Abstract
Chrysanthemum chlorotic mottle viroid (CChMVd) is an RNA of 398–399 nt that can adopt hammerhead structures in both polarity strands. We have identified by Northern-blot hybridization a nonsymptomatic strain (CChMVd-NS) that protects against challenge inoculation with the symptomatic strain (CChMVd-S). Analysis of CChMVd-NS cDNA clones has revealed a size and sequence very similar to those of the CChMVd-S strain. Some of the mutations observed in CChMVd-NS molecular variants were previously identified in CChMVd-S RNA, but others were never found in this RNA. When bioassayed in chrysanthemum, cDNA clones containing the CChMVd-NS specific mutations were infectious but nonsymptomatic. Site-directed mutagenesis showed that one of the CChMVd-NS-specific mutations, a UUUC → GAAA substitution, was sufficient to change the symptomatic phenotype into the nonsymptomatic one without altering the final accumulation level of the viroid RNA. The pathogenicity determinant–to our knowledge, a determinant of this class has not been described previously in hammerhead viroids–is located in a tetraloop of the computer-predicted branched conformation for CChMVd RNA. Analysis of the sequence heterogeneity found in CChMVd-S and -NS variants strongly supports the existence of such a conformation in vivo, showing that the rod-like or quasi-rod-like secondary structure is not a universal paradigm for viroids.
Keywords: catalytic RNAs, ribozymes
Viroids, the smallest pathogenic RNAs endowed with autonomous replication, can induce a series of diseases in their host plants (1). Their minimal genomic size (between 246 and 399 nt) (2), lack of messenger RNA activity implying direct RNA–cell component interactions in viroid replication and pathogenesis (1), and the presence in some members of the group of ribozymatic domains (3, 4) make them unique and very attractive systems for the study of RNA structure–function relationships.
The causal agent of chrysanthemum chlorotic mottle (CChM) disease has recently been identified and characterized as a new viroid of 398–399 nt (CChMVd), the largest one described thus far, excluding those with sequence duplications (5). CChMVd RNA does not contain the central conserved region characteristic of the viroid genera that form the Pospiviroidae family (2). However, plus and minus CChMVd strands do contain the conserved sequences and structural elements characteristic of the hammerhead structures and self-cleave in vitro, and at least the plus strand also in vivo, as predicted by these ribozymatic domains. Hammerhead structures have also been found in two other viroids, avocado sunblotch viroid (6) and peach latent mosaic viroid (PLMVd) (7), that with CChMVd form the second viroid family, Avsunviroidae (2). Avocado sunblotch viroid replicates through a symmetric pathway with two rolling circles and hammerhead ribozyme processing (8, 9), as probably do PLMVd and CChMVd. The branched secondary structure of lowest free energy proposed for CChMVd differs remarkably from the rod-like or quasi-rod like models proposed for most other viroids (10, 11), with the exception of PLMVd, whose most stable folding is also branched (7).
The existence of an infectious but nonsymptomatic strain of the CChM disease agent was previously postulated to explain why some plants of a chrysanthemum cultivar sensitive to the disease were unable to develop the characteristic symptoms when inoculated with extracts from symptomatic leaves (12). This observation was assumed to result from a cross-protection phenomenon: plants infected with a latent or nonsymptomatic strain would be protected against the challenge inoculation by a severe strain of the same or very similar agent. In fact, by using cross-protection assays, the existence of a transmissible agent that protected against CChM disease was shown (12), but direct physical evidence in support of this contention has remained elusive for more than 20 years. We reasoned that the identification and characterization of this putative RNA could provide some insight into the molecular determinants of pathogenicity of a member of the Avsunviroidae family, in which structure–function relationships are poorly understood.
We report here on the sequence of an RNA with the predicted properties, which we shall term CChMVd-NS (hereafter, the suffixes NS and S will refer to the nonsymptomatic and symptomatic CChMVd RNAs, respectively). From sequence analyses of CChMVd-S and -NS variants, bioassays with individual CChMVd-NS cDNA clones, and site-directed mutagenesis, we have identified a tetraloop in the most stable branched conformation as the determinant of the different pathogenic effects induced by CChMVd-S and -NS strains. Moreover, the sequence heterogeneity observed in variants from both strains strongly supports a branched conformation for CCMVd RNA in vivo.
MATERIALS AND METHODS
CChMVd Strains and Extraction of Viroid RNA.
The CChMVd-S strain has been described (13) and sequenced (5), and the CChMVd-NS strain was obtained from a chrysanthemum clone of the cultivar “Yellow Delaware.” Preparations enriched in viroid RNA were generated by extraction of leaves with buffer-saturated phenol, fractionation with 2 M LiCl (recovering the insoluble RNAs), and removal of polysaccharides with methoxyethanol (5). Preparations were used for bioassay and for PAGE and Northern-blot RNA analysis (5). For dot-blot hybridization, RNAs from chrysanthemum leaves (2 g) were extracted with buffer-saturated phenol and fractionated on nonionic cellulose (CF11, Whatman), which was washed with STE (50 mM Tris⋅HCl/100 mM NaCl/1 mM EDTA, pH 7.2) containing 35% ethanol and then with STE (14).
Cross-Protection and Infectivity Bioassays.
Chrysanthemum (Dendranthema grandiflora Tzvelez, cv. “Bonnie Jean”), was propagated in growth chambers (5). For cross-protection bioassays, plants were mechanically inoculated with an RNA preparation from the CChMVd-NS strain; 1 month later, part was challenge-inoculated with an RNA preparation from the CChMVd-S strain and the rest with a similar preparation from healthy plants. For bioassay of CChMVd-NS variants, recombinant plasmids with head-to-tail dimeric cDNA inserts were mechanically inoculated (2 μg of plasmid per plant).
RNA Analysis.
Nucleic acids from CChMVd-infected plants, enriched in RNAs of 300–400 nt, were obtained by cutting a section from nondenaturing 5% polyacrylamide gels delimited by appropriate markers. These RNAs, and parallel preparations from healthy plants, were examined in 5% polyacrylamide gels containing 8 M urea and 1× TBE (89 mM Tris/89 mM boric acid/2.5 mM EDTA, pH 8.3). After ethidium bromide staining, RNAs were electroblotted to nylon membranes (Hybond N+, Amersham Pharmacia) and UV-fixed with a Stratalinker (Stratagene). For dot-blot hybridization, 5 μl of the extracts and 1:10 dilutions were applied onto membranes that were UV-irradiated. Prehybridization, hybridization (at 70°C in 50% formamide), and washing were as reported (15). The probe, a radioactive full-length RNA complementary to the CChMVd-S variant CM 20, was transcribed with T7 RNA polymerase from a linearized recombinant pBluescript II KS+ plasmid (Stratagene). Membranes were scanned with a Bioimage analyzer (Fuji BAS1500).
Reverse Transcription–PCR Amplification, Cloning, and Sequencing.
Viroid circular forms of the CMNS strain, purified by two consecutive PAGE steps (16), were reverse-transcribed and PCR-amplified with two pairs of adjacent primers derived from the CM 20 sequence of CChMVd-S strain (5). Primer PI (complementary to nucleotides 294–269) was used with primer PII (homologous to nucleotides 295–320), and primer PIII (complementary to nucleotides 133–108) was used with primer PIV (homologous to nucleotides 134–159) (Fig. 3). Reverse transcription, PCR amplification (with Pfu DNA polymerase, Stratagene), and cloning were performed as described (5). Inserts were sequenced automatically with an ABI Prism DNA sequencer (Perkin–Elmer).
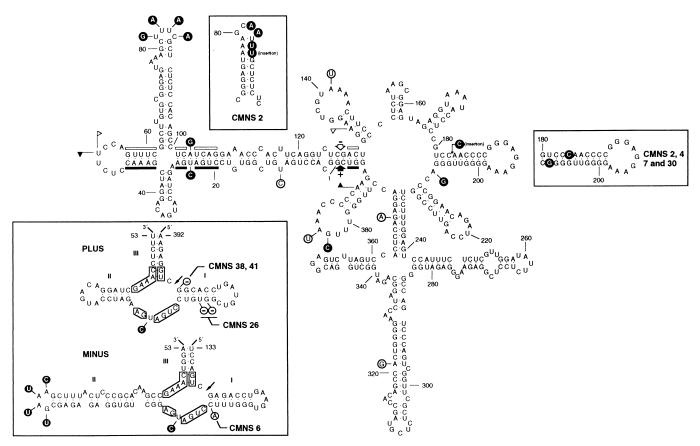
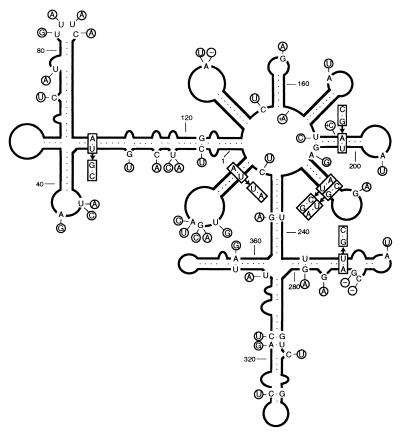
Figure 3.
Secondary structure of lowest free energy of the symptomatic CM 20 variant. Numbering does not coincide exactly with that of the alignment because of the introduced gaps. The structure of the region between positions 70 and 93 is slightly different from that reported previously (5), because an updated version of the mfold program has been used. The most frequent mutations detected in CChMVd-NS variants are shown; those on a black background have never been found in CChMVd-S variants, whereas those on a white background were previously found in CChMVd-S variants. Sequences forming plus and minus hammerhead structures are indicated by flags, nucleotides conserved in most natural hammerhead structures by bars, and self-cleavage sites by arrows. Solid and open symbols refer to plus and minus polarities, respectively. (Upper Left Inset) Most stable secondary structure of the hairpin delimited by positions 70 to 94 in variant CMNS 2. (Right Inset) Most stable secondary structure of the hairpin delimited by positions 180–210 in variants CMNS 2, 4, 7, and 30. (Lower Left Inset) Hammerhead structures of plus and minus strands of the CM 20 variant. The most frequent mutations found in CChMVd-NS variants are on a black background and do not alter the stability of the hammerhead structures. Less frequent mutations of other CChMVd-NS variants, which affect the stability of their hammerhead structures, are on a white background with deletions indicated by −. Nucleotides conserved in most natural hammerhead structures are boxed, and self-cleavage sites are shown by arrows. The same numbering is used in both polarities and does not coincide exactly with numbering of the alignment because of the introduced gaps.
RNA Self-Cleavage During in Vitro Transcription.
The EcoRI–BamHI fragments from recombinant pUC18 plasmids with full-length CChMVd-NS cDNA inserts were subcloned in pBluescript II KS+, and radioactive transcripts of both polarities were obtained with T3 and T7 RNA polymerases (17). Primary transcripts and their self-cleavage products were separated by PAGE in 5% gels containing 8 M urea and 40% formamide that were quantitatively scanned with a Bioimage analyzer.
Site-Directed Mutagenesis.
A PCR-based protocol (18) was followed with minor modifications. Plasmid pCM20 (5 ng) (5) was amplified with 250 ng each of the phosphorylated primers PV (5′-TCGCTTTACTCCCGCACAAGCCGAAAC-3′) complementary to nucleotides 57–83 and PVI (5′-AAGCTCTCTCCACAGCCTCATCAGGAAAC-3′) homologous to nucleotides 84–112 of the CM 20 sequence (Fig. 3), except in the residues in bold introduced to change UUUC82–85 into GAAA. Plasmid pCM20 was also amplified with the phosphorylated primers PVII (5′-GGGTTTCCTGACGAGGCTGTGGAGAG-3′) complementary to nucleotides 11–35 and PVIII (5′-ACTTCAGGTCTCGACTGGAAGGTC-3′) homologous to nucleotides 36–59 of the CM 20 sequence (Fig. 3), except in the residue of PVII in bold introduced to change A103 into a G. The PCR cycling profile, designed to amplify the complete plasmid with Pfu DNA polymerase, consisted of a hot start of 95°C for 2 min, four cycles of 95°C for 40 s, 50°C for 30 s, and 72°C for 4 min; and 25 cycles of 95°C for 40 s, 65°C for 30 s, and 72°C for 4 min, with a final extension at 72°C for 10 min. After electrophoresis in agarose gels, PCR products of plasmid length were eluted, circularized with T4 DNA ligase, and used for transformation. Sequencing confirmed that the new plasmids contained only the expected mutations.
Computer Analysis.
CChMVd sequences were aligned by the pileup program (Genetics Computer Group, Madison, WI). RNA secondary structures of lowest free energy at 37°C were determined by the circular version of the mfold program from the same package.
RESULTS
Detection of a Nonsymptomatic Strain of CChMVd.
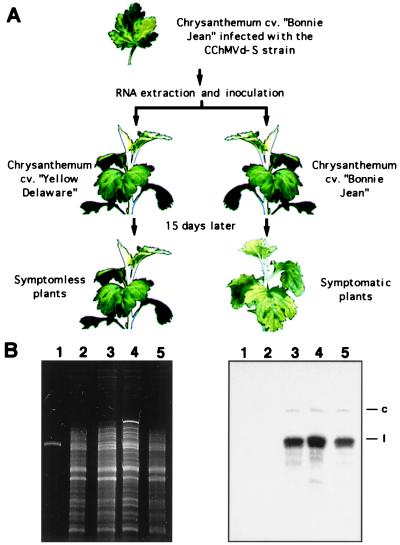
To study the symptoms induced by the severe CChMVd-S strain in a chrysanthemum cultivar different to “Bonnie Jean,” which was used in the initial characterization of this viroid (5), apparently healthy plants of the cultivar “Yellow Delaware” were inoculated with extracts of “Bonnie Jean” leaves expressing the CChMVd-S symptoms. Thirty days after inoculation, the “Yellow Delaware” plants remained symptomless, in contrast to “Bonnie Jean” controls inoculated with the same extract, which showed the typical chlorosis 10–15 days postinoculation (Fig. 1A). Because cultivar “Yellow Delaware” is sensitive to the CChM disease (19), we suspected that the lack of symptoms might result from a previous infection with CChMVd-NS, whose existence had been postulated before CChMVd was recognized as a viroid on the basis of similar experiments (12).
Figure 1.
Identification of a nonsymptomatic CChMVd RNA. (A) Differential response of two chrysanthemum cultivars after inoculation with a nucleic acid preparation of the severe CChMVd-S strain. (B) Analysis of chrysanthemum RNAs by denaturing PAGE and ethidium bromide staining (Left) or Northern-blot hybridization with a probe complementary to CChMVd-S RNA (Right). Lane 1, DNA marker of 392 nt; lanes 2 and 3, RNAs from healthy and symptomatic “Bonnie Jean” plants, respectively; lanes 4 and 5, RNAs from symptomless “Yellow Delaware” plants uninoculated and inoculated with a CChMVd-S RNA preparation, respectively. The migration of the circular (c) and linear (l) viroid forms is indicated at the right.
To test this hypothesis, RNAs within a size window of 300–400 nt from “Yellow Delaware” and “Bonnie Jean” plants uninoculated and inoculated with the CChMVd-S strain were analyzed by denaturing PAGE and Northern-blot hybridization with a probe complementary to CChMVd-S RNA. Hybridization signals in the same position and with similar intensity as those produced in “Bonnie Jean” by CChMVd-S RNA were observed in the inoculated and uninoculated “Yellow Delaware” plants, whereas no signal was detected in the uninoculated “Bonnie Jean” plants (Fig. 1B). These results showed that the “Yellow Delaware” chrysanthemums used in our experiments were infected with a CChMVd-NS strain, very similar in sequence to CChMVd-S, that presumably prevented the appearance of the symptoms incited by the latter.
Cross-Protection Bioassays.
To corroborate that the newly identified CChMVd-NS strain was infectious and nonpathogenic, “Bonnie Jean” plants were inoculated with extracts from CChMVd-NS-infected “Yellow Delaware” chrysanthemums. The “Bonnie Jean” plants remained symptomless, although Northern-blot analysis with a probe specific for CChMVd-S RNA revealed a signal in the position of the viroid RNA (data not shown) indicating, therefore, that they were infected by the CChMVd-NS strain. When cuttings of these plants, as well as parallel control plants of the same cultivar not exposed to CChMVd-NS, were inoculated with CChMVd-S RNA, symptoms of the CChM disease appeared only in the control plants. This confirmed the ability of CChMVd-NS RNA to replicate in “Bonnie Jean” and to induce a cross-protection effect against the CChMVd-S strain.
Molecular Characterization of CChMVd-NS RNA.
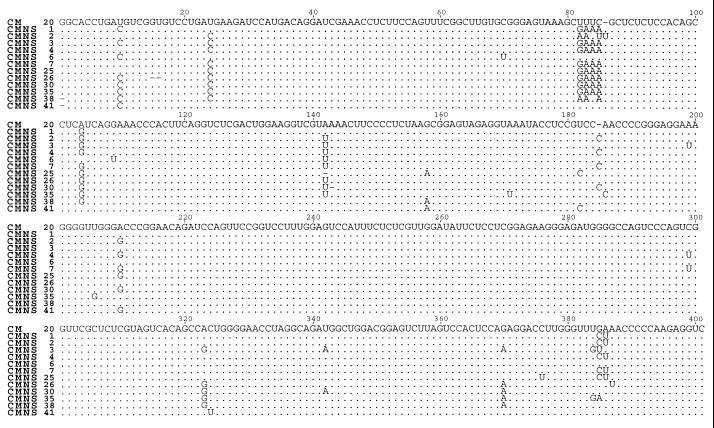
Sequence alignment of 12 randomly chosen CChMVd-NS cDNA clones with the CM 20 cDNA clone of the CChMVd-S strain (5) revealed that variants from CChMVd-NS and -S strains were very similar in size and sequence (Fig. 2). Part of the CChMVd-NS polymorphism was previously observed in CChMVd-S variants (ref. 5; data not shown), but other changes were not found before in CChMVd-S variants. Among the latter, those affecting positions 24 (U → C), 82 to 85 (UUUC → GAAA), and 104 (A → G) in the alignment were the most frequent, appearing in 73, 64, and 82% of the sequences, respectively (Fig. 2).
Figure 2.
Sequence alignment of 12 molecular variants from “Bonnie Jean” plants inoculated with the CChMVd-NS strain. For comparative purposes, the sequence of the symptomatic CM 20 variant is shown on the top. Dots indicate residues identical to the CM 20 sequence and dashes denote gaps. Variants CMNS 1, 6, 25, 26, 30, 35, 38, and 41 were synthesized with primers PI and PII, and the rest with PIII and PIV.
The most stable secondary structure of the CM 20 RNA (5) was unaffected by most of the mutations found in CChMVd-NS (CMNS) variants (Fig. 3). In CMNS 2 variant, a slightly different hairpin was formed in the region delimited by positions 70–93 in the reference sequence (Fig. 3). A similar situation was observed in variants CMNS 2, 4, 7, and 30, in which a rearranged hairpin was formed in the region between positions 180–209 in the reference sequence (Fig. 3). The substitution G → U in position 109 (CMNS 6) and deletions in positions 1 (CMNS 38 and 41) and 15 and 16 (CMNS 26) in the alignment are probably artifactual (see below).
Hammerhead Ribozymes and in Vitro Self-Cleavage of CChMVd-NS RNAs.
The three most frequent changes found in CChMVd-NS variants (positions 24, 82–85, and 104 in the alignment) are located in the hammerhead structures, but their stability remains unaltered. Interestingly, those affecting residues 24 and 104 map at position 7 according to the standard numbering for hammerhead structures (20), which resides between the conserved CUGA and GA of the plus and minus hammerhead structures, respectively (Fig. 3). Similar U → C transitions in this position 7 have been observed between variants of PLMVd (7, 21). The third mutation, affecting positions 82–85, is located in loop 2 (20) of the minus hammerhead structure (Fig. 3). The extent of self-cleavage during in vitro transcription of the RNAs with all or part of these mutations (CMNS 1, 25, 30, and 35; CMNS 2 has a different mutation in positions 82–85) was comparable to that of the CM 20 RNA (Fig. 4), as anticipated from the stability of their hammerhead structures. On the other hand, changes in variants CMNS 6, 26, 38, and 41 decreased the stability of their hammerhead structures by disrupting the first base pair of helix I of the plus (CMNS 38 and 41) and minus (CMNS 6) ribozymes or by shortening the length of helix I of the plus hammerhead structure (CMNS 26) (Fig. 3). Accordingly, the self-cleavage of these RNAs was considerably reduced (Fig. 4).
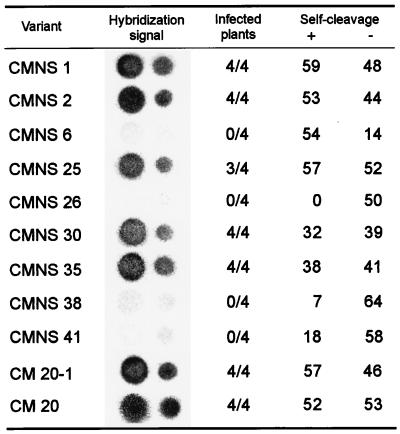
Figure 4.
Infectivity of some representative CChMVd-NS variants and self-cleavage of their monomeric plus (+) and minus (−) RNAs. Dot-blot hybridization signals correspond to undiluted extracts and 1:10 dilutions (left and right spot columns, respectively). The extent of self-cleavage is expressed as the fraction (%) of the primary transcript that self-cleaved during in vitro transcription. The nonsymptomatic CM 20–1 variant was obtained from the symptomatic CM 20 variant by introducing the change UUUC82–85 → GAAA.
Biological Properties of CChMVd-NS Variants.
The presence of specific changes in most CChMVd-NS sequences suggested that they might determine the symptomless phenotype induced by the CChMVd-NS strain. To establish a causal relationship, plasmids containing dimeric head-to-tail cDNA inserts of CMNS 1, 2, 6, 25, 26, 30, 35, 38, and 41 were inoculated into “Bonnie Jean” plants. Previous experiments had shown that plasmids with CChMVd-S cDNA dimeric inserts are infectious (5). CMNS 4 and 7 on the one hand, and CMNS 3 on the other, were not bioassayed because of their close similarity to CMNS 25 and CMNS 35, respectively (Fig. 2). Although all of the inoculated plants remained symptomless, in contrast with those inoculated with the CM 20 variant of the CChMVd-S strain that displayed the typical symptoms, dot-blot analysis showed that variants CMNS 1, 2, 25, 30, and 35 were highly infectious (Fig. 4). The lack of infectivity of CMNS 6, 26, 38, and 41, was consistent with the reduced self-cleavage of their hammerhead structures (Fig. 4).
Identification of a Pathogenicity Determinant in CChMVd.
As inferred from bioassays, only changes affecting positions 82–85 and 104 in the alignment were common to the infectious CChMVd-NS cDNA clones. The third most frequent substitution in CChMVd-NS variants, U24 → C, was discarded as a determinant of pathogenicity because CMNS 1 (infectious but nonsymptomatic) has a U at this position, as do symptomatic variants of the CChMVd-S strain. To provide more precise and direct evidence of the role of substitutions UUUC82–85 → GAAA and A104 → G in pathogenicity, they were independently introduced by site-directed mutagenesis into a plasmid with the CM 20 insert of the CChMVd-S strain to generate plasmids pCM20–1 and pCM20–2, respectively. Plasmids with the corresponding dimeric head-to-tail inserts, pCM20–1d and pCM20–2d, respectively, were bioassayed in “Bonnie Jean” chrysanthemum. All plants inoculated with pCM20–2d developed symptoms within 15 days, as opposed to those inoculated with pCM20–1d, in which no symptoms were observed, although dot-blot hybridization revealed that they were infected (Fig. 4). These results demonstrated that a symptomatic variant could be converted in a nonsymptomatic one by introducing the substitution UUUC → GAAA in positions 82–85 which, therefore, delimit this pathogenic determinant.
DISCUSSION
Understanding structure–function relationships in the Avsunviroidae family, which differs in fundamental aspects from the better known Pospiviroidae family (2), has been hampered by lack of appropriate experimental systems. The recent characterization of CChMVd (5) [whose natural host, chrysanthemum, is also a convenient experimental host because of the short time (12–15 days) that elapses between inoculation and onset of symptoms (13)] allows the study of functional determinants in CChMVd. Among them, determinants responsible for pathogenesis are particularly intriguing. The observation that plants of the chrysanthemum cultivar “Yellow Delaware” remained symptomless after inoculation with the severe CChMVd-S strain suggested to us the possibility that these were infected with a nonsymptomatic strain of the same agent (CChMVd-NS), which protected them against a CChMVd-S challenge inoculation. Additional support for such a cross-protection effect was obtained by experimentally reproducing the same result in the “Bonnie Jean” cultivar. Northern-blot hybridization provided the first direct proof for the existence of a CChMVd-NS RNA in chrysanthemum plants. Interestingly, the intensity of the hybridization signals generated by CChMVd-S and -NS RNAs was similar, indicating that their associated phenotypes were not the consequence of different accumulation levels in the infected tissue.
Sequence analysis of a series of CChMVd-NS cDNA clones showed, as in the CChMVd-S strain (5), a distribution of closely related variants forming a quasi-species (22). Distributions of this kind have been found in other members of both families, including potato spindle tuber viroid (PSTVd) (23), citrus exocortis viroid (24), avocado sunblotch viroid (25), and PLMVd (7, 21). Mutations observed in CChMVd-NS variants were not randomly scattered along the RNA molecule. This, together with the observation of several clones with the same changes, was a solid indication that they were present in the viroid RNA population.
CChMVd-NS cDNA clones of variants CMNS 6, 26, 38, and 41 were not infectious, probably because the deficient self-cleavage activity of their RNAs (Fig. 4) impaired replication. In particular, deletion of the residue located 3′ to the self-cleavage site of the plus hammerhead structure (as in CMNS 38 and 41), has been reported in PLMVd variants with a much reduced RNA self-cleavage and infectivity (21). We assume this deletion is a cloning artifact introduced by the reverse transcriptase because of the extra 2′-phosphate at the nucleotide preceding the self-cleavage/ligation site, as was the case with a viroid-like satellite RNA (26). Variants CMNS 1, 2, 25, 30, and 35, containing the mutations specific to the CChMVd-NS strain, caused symptomless infections. When the contribution of each mutation to the nonsymptomatic phenotype was assessed, only the tetranucleotide change at positions 82–85 was found to be directly involved. Indeed, introduction of the change UUUC82–85 → GAAA in the symptomatic variant CM 20 by site-directed mutagenesis led to a symptomless infection without detectable alterations of the accumulation level of the viroid progeny.
The CChMVd-NS strain appears therefore to be composed essentially of two types of variants: a minor fraction with the UUUC82–85 sequence characteristic of the severe CChMVd-S strain (5) and a major fraction with the GAAA82–85 motif that confers the nonsymptomatic phenotype. The coexistence of sequence variants with different pathogenicity within a viroid isolate has also been reported in PLMVd (21) and in some members of the Pospiviroidae family such as PSTVd and citrus exocortis viroid (23, 24). Taking advantage of the fact that in the cDNA, the residues UU82–83 form part of a HindIII site (AAGCUU78–83), we explored with restriction analysis 28 and 32 additional cDNA clones from two other reverse transcription–PCR amplifications of CChMVd-NS RNA preparations of “Bonnie Jean” and “Yellow Delaware” plants, respectively. The 60 clones were resistant to digestion, and complete sequencing of 12 confirmed the presence of the GAAA82–85 motif (data not shown), indicating a clear predominance, if not a total dominance, of the nonpathogenic variants in these two CChMVd-NS populations.
Looking at the mechanism linking the tetranucleotide change in CChMVd with the phenotypic effect, it is interesting to consider what is known in this respect in the Pospiviroidae family. Mutations of 3–4 nt with dramatic effects on pathogenicity (27) have been mapped in PSTVd at a “virulence-modulating region” that overlaps a premelting region within a domain of the rod-like structure (reviewed in ref. 28). A similar situation was observed in citrus exocortis viroid (29) and, accordingly, this was termed the pathogenic domain (30), although more recent data indicate the participation of additional domains in symptom expression (31). Moreover, pathogenicity of hop stunt viroid is regulated by specific changes in another domain of the rod-like structure (32). For some PSTVd strains, an inverse correlation between the thermodynamic stability of the premelting region and virulence in tomato was found (33), but other data obtained with this viroid (34) and with citrus exocortis viroid (24) do not support this correlation. Taking into account bending of RNA helices, an alternative picture that emerges from comparison of the most stable secondary structure of PSTVd variants of different pathogenicity points to major differences in the geometry of their virulence-modulating regions that, together with concomitant alterations in RNA–protein interactions, may be the primary cause of viroid pathogenicity (34, 35).
The situation with CChMVd, however, appears very different because this viroid, together with PLMVd, does not adopt a rod-like secondary structure of minimum free energy but rather a highly branched conformation that is supported by studies of solubility in 2 M LiCl (5, 7, 21). In CChMVd particularly, the sequence heterogeneity detected in the variants from the S and NS strains characterized so far (ref. 5; this work and unpublished data), provides strong support for the biological significance of the proposed branched conformation, either because the changes are found in loops or because when affecting a base pair, the substitutions are compensatory (Fig. 5). This is for example the case of the very stable hairpins delimited by positions 181–207, 210–230, and 374–394 in which, additionally, the covariations are generally double transversions. The existence of the first of these hairpins is further supported by the change CC182–183 → CCC and the substitution A208 → G found in CMNS 2, 4, 7, and 30, that lead to an extension and rearrangement of the basal portion of the stem (Fig. 3 and 5). Other mutations in double-stranded regions of CChMVd not affecting the computer-predicted secondary structure are the covariations U251 → C and A271 → G, A103 → G and U24 → C, and other point mutations converting canonical pairs into G-U wobble pairs or the opposite (G279 → A, A321 → G, C322 → U, A357 → G, and G368 → A) (Fig. 5). Because covariations or compensatory mutations are considered the most powerful approach for testing computer-predicted structures in RNA (36), our results show that the rod-like or quasi-rod-like secondary structure is not a universal property of viroid RNAs because some of them, as illustrated by CChMVd, present a clearly branched conformation in vivo.
Figure 5.
Effects of the sequence heterogeneity found in variants characterized so far from CChMVd-S and -NS strains on the secondary structure predicted for CChMVd RNA. Mutations of CMNS 6, 26, 38, and 41, which are presumably artifactual for the reasons stated in the text, are not included. Insertions and deletions are denoted by symbols + and −, respectively. For other details of the CChMVd reference variant, see Fig. 3.
The substitution UUUC82–85 → GAAA responsible for changing the induced phenotype from symptomatic to nonsymptomatic forms a tetraloop capping the stem of a hairpin (Fig. 3). The GAAA hairpin loop belongs to the GNRA tetraloop family (where R stands for a purine and N for any base) found frequently in different RNAs (37) and endowed with an unusually high thermodynamic stability resulting from a network of heterogeneous hydrogen bonds (38). It has been proposed that these tetraloops play an important function in RNA folding (39), in RNA–RNA tertiary interactions (40), and as protein-binding sites (41). A GAAA tetraloop has also been implicated in processing of PSTVd (42), but a similar role for the CChMVd tetraloop at positions 82–85 can be dismissed, because processing in this viroid is mediated by hammerhead ribozymes and does not occur in this region (5). That the CChMVd mutation UUUC82–85 → GAAA indeed maps at a tetraloop is not only supported by the concurrent change of four consecutive nucleotides but also by the observation that in variant CMNS 2, the UUUC82–85 mutates into AAUUU, which leads in the most stable secondary structure to a rearrangement of the hairpin and to a new GCAA tetraloop of the same GNRA family (Fig. 3).
The finding that some chrysanthemum plants are resistant to a severe CChMVd strain because they are infected with a closely related nonsymptomatic CChMVd strain, along with parallel results in PLMVd (21) extends to the Avsunviroidae family the cross-protection phenomena reported previously in the Pospiviroidae family (reviewed in ref. 3). Considering the very different properties of the two viroid families, distinct underlying mechanisms must necessarily be involved in the repeated appearance of phenomena of this class in the interactions between plants and these subviral pathogens.
Acknowledgments
We thank A. Ahuir for technical assistance, Drs C. Hernández and J.A. Daròs for reading the manuscript, and D. Donnellan for English revision. M.D. and B.N. were recipients of predoctoral fellowships from the Ministerio de Educación y Cultura. This work was supported by grant PB95–0139 (to R.F.) from the Dirección General de Investigación Científica y Técnica.
ABBREVIATIONS
- CChMVd
chrysanthemum chlorotic mottle viroid
- PLMVd
peach latent mosaic viroid
- PSTVd
potato spindle tuber viroid
- S
symptomatic
- NS
nonsymptomatic
- CMNS
cchMVd-NS variant
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ247112–247123).
References
- 1.Diener T O. The Viroids. New York: Plenum; 1987. [Google Scholar]
- 2.Flores R, Randles J W, Bar-Joseph M, Diener T O. Arch Virol. 1998;143:623–629. doi: 10.1007/s007050050318. [DOI] [PubMed] [Google Scholar]
- 3.Flores R, Di Serio F, Hernández C. Semin Virol. 1997;8:65–73. [Google Scholar]
- 4.Symons R H. Nucleic Acids Res. 1997;20:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navarro B, Flores R. Proc Natl Acad Sci USA. 1997;94:11262–11267. doi: 10.1073/pnas.94.21.11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutchins C J, Rathjen P D, Forster A C, Symons R H. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernández C, Flores R. Proc Natl Acad Sci USA. 1992;89:3711–3715. doi: 10.1073/pnas.89.9.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daròs J A, Marcos J F, Hernàndez C, Flores R. Proc Natl Acad Sci USA. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchins C J, Keese P, Visvader J E, Rathjen P D, McInnes J L, Symons R H. Plant Mol Biol. 1985;4:293–304. doi: 10.1007/BF02418248. [DOI] [PubMed] [Google Scholar]
- 10.Gross H J, Domdey H, Lossow C, Jank P, Raba M, Alberty H, Sänger H L. Nature (London) 1978;273:203–208. doi: 10.1038/273203a0. [DOI] [PubMed] [Google Scholar]
- 11.Gast F U, Kempe D, Spieker R L, Sänger H L. J Mol Biol. 1996;263:652–670. doi: 10.1006/jmbi.1996.0543. [DOI] [PubMed] [Google Scholar]
- 12.Horst R K. Phytopathology. 1975;65:1000–1003. [Google Scholar]
- 13.Romaine C P, Horst R K. Virology. 1975;64:86–95. doi: 10.1016/0042-6822(75)90081-1. [DOI] [PubMed] [Google Scholar]
- 14.Pallás V, Navarro A, Flores R. J Gen Virol. 1987;68:2095–2102. [Google Scholar]
- 15.Flores R. J Virol Methods. 1986;13:161–169. doi: 10.1016/0166-0934(86)90084-4. [DOI] [PubMed] [Google Scholar]
- 16.Flores R, Durán-Vila N, Pallás V, Semancik J S. J Gen Virol. 1985;66:2095–2102. [Google Scholar]
- 17.Forster A C, Davies C, Hutchins C J, Symons R H. Methods Enzymol. 1990;181:583–607. doi: 10.1016/0076-6879(90)81153-l. [DOI] [PubMed] [Google Scholar]
- 18.Byrappa S, Gavin D K, Gupta K C. PCR Methods Appl. 1995;5:404–407. [Google Scholar]
- 19.Dimock A W, Geissinger C M, Horst R K. Phytopathology. 1971;61:415–419. [Google Scholar]
- 20.Hertel K J, Pardi A, Uhlenbeck O C, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach W L, Hodgson R, Symons R H. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambrós S, Hernández C, Desvignes J C, Flores R. J Virol. 1998;72:7397–7406. doi: 10.1128/jvi.72.9.7397-7406.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domingo E, Holland J, Biebricher C, Eigen M. In: Molecular Basis of Virus Evolution. Gibbs A, Calisher C H, García-Arenal F, editors. Cambridge, U.K.: Cambridge Univ. Press; 1995. pp. 181–191. [Google Scholar]
- 23.Góra A, Candresse T, Zagórski W. Arch Virol. 1994;138:223–245. doi: 10.1007/BF01379128. [DOI] [PubMed] [Google Scholar]
- 24.Visvader J E, Symons R H. Nucleic Acids Res. 1985;5:2907–2920. doi: 10.1093/nar/13.8.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakowski A G, Symons R H. Virology. 1989;173:352–356. doi: 10.1016/0042-6822(89)90256-0. [DOI] [PubMed] [Google Scholar]
- 26.Kiberstis P A, Haseloff J, Zimmern D. EMBO J. 1985;4:817–822. doi: 10.1002/j.1460-2075.1985.tb03703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross H J, Liebl U, Alberty H, Krupp G, Domdey H, Ramm K, Sänger H L. Biosci Rep. 1981;1:235–241. doi: 10.1007/BF01114910. [DOI] [PubMed] [Google Scholar]
- 28.Riesner D. Semin Virol. 1990;1:83–99. [Google Scholar]
- 29.Visvader J E, Symons R H. EMBO J. 1986;13:2051–2055. doi: 10.1002/j.1460-2075.1986.tb04465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keese P, Symons R H. Proc Natl Acad Sci USA. 1985;82:4582–4586. doi: 10.1073/pnas.82.14.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sano T, Candresse T, Hammond R W, Diener T O, Owens R A. Proc Natl Acad Sci USA. 1992;89:10104–10108. doi: 10.1073/pnas.89.21.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reanwarakorn K, Semancik J S. J Gen Virol. 1998;79:3163–3171. doi: 10.1099/0022-1317-79-12-3163. [DOI] [PubMed] [Google Scholar]
- 33.Schnölzer M, Haas B, Ramm K, Hofmann H, Sänger H L. EMBO J. 1985;4:2181–2190. doi: 10.1002/j.1460-2075.1985.tb03913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owens R A, Steger G, Hu Y, Fels A, Hammond R W, Riesner D. Virology. 1996;222:144–158. doi: 10.1006/viro.1996.0405. [DOI] [PubMed] [Google Scholar]
- 35.Schmitz A, Riesner D. RNA. 1998;4:1295–1303. doi: 10.1017/s1355838298980815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutell R R, Larsen N, Woese C R. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woese C R, Winker S, Gutell R R. Proc Natl Acad Sci USA. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jucker F N, Heus H A, Yip P F, Moors E H M, Pardi A. J Mol Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 39.Uhlenbeck O C. Nature (London) 1990;346:613–614. doi: 10.1038/346613a0. [DOI] [PubMed] [Google Scholar]
- 40.Pley H, Flaherty K M, McKay D B. Nature (London) 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- 41.Gluck A, Endo Y, Wool I G. J Mol Biol. 1992;226:411–424. doi: 10.1016/0022-2836(92)90956-k. [DOI] [PubMed] [Google Scholar]
- 42.Baumstark T, Schröder A R W, Riesner D. EMBO J. 1997;16:599–610. doi: 10.1093/emboj/16.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]