.1002/bies.950180609. [DOI] [PubMed] [Google Scholar]
- Sundaram MV. The love-hate relationship between Ras and Notch. Genes Dev. 2005;19:1825–39. doi: 10.1101/gad.1330605. [DOI] [PubMed] [Google Scholar]
- Tan PB, Kim SK. Signaling specificity: the RTK/RAS/MAP kinase pathway in metazoans. Trends Genet. 1999;15:145–9. doi: 10.1016/s0168-9525(99)01694-7. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Lee PS, Rebay I. CRM1-mediated nuclear export and regulated activity of the Receptor Tyrosine Kinase antagonist YAN require specific interactions with MAE. Development. 2003;130:845–57. doi: 10.1242/dev.00312. [DOI] [PubMed] [Google Scholar]
- Tsuji M, Shinkura R, Kuroda K, Yabe D, Honjo T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc Natl Acad Sci U S A. 2007;104:1610–5. doi: 10.1073/pnas.0610520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–82. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M. A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. Embo J. 2002;21:4277–86. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Manavathi B, Singh RR, Nguyen D, Li F, Kumar R. An essential role of Pak1 phosphorylation of SHARP in Notch signaling. Oncogene. 2005;24:4591–6. doi: 10.1038/sj.onc.1208672. [DOI] [PubMed] [Google Scholar]
- Voas M, Rebay I. Signal integration during development: insights from the Drosophila eye. Mech Dev. 2003 doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Wiellette EL, Harding KW, Mace KA, Ronshaugen MR, Wang FY, McGinnis W. spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development. 1999;126:5373–85. doi: 10.1242/dev.126.23.5373. [DOI] [PubMed] [Google Scholar]
- Wolff T. Histological techniques for the Drosophila eye part I: larva and pupa. In: Sullivan W, et al., editors. Drosophila Protocols. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 201–227. [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–50. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–37. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev Cell. 2003;4:359–69. doi: 10.1016/s1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- Yang X, Li J, Qin H, Yang H, Zhou P, Liang Y, Han H. Mint represses transactivation of the type II collagen gene enhancer through interaction with alpha A-crystallin-binding protein 1. J Biol Chem. 2005;280:18710–6. doi: 10.1074/jbc.M500859200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Up-regulation of Notch pathway components in spen clones. Confocal micrograph of spen eye discs clones stained (in red) for (A,B) Scabrous, (C,D) extracellular Notch (NotchEC, 1:200 C458.2H, DSHB) and (E,F) E(spl)-bHLH (mAb323). spen mutant tissue lacks GFP (green). Sca expression is broadened in spen clones (A,B, brackets) as compared to its restricted expression in IG cells. Notch expression is elevated in clones (C,D, arrows). E(spl)-bHLH expression is broadened in spen clones (E,F, brackets) and elevated posterior to the MF. E(spl)-bHLH is normally restricted to alternating groups of cells (E, arrowheads). Discs are oriented anterior to the left. Scale bars = 20 μm.
Fig. S2. E(spl)-bHLH transcripts are elevated in spen mutant discs. qRT-PCR was performed to measure relative (A) E(spl)-m3 and (B) E(spl)-mβ mRNA levels in wild-type and spen/cell lethal eye discs (three samples, mean + st. dev.). Elevated levels are significant (*), p = 0.0026 and 0.018, respectively.
Fig. S3. spen loss affects EGFR signaling pathway components. (A,B) Confocal micrographs of third instar eye imaginal discs oriented with anterior to the left in spen clones stained (in red) for (A,B) dpERK or (C,D) Yan. spen mutant tissue lacks GFP (green). dpERK is lost at the MF in spen clones (A,B, asterisks *) but appears unchanged posterior to the MF. Yan is up-regulated in spen clones both in and posterior to the MF (B,D, arrows). Scale bars = 20 μm.
Fig. S4. Yan is up-regulated in spen wing clones that express ectopic Yan from a cDNA transgene. (A,B) confocal migraphs of third instar wing imaginal discs that co-expression NLS-tagged GFP with (A,B) YanWT or (B) YanACT under the control of the dpp-GAL4 driver. dpp>YanWT (A,B, arrows) is expressed in a subset of cells where dpp>GFPNLS is driven (A,B, arrowheads). However, dpp>YanACT co-localizes well (C,D, arrows) with dpp>GFPNLS. (E,F) dpp>YanWT is expressed in a spen clones background. spen clones lack GFP (green). More red Yan-positive cells are seen in spen mutant tissue (E,F, arrowheads) than in control tissue (E,F, arrow). Discs are oriented dorsal up. Scale bars = 20 μm.
Fig. S5. Loss of organization and cell types in spen mutant pupal tissue. Confocal micrographs of eye imaginal discs 42 hr APF (after puparium formation) in wild-type (A,C,E) and spen clones (B,D,F). Discs were stained for (A,B) DE-cadherin (DE-cad, 1:50 DCAD2, DSHB), (C,D) Elav, and (E,F) Cut. spen mutant tissue is marked by lack of GFP (green). C,E and D,F correspond to the same sample, respectively. DE-cad (A,B) is a cell boundary marker that especially marks cone, pigment, and bristle cells. spen mutant tissue (B) shows loss of these cell types and loss of the regular ommatidial organization found in wild-type (A). In spen clones, there are fewer Elav-positive R cells (D) and Cut-positive cone cells (F). Scale bars = 10 μm.


