Summary
The past few years have seen a shift in the use of Drosophila, from studies of growth and development toward genetic characterization of carbohydrate, sterol, and lipid metabolism. This research, reviewed below, establishes a new foundation for using this simple genetic model system to define the basic regulatory mechanisms that underlie metabolic homeostasis, and holds the promise of providing new insights into the causes and treatments of critical human disorders such as diabetes and obesity.
Drosophila as a system for studies of metabolism
The multicellular complexity of higher organisms imposes unique demands, requiring that animals sense their nutritional status and respond in a concerted manner to coordinate growth and maintain energy homeostasis. As a result, metabolic regulation and physiological feedback systems are central to all aspects of postembryonic life, balancing energy needs with dietary input, contributing to the timing of sexual maturation, influencing adult fertility, and acting as a key determinate of aging. Consistent with these pervasive roles in development and physiology, metabolic dysfunction is associated with major human diseases, including diabetes, cardiovascular disease, and some forms of cancer. Although largely assumed to be an affliction of wealthy societies with abundant food supplies, obesity has expanded to worldwide proportions, with the World Health Organization estimating that at least 300 million adults are clinically obese. This increasing impact on human health has resulted in a resurgence of interest in understanding the mechanisms that underlie metabolic control, often depending on mouse models to study key metabolic disorders. Remarkably, however, relatively little has been done to exploit the power of simple genetic model systems to define the central mechanisms that coordinate metabolic responses. In this review, we focus on the status of such studies in the fruit fly, Drosophila melanogaster, with an emphasis on how Drosophila has been used to define key aspects of metabolic control that are conserved through evolution – providing new insights that were not evident from studies of more complex vertebrate systems.
Drosophila share most of the same basic metabolic functions found in vertebrates. As discussed in more detail below, the fly maintains appropriate circulating sugar levels, compensating for changing environmental conditions and storing excess energy in the forms of glycogen and lipid. These reserves are mobilized during periods of energy need, such as exercise or nutrient depletion (Rusten et al., 2004; Scott et al., 2004; Wigglesworth, 1949). Many of the analogous organ systems that control nutrient uptake, storage, and metabolism in humans are present in the fruit fly. Digestion and nutrient absorption occur in the Drosophila midgut, the functional equivalent of the stomach and intestine. The fat body acts like the mammalian liver and white adipose tissue, metabolizing nutrients and storing large reserves of glycogen and lipid. Lipids are carried through the circulatory system as either high density or low density lipophorin particles (Canavoso et al., 2001). Specialized clusters of Drosophila cells, the oenocytes, accumulate lipids upon starvation and are proposed to perform hepatocyte-like functions in lipid processing (Gutierrez et al., 2007). In addition, separate, discrete clusters of cells maintain insect carbohydrate homeostasis in a manner analogous to the pancreatic α and β cells (Lee and Park, 2004; Rulifson et al., 2002). The central pathways of intermediary metabolism and regulators of homeostasis are present in the fly, demonstrating that most essential metabolic functions have been conserved through evolution (Suppl. Table 1). A key difference between mammalian and insect metabolism is the inability of insects to synthesize cholesterol, rendering them cholesterol auxotrophs (Gilbert et al., 2002). Importantly, however, this divergence does not mean that Drosophila cannot be used for studies of cholesterol metabolism, as described in more detail below for insights into the causes of Niemann Pick type C disease in humans.
The relative ease of growing large numbers of Drosophila larvae or adults overcomes the disadvantage of their small size, allowing researchers to use many of the same basic assays to score metabolic function. These include measurements of mitochondrial activity, ATP assays, lipid metabolic profiling, insulin tolerance tests, assays for the main stored form of lipid, triacylglycerol (TAG), as well as both whole animal and circulating sugar levels. Elegant assays for specific metabolic responses are also possible in Drosophila that cannot be performed in more complex vertebrate systems, such as a GFP assay for membrane-associated PIP3 in intact tissues, a hallmark of activated phosphoinositide-3-kinase (PI3K) signaling (Britton et al., 2002), or a GFP reporter that can be used in whole animal studies to follow the temporal and spatial patterns of SREBP activation (Kunte et al., 2006).
In this review, we survey major metabolic responses that are conserved between flies and humans, emphasizing how studies in Drosophila have provided new insights into how these pathways are regulated. To restrict our survey, we focus on papers that use direct assays of metabolic function to characterize mutant phenotypes, and exclude papers that cover the regulation of growth by insulin signaling or genetic studies of aging, both of which have been reviewed elsewhere (e.g. Edgar, 2006; Helfand and Rogina, 2003; Partridge and Gems, 2002). The picture that emerges from the papers described below is the dawn of a new era in Drosophila biology – where this simple genetic system is being increasingly exploited to define the central pathways that control metabolism and physiology, with implications for improving our understanding of how homeostasis is maintained in all higher organisms and the causes of metabolic disorders in humans.
Drosophila as a new genetic model for diabetes
The conserved insulin/IGF pathways play a central role in growth and metabolism in higher organisms. In mammals, IGFs primarily regulate growth, while insulin functions mainly in glucose homeostasis. These two activities are unified in the fly into a single insulin/IGF pathway. Seven insulin-like peptides (DILP1-7), the functions of which have not been completely elucidated (Brogiolo et al., 2001; Ikeya et al., 2002), act through the Drosophila insulin-like receptor (InR) to initiate a cascade of intracellular events mediated by conserved components of the insulin/IGF pathway. These include the insulin receptor substrate (IRS) Chico, the insulin signaling antagonist PTEN, PI3K, PKB/Akt kinase, and the single FOXO ortholog dFOXO (reviewed by Oldham and Hafen, 2003) (Figure 1).
Figure 1.
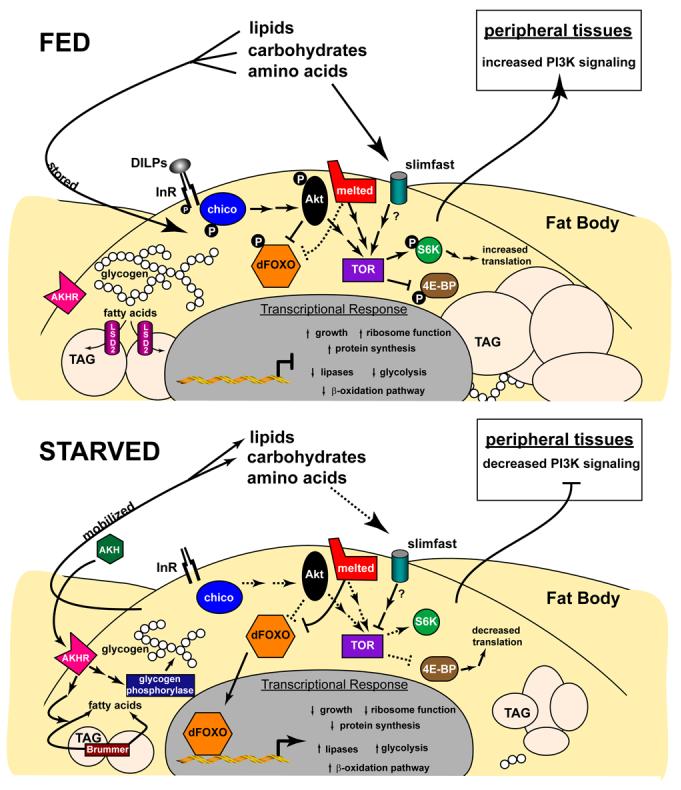
Schematic representation of signaling pathways that regulate Drosophila metabolism. Functional interactions described in the text are depicted for a fat body cell under both fed and starved conditions. Solid lines and arrows represent signaling interactions, while dotted lines and arrows represent regulatory effects that occur in the absence of that signaling pathway. In the fed condition (top), DILPs and nutrients signal through the insulin and TOR pathways, respectively, resulting in retention of FOXO in the cytoplasm and increased translation, driving growth. Sugars and fatty acids are stored as glycogen and TAG, while energy-producing metabolic pathways are down-regulated. In starved animals (bottom), insulin and TOR signaling is attenuated, directing FOXO nuclear translocation and reducing protein synthesis, restricting growth. Glycogen and TAG are mobilized by AKH and lipases such as Brummer, while energy producing metabolic pathways are up-regulated. See text for more details.
Pioneering studies from a few labs have provided exciting new insights into how DILPs regulate carbohydrate metabolism in Drosophila. Under normal feeding conditions, three dilp genes (dilp2,3,5) are expressed in small clusters of median neurosecretory cells within the brain (Brogiolo et al., 2001; Broughton et al., 2005; Ikeya et al., 2002). The expression of dilp3 and dilp5 is reduced in these insulin-producing cells (IPCs) in response to lower dietary carbohydrate levels but not amino acid starvation, indicating that dilp levels can respond to specific nutritional cues much like insulin in humans (Colombani et al., 2003; Ikeya et al., 2002). Moreover, IPC ablation results in diabetic phenotypes, with animals exhibiting a significant increase in circulating glucose and trehalose (a disaccharide that is the primary blood sugar in insects) as well as a moderate increase in stored lipid (Broughton et al., 2005; Rulifson et al., 2002). The IPCs appear to function like pancreatic β cells in that they directly contact the heart and thus could release DILPs into the circulatory system to maintain appropriate levels of circulating sugars, although the regulation of DILP secretion by IPCs remains unknown (Rulifson et al., 2002).
Similar to the release of glucagon from pancreatic α cells in mammals, insect adipokinetic hormone (AKH) counterbalances the actions of insulin by activating glycogen phosphorylase, reducing fat body glycogen, and increasing circulating sugars (Gade and Auerswald, 2003; Kim and Rulifson, 2004; Lee and Park, 2004). Akh is expressed exclusively in the corpora cardiaca (CC) region of the ring gland, the main endocrine organ of the insect (Lee and Park, 2004; Noyes et al., 1995), with direct contacts to the IPCs and heart (Kim and Rulifson, 2004). Removal of Akh function by CC-specific cell ablation results in a dramatic decrease in circulating trehalose, with no significant effect on glucose or stored lipid levels (Isabel et al., 2005; Kim and Rulifson, 2004; Lee and Park, 2004). In addition, ectopic Akh expression in its primary target tissue, the fat body, results in hypertrehalosemia and a reduction of stored lipid through increased lipolysis (Lee and Park, 2004). Stimulated release of AKH by the CC is intimately linked to circulating sugar levels through an inverse change in intracellular calcium stores that is mediated by ATP-sensitive potassium (KATP) channels, similar to the mechanisms that control glucagon secretion by pancreatic α cells (Kim and Rulifson, 2004). Importantly, this regulated process is sensitive to treatment with sulphonylureas, drugs that are used for treating type 2 diabetes through their effects on KATP channels. These studies demonstrate that the central regulatory functions of insulin and glucagon are conserved through evolution, and establish Drosophila as a valid model system for functional studies of glucose homeostasis and the mechanisms that underlie the onset of diabetes.
Metabolic functions of TOR signaling
The Target of Rapamycin (TOR) signaling pathway responds to cellular levels of amino acids and ATP through its upstream effectors, the Tuberous Sclerosis Complex (TSC1 and TSC2) and Rheb GTPase. TOR signaling, together with complex regulatory interactions with the insulin pathway, directs critical changes in cellular physiology that link growth, translation, and autophagy to the nutrient status of the animal (for reviews, see Edgar, 2006; Hay and Sonenberg, 2004). For example, TOR kinase is active in the presence of sufficient nutrients, phosphorylating S6 kinase (S6K), driving protein synthesis and facilitating growth (Figure 1). In addition, Akt-mediated phosphorylation of a key transcriptional effector of insulin signaling, FOXO, renders it inactive by restricting it to the cytoplasm (Figure 1). In contrast, TOR activity is reduced under starvation conditions, leading to decreased translational capacity. Non-phosphorylated FOXO translocates into the nucleus, triggering a transcriptional program that includes the up-regulation of 4E-BP, which controls lipid mobilization (Figure 1) (Teleman et al., 2005a). A recently described hypomorphic dTOR allele results in reduced fat body lipid levels, increased β-hydroxybutyrate (indicative of increased conversion of lipids into ketone bodies), and reduced glucose levels, providing a new genetic system to better characterize the effects of TOR signaling on energy metabolism (Luong et al., 2006).
Although amino acid levels are known to modulate TOR activity, the mechanisms that underlie this response remain poorly understood. Genetic characterization of the Slimfast amino acid transporter has provided some initial clues into this pathway, indicating that it functions as a nutrient sensor in the larval fat body, controlling a systemic response that links amino acid levels with organismal growth (Colombani et al., 2003). The observation that fat body-specific inactivation of either slimfast or dTOR leads to similar phenotypes supports the proposal that Slimfast can signal through dTOR in the fat body to globally regulate growth and metabolism in response to amino acid levels. This fat body amino acid sensor pathway can override insulin signaling in peripheral tissues through inhibition of PI3K activity, apparently through one or more unidentified factors that emanate from the fat body (Figure 1).
A genetic screen for growth regulators in Drosophila revealed a new member of the TOR signaling pathway, melted (Teleman et al., 2005b). This gene encodes a protein with a pleckstrin homology (PH) domain that is essential for its function. Melted can bind TSC1 and recruit the TSC1/2 complex to cell membranes, suggesting it can act upstream of TOR. melted mutants have reduced lipids, but display no apparent defects in circulating sugar levels. Although the mechanisms remain unclear, Melted may help to facilitate the phosphorylation of dFOXO and TSC2 by Akt in response to insulin input. Consistent with the proposal that Melted limits the dFOXO response, the dFOXO target 4E-BP is highly up-regulated in starved melted mutants, and reduced dFOXO levels can suppress the lipid metabolic defects of melted mutants. The identification of melted homologs in C. elegans, mice and humans sets the stage for further studies to understand its role in TOR and FOXO signaling as well as lipid metabolism.
Based largely on studies in cultured cells, 4E-BP has been thought to mediate the growth effects of TOR through direct inhibition of protein synthesis (Hay and Sonenberg, 2004; Oldham and Hafen, 2003). Recent Drosophila genetic studies, however, have challenged this model. Loss-of-function and gain-of-function studies have demonstrated that 4E-BP has no effect on growth (Teleman et al., 2005a). Rather, 4E-BP mutant flies are sensitive to starvation and display a significant decrease in stored lipids upon prolonged starvation. Conversely, animals expressing a constitutively active form of 4E-BP display increased total lipid levels. This role for 4E-BP is consistent with the lipid metabolic defects seen in 4E-BP mutant mice (Tsukiyama-Kohara et al., 2001). In addition, Drosophila Lk6 mutants display elevated lipid levels, consistent with the role of Lk6 in opposing the inhibitory effect of 4E-BP (Reiling et al., 2005). These studies confirm the critical importance of genetic studies in animals to test models derived from cultured cells, and provide a basis for characterizing the mechanisms by which FOXO and TOR signaling regulate homeostasis.
Regulation of metabolism by SREBP and microRNAs
A few studies, including those on the sterol regulatory element binding proteins (SREBPs) and two miRNAs discussed below, have begun to address the mechanisms by which trans-acting factors control Drosophila metabolism. SREBPs play a critical role in maintaining unsaturated fatty acid and cholesterol levels in mammalian cells. These transcription factors reside as integral membrane proteins in the endoplasmic reticulum. When sterol concentrations drop inside the cell, the sterol-binding protein SCAP directly facilitates SREBP transport to the Golgi complex, where SREBP is cleaved. The DNA binding component of SREBP can then translocate to the nucleus and induce cholesterol biosynthetic gene expression, defining an elegant feedback loop for maintaining cholesterol homeostasis (Brown and Goldstein, 1997). The discovery of an SREBP ortholog in flies, dSREBP, along with dSCAP and two SREBP proteases raises the interesting possibility that aspects of this feedback circuit have been maintained through evolution (Seegmiller et al., 2002). Studies in Drosophila tissue culture cells confirmed this proposal, showing that the major phospholipid in Drosophila, phosphatidylethanolamine, regulates dSREBP processing and controls membrane lipid production (Dobrosotskaya et al., 2002). Significantly, however, dSREBP does not respond to cholesterol, consistent with the inability of Drosophila to synthesize cholesterol.
Genetic studies of a dSREBP mutant have provided insights into its roles in the animal (Kunte et al., 2006). dSREBP null mutants die as undersized second instar larvae and display reduced levels of fatty acids, although the relative proportions of major long chain fatty acids remain unchanged. Consistent with this phenotype, the authors found reduced expression of three fatty acid synthesis genes in dSREBP mutants, and adding soy lipids or specific long-chain fatty acids to the growth medium was sufficient to rescue lethality. The authors designed a dSREBP-GFP reporter system to monitor the spatial and temporal patterns of dSREBP transcriptional activity in the animal, showing that it is normally active in the fat body, midgut, and oenocytes of feeding larvae. Importantly, this activity can be suppressed by adding excess lipid to the diet, demonstrating that the reporter is subject to normal feedback control. It will be interesting to examine other metabolic parameters in dSREBP mutants, such as glucose and lipid levels, as well as identify direct transcriptional targets that provide a better mechanistic framework for understanding its roles in lipid physiology.
Two papers on Drosophila miRNAs have demonstrated important roles for these small RNAs in the post-transcriptional control of lipid metabolism. In addition to its role in cell death, miR-14 is required for a normal adult lifespan and proper lipid levels (Xu et al., 2003). miR-14 mutants have increased levels of TAG and the main circulating lipid diacylglycerol (DAG), and have enlarged lipid droplets in their adult fat, while animals carrying four copies of miR-14 display an opposite phenotype. Levels of the major fatty acid classes do not appear to be altered in miR-14 mutants, suggesting that it is specific for lipid mobilization from the fat body. In contrast, miR-278 mutants display an opposite phenotype – lean animals with significantly reduced TAG levels (Teleman et al., 2006). The authors observed that dilp-2, dilp-3, and dilp-5 are increased in miR-278 mutants and found that removing one copy of Dp110 (encoding the catalytic PI3K subunit) is sufficient to alleviate most of the leanness associated with the miR-278 mutation, suggesting that this phenotype is at least partly due to increased insulin signaling. The miR-278 mutants, however, also display increased levels of circulating trehalose, suggesting they are insulin resistant, similar to what is seen in patients with type 2 diabetes. Consistent with this, insulin injection resulted in only modest reduction of 4E-BP mRNA in the fat body (as a readout of insulin signaling through FOXO), compared to a 3-fold reduction seen in wild type larvae. miR-278 thus appears to play a role in appropriate insulin responsiveness. Our understanding of these miRNAs, however, requires the identification of their target transcripts, and Teleman et al. (2006) provide evidence that miR-278 may act through expanded, which encodes part of a membrane-associated cell signaling complex. The observation that miRNAs regulate metabolism in other higher organisms suggests that their ability to fine-tune metabolic responses at the post-transcriptional level has been conserved through evolution (Wilfred et al., 2007).
New insights into the regulation of lipolysis
The larval fat body serves as a dynamic source for maintaining energy homeostasis and as a requisite reservoir for stored lipid during the prolonged period of non-feeding during metamorphosis. Accordingly, fat body lipid mass rises to ∼15% the total weight of the animal in the third instar, from ∼6% as a newly hatched first instar, most of which can be attributed to TAG (Church and Robertson, 1966a; Church and Robertson, 1966b). The larval fat body in newly emerged adults is replaced during the first few days by adult fat cells, with adults maintaining ∼6.5% of their body weight as lipid, similar to that seen in the first instar (Aguila et al., 2007; Rizki, 1978; Teague et al., 1986). This shift in lipid load is indicative of the change in fat body function, from directing organismal growth and TAG storage during larval stages to maintaining energy homeostasis in the adult (Colombani et al., 2003; Rizki, 1978). Consistent with this, the adult fat body is subject to diet-induced lipid overload, unlike the larval fat body, an observation that establishes the adult fly as an ideal context for functional studies of diet-induced obesity, a critical risk factor for human disease (Bross et al., 2005; Sanchez-Blanco et al., 2006).
One example of this is the naturally-occurring adipose60 mutant, which displays increased levels of total lipid under normal feeding conditions in the adult, with no effects at earlier stages, along with significant starvation resistance (Hader et al., 2003; Teague et al., 1986). The adipose gene encodes a protein with WD40 repeats that is widely expressed at all developmental stages, and which can decrease TAG when overexpressed in the larval fat body (Hader et al., 2003). This study also noted that orthologs of Adipose are encoded by both the mouse and human genomes, and speculated that its role in lipid metabolism may be conserved through evolution. Importantly, this prediction was recently confirmed by Suh et al. (2007), who showed that mice with one mutant copy of adipose are obese and insulin resistant, while transgenic adipose overexpression in fat results in mice that are lean and insulin sensitive. Biochemical and cell culture studies indicate that Adipose protein can bind histones and HDAC3, and may function as part of a transcriptional corepressor complex. Taken together, these studies demonstrate the powerful predictive function of Drosophila genetics, where genes discovered in the fly reveal new levels of metabolic control that are conserved through evolution to mice and humans.
Genetic analysis of two central players in lipid deposition and mobilization have provided significant new insights into the control of lipolysis. Characterization of the Drosophila perilipin-like protein LSD2 has demonstrated an evolutionary conserved role for the lipid droplet-associated PAT-domain proteins in promoting lipid storage (Grönke et al., 2003; Teixeira et al., 2003). Similarly, studies of the TAG lipase Brummer foreshadowed the essential role of its vertebrate ortholog ATGL in energy metabolism (Grönke et al., 2005; Haemmerle et al., 2006). Both LSD2 and Brummer reside on the outer membrane of lipid droplets (Grönke et al., 2003; Grönke et al., 2005). Mutant lsd2 animals exhibit a starvation-sensitive lean phenotype, due to a severe reduction in TAG, while lsd2 overexpression in the fat body increases TAG storage in a dose-dependent manner, leading to starvation resistance (Grönke et al., 2003; Teixeira et al., 2003). Conversely, the lipase activity of Brummer mobilizes stored TAG. Mutants of brummer display progressive obesity during early adult development, with no effects on glycogen levels, while ectopic fat body expression significantly depletes stored TAG (Grönke et al., 2005). These mutants are also starvation resistant, indicating that other pathways exist to mobilize the stored energy in these obese animals. The counterbalancing actions of these two lipid droplet proteins can be seen in lsd2; brummer double mutants, which display normal TAG levels, demonstrating their central role in modulating lipolysis (Grönke et al., 2005).
A recent study has addressed how insect AKH acts in parallel with Brummer to regulate lipolysis, in addition to the effects of AKH on sugar homeostasis described above (Grönke et al., 2007; Lee and Park, 2004). Similar to β-adrenergic signaling in mammalian lipid mobilization, AKH acts in an endocrine manner via the AKH receptor (AKHR) to activate protein kinase A and initiate lipolysis. Like lsd2 and brummer, akhr is expressed highly in the fat body (Grönke et al., 2007). Fat body-specific akhr overexpression results in a significant reduction in TAG, while akhr mutants accumulate lipid storage droplets and display increased TAG (Grönke et al., 2007). Importantly, the ability of chronic AKH overexpression to deplete lipid stores has no effect in akhr mutants, indicating that all lipid regulatory functions of AKH go through this receptor. Double mutants for brummer and akhr show an additive effect on lipid accumulation, but are also starvation sensitive. Measurement of TAG levels in these double mutants demonstrated that their excess lipid reserves do not change upon starving them to death. These starved mutants can, however, access their carbohydrate stores, demonstrating that this is not a general metabolic block but, rather, that brummer and AKH define the major lipolytic pathways in the animal. Further studies of starvation-induced lipolysis in akhr and brummer mutants revealed at least two distinct temporal phases, suggesting that AKH acts early upon food deprivation while brummer acts as the primary basal lipolytic factor and also upon prolonged starvation.
Taken together, these pioneering genetic studies of Drosophila lipid metabolism reveal the evolutionary conservation of perilipin and ATGL function as well as the distinct mechanisms by which Brummer and AKHR control lipid mobilization in Drosophila. The relatively large number of putative lipases encoded by the fly genome (Suppl. Table 1), the regulation of many of these genes by starvation (Grönke et al., 2005; Zinke et al., 2002), and the similar proteomic composition of fly and mammalian lipid droplets (Cermelli et al., 2006), establish parallels between fly and human lipid physiology and indicate that future studies in Drosophila will provide new insights into how lipid homeostasis is maintained.
Sterol absorption and trafficking defects in Niemann-Pick type C pathology
Aside from its essential role in cell membranes, cholesterol acts as the precursor for steroid hormones such as the insect steroid ecdysone, which triggers the major developmental transitions in the life cycle (Thummel, 2001). Although recent studies have defined many of the biochemical steps by which cholesterol is converted into ecdysone (Rewitz et al., 2006), the mechanisms that control cholesterol sensing, absorption, intracellular trafficking, and homeostasis, have remained unclear. Recent work on the Niemann-Pick type C (NPC) proteins has offered insights into these processes in the fly, with implications for understanding sterol homeostasis in humans.
The NPC proteins were identified based on the abnormal intracellular accumulation of lipids and cholesterol seen in patients with mutations in NPC genes, defects that eventually lead to neurodegeneration and death during adolescence (Vance, 2006). Two genes in Drosophila, npc1a and npc1b, are similar to human NPC1 and its paralog NPC1L1, encoding proteins with a 13-pass transmembrane domain and a putative sterol sensing region (Fluegel et al., 2006; Huang et al., 2005). The midgut-restricted expression pattern and function of npc1b mirror that of NPC1L1 in the mouse, which is required specifically in the intestine for sterol absorption (Ioannou, 2007). Drosophila npc1b mutants die following a prolonged second instar larval stage and can be effectively rescued by midgut-specific expression of a wild type transgene, supporting an essential role for npc1b in this tissue (Voght et al., 2007). Radiolabeled cholesterol feeding experiments show that cholesterol absorption is reduced dramatically in these mutants. Consistent with this, npc1b mutant midguts display virtually no staining with the cholesterol-binding compound filipin. Total cholesterol levels in npc1b mutants and filipin staining in peripheral tissues, however, both appear normal, indicating that the critical function for this gene is in dietary cholesterol absorption – an essential role for a cholesterol auxotroph such as Drosophila.
Studies of npc1a have shown that it is required for intracellular sterol trafficking (Fluegel et al., 2006; Huang et al., 2005). Null npc1a mutants display abnormal punctate filipin staining throughout the animal, indicating subcellular sterol accumulation and reflecting the widespread expression of npc1a. As a result, npc1a mutants die following a prolonged first instar larval stage, a phenotype that can be rescued by feeding excess cholesterol. Partial rescue can also be achieved by feeding ecdysone to mutant larvae, or expressing wild type npc1a specifically in the ring gland, demonstrating an essential role for this gene in providing sufficient sterol precursors for steroidogenesis. Interestingly, although punctate filipin staining is seen in human NPC patients, npc1a mutants display no evidence of another hallmark of the disease, neurodegeneration, suggesting that this phenotype is not a primary defect but rather may be a secondary consequence due to low levels of neurosteroids – a proposal supported by genetic studies of NPC1 in mice (Griffin et al., 2004). Moreover, while molecular mechanisms for NPC1L1 have remained unclear, studies of Drosophila npc1b mutants suggest that the primary defect is sterol absorption by the intestinal epithelium, although it may also play roles in intracellular sterol trafficking (Voght et al., 2007). In addition to providing insights into NPC function in humans, these genetic studies in Drosophila have established a framework for studies of sterol homeostasis. For example, larvae lacking both npc1a and npc1b retain the ability to effectively absorb cholesterol, raising the interesting possibility that there are alternate modes for sterol uptake, and suggesting that other factors may be discovered that could impact NPC disease (Voght et al., 2007). In addition, these Drosophila mutants provide invaluable tools to manipulate cholesterol levels in vivo, and thus could be useful in future studies to uncover how sterol levels are sensed in the animal and maintained during development.
Why study metabolism in Drosophila?
Over the past two decades, genetic studies in Drosophila have established the basis for our understanding of the central regulatory mechanisms that control animal development. Well known signaling pathways, such as Notch, wingless, and hedgehog, owe their discovery to Drosophila genetic screens, and have had profound implications for our understanding of vertebrate development as well as the origins of human disease (Bier, 2005). The past few years have seen a shift in direction, in which Drosophila is being increasingly exploited to understand the fundamental mechanisms that control metabolism. As with our use of the fly to study development, these genetic efforts provide a unique opportunity to uncover critical new insights into central regulatory pathways that are conserved through evolution, and have direct implications for the origin and treatment of human disease risk factors such as diabetes and obesity. Indeed, even the relatively few Drosophila papers described in this review have changed our understanding of vertebrate metabolic control. For example, contrary to studies in mammalian cell culture, 4E-BP regulates organismal lipid homeostasis rather than growth (Teleman et al., 2005a). Similarly, the discovery of evolutionarily-conserved essential regulators of lipid metabolism in Drosophila, such as melted and adipose, provide new directions for understanding the control of these pathways in humans, and also raise the possibility that mutations in these genes may be risk factors for human metabolic disorders. The first genetic study of an essential enzyme in organismal lipolysis, the ATGL homolog Brummer, foreshadowed genetic studies of its vertebrate counterpart in mice (Grönke et al., 2005; Haemmerle et al., 2006). Similarly, the extreme lipolytic disorder seen in brummer, akhr double mutants has not yet been approached in mouse models (Grönke et al., 2007). In addition, as pointed out by Huang et al. (2005), contrary to the proposal that lipid accumulation is a cause of NPC pathology, sterol shortage should instead be considered as a possible primary defect, a conclusion that has radical implications for the current use of lower cholesterol levels as part of NPC disease treatment.
The ability to manipulate the fly genome in virtually any way desired, combined with a range of newly available genomic resources, allow the researcher to define the molecular mechanisms of gene function at a level of resolution not possible in more complex organisms. In addition, no studies to date have exploited one of the main strengths of Drosophila – the ability to conduct large-scale open-ended genetic screens – an approach that holds the promise of extending our understanding of metabolic regulation in new and unexpected directions. It is likely that we have much to look forward to in the next few years, as further studies exploit the humble fruit fly to reveal new insights into the regulation of metabolism and homeostasis.
Supplementary Material
Acknowledgements
We thank M. Horner, A.-F. Ruaud, and M. Sieber for critical comments on the manuscript. Research in the Thummel lab is supported by the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguila JR, Suszko J, Gibbs AG, Hoshizaki DK. The role of larval fat cells in adult Drosophila melanogaster. The Journal of experimental biology. 2007;210:956–963. doi: 10.1242/jeb.001586. [DOI] [PubMed] [Google Scholar]
- Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nature reviews. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Developmental cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Bross TG, Rogina B, Helfand SL. Behavioral, physical, and demographic changes in Drosophila populations through dietary restriction. Aging cell. 2005;4:309–317. doi: 10.1111/j.1474-9726.2005.00181.x. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Canavoso LE, Jouni ZE, Karnas KJ, Pennington JE, Wells MA. Fat metabolism in insects. Annual review of nutrition. 2001;21:23–46. doi: 10.1146/annurev.nutr.21.1.23. [DOI] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- Church RB, Robertson FW. A biochemical study of the growth of Drosophila melanogaster. J Exp Zool. 1966a;162:337–351. [Google Scholar]
- Church RB, Robertson FW. Biochemical analysis of genetic differences in the growth of Drosophila. Genet Res. 1966b;7:383–407. doi: 10.1017/s0016672300009836. [DOI] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Science. Vol. 296. New York, NY: 2002. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila; pp. 879–883. [DOI] [PubMed] [Google Scholar]
- Edgar BA. How flies get their size: genetics meets physiology. Nature reviews. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Fluegel ML, Parker TJ, Pallanck LJ. Mutations of a Drosophila NPC1 gene confer sterol and ecdysone metabolic defects. Genetics. 2006;172:185–196. doi: 10.1534/genetics.105.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gade G, Auerswald L. Mode of action of neuropeptides from the adipokinetic hormone family. Gen Comp Endocrinol. 2003;132:10–20. doi: 10.1016/s0016-6480(03)00159-x. [DOI] [PubMed] [Google Scholar]
- Gilbert LI, Rybczynski R, Warren JT. Control and biochemical nature of the ecdysteroidogenic pathway. Annu Rev Entomol. 2002;47:883–916. doi: 10.1146/annurev.ento.47.091201.145302. [DOI] [PubMed] [Google Scholar]
- Griffin LD, Gong W, Verot L, Mellon SH. Niemann-Pick type C disease involves disrupted neurosteroidogenesis and responds to allopregnanolone. Nature medicine. 2004;10:704–711. doi: 10.1038/nm1073. [DOI] [PubMed] [Google Scholar]
- Grönke S, Beller M, Fellert S, Ramakrishnan H, Jackle H, Kuhnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr Biol. 2003;13:603–606. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell metabolism. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Grönke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, Jackle H, Kuhnlein RP. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5:e137. doi: 10.1371/journal.pbio.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Hader T, Muller S, Aguilera M, Eulenberg KG, Steuernagel A, Ciossek T, Kuhnlein RP, Lemaire L, Fritsch R, Dohrmann C, et al. Control of triglyceride storage by a WD40/TPR-domain protein. EMBO Rep. 2003;4:511–516. doi: 10.1038/sj.embor.embor837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, et al. Science. Vol. 312. New York, NY: 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase; pp. 734–737. [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes & development. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Helfand SL, Rogina B. Genetics of aging in the fruit fly, Drosophila melanogaster. Annual review of genetics. 2003;37:329–348. doi: 10.1146/annurev.genet.37.040103.095211. [DOI] [PubMed] [Google Scholar]
- Huang X, Suyama K, Buchanan J, Zhu AJ, Scott MP. A Drosophila model of the Niemann-Pick type C lysosome storage disease: dnpc1a is required for molting and sterol homeostasis. Development. 2005;132:5115–5124. doi: 10.1242/dev.02079. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Ioannou YA. Niemann-Pick C proteins in sterol transport and absorption: flies in the ointment. Developmental cell. 2007;12:481–483. doi: 10.1016/j.devcel.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Isabel G, Martin JR, Chidami S, Veenstra JA, Rosay P. AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. Am J Physiol Regul Integr Comp Physiol. 2005;288:R531–538. doi: 10.1152/ajpregu.00158.2004. [DOI] [PubMed] [Google Scholar]
- Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- Kunte AS, Matthews KA, Rawson RB. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell metabolism. 2006;3:439–448. doi: 10.1016/j.cmet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong N, Davies CR, Wessells RJ, Graham SM, King MT, Veech R, Bodmer R, Oldham SM. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell metabolism. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Min KT, Benzer S. Science. Vol. 284. New York, NY: 1999. Preventing neurodegeneration in the Drosophila mutant bubblegum; pp. 1985–1988. [DOI] [PubMed] [Google Scholar]
- Mourikis P, Hurlbut GD, Artavanis-Tsakonas S. Enigma, a mitochondrial protein affecting lifespan and oxidative stress response in Drosophila. Proc Natl Acad Sci U S A. 2006;103:1307–1312. doi: 10.1073/pnas.0510564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlig-Versen M, da Cruz AB, Tschape JA, Moser M, Buttner R, Athenstaedt K, Glynn P, Kretzschmar D. Loss of Swiss cheese/neuropathy target esterase activity causes disruption of phosphatidylcholine homeostasis and neuronal and glial death in adult Drosophila. J Neurosci. 2005;25:2865–2873. doi: 10.1523/JNEUROSCI.5097-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes BE, Katz FN, Schaffer MH. Identification and expression of the Drosophila adipokinetic hormone gene. Molecular and cellular endocrinology. 1995;109:133–141. doi: 10.1016/0303-7207(95)03492-p. [DOI] [PubMed] [Google Scholar]
- Okamura T, Shimizu H, Nagao T, Ueda R, Ishii S. ATF-2 regulates fat metabolism in Drosophila. Molecular biology of the cell. 2007;18:1519–1529. doi: 10.1091/mbc.E06-10-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- Partridge L, Gems D. Mechanisms of ageing: public or private? Nature reviews. 2002;3:165–175. doi: 10.1038/nrg753. [DOI] [PubMed] [Google Scholar]
- Reiling JH, Doepfner KT, Hafen E, Stocker H. Diet-dependent effects of the Drosophila Mnk1/Mnk2 homolog Lk6 on growth via eIF4E. Curr Biol. 2005;15:24–30. doi: 10.1016/j.cub.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Rewitz KF, Rybczynski R, Warren JT, Gilbert LI. The Halloween genes code for cytochrome P450 enzymes mediating synthesis of the insect moulting hormone. Biochemical Society transactions. 2006;34:1256–1260. doi: 10.1042/BST0341256. [DOI] [PubMed] [Google Scholar]
- Rizki TM. Fat Body. In: Ashburner M, Wright TR, editors. The Genetics and Biology of Drosophila. Academic Press; New York: 1978. pp. 561–601. [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Science. Vol. 296. New York, NY: 2002. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes; pp. 1118–1120. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Developmental cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Sanchez-Blanco A, Fridell YW, Helfand SL. Involvement of Drosophila uncoupling protein 5 in metabolism and aging. Genetics. 2006;172:1699–1710. doi: 10.1534/genetics.105.053389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Developmental cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Developmental cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Suh JM, Zeve D, McKay R, Seo J, Salo Z, Li R, Wang M, Graff JM. Adipose is a conserved dosage-sensitive antiobesity gene. Cell metabolism. 2007;6:195–207. doi: 10.1016/j.cmet.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague BD, Clark AG, Doane WW. Developmental analysis of lipids from wild-type and adipose60 mutants of Drosophila melanogaster. J Exp Zool. 1986;240:95–104. doi: 10.1002/jez.1402400112. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Rabouille C, Rorth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech Dev. 2003;120:1071–1081. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Chen YW, Cohen SM. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes & development. 2005a;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Chen YW, Cohen SM. Drosophila Melted modulates FOXO and TOR activity. Developmental cell. 2005b;9:271–281. doi: 10.1016/j.devcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes & development. 2006;20:417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Developmental cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM, et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nature medicine. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- Ueyama M, Chertemps T, Labeur C, Wicker-Thomas C. Mutations in the desat1 gene reduces the production of courtship stimulatory pheromones through a marked effect on fatty acids in Drosophila melanogaster. Insect biochemistry and molecular biology. 2005;35:911–920. doi: 10.1016/j.ibmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Vance JE. Lipid imbalance in the neurological disorder, Niemann-Pick C disease. FEBS Lett. 2006;580:5518–5524. doi: 10.1016/j.febslet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Voght SP, Fluegel ML, Andrews LA, Pallanck LJ. Drosophila NPC1b promotes an early step in sterol absorption from the midgut epithelium. Cell metabolism. 2007;5:195–205. doi: 10.1016/j.cmet.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Wigglesworth VB. The utilization of reserve substances in Drosophila during flight. The Journal of experimental biology. 1949;26:150–163. doi: 10.1242/jeb.26.2.150. [DOI] [PubMed] [Google Scholar]
- Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: A review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Molecular genetics and metabolism. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. The EMBO journal. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


