Abstract
The paired-related homeobox genes, Prx1 and Prx2, encode transcription factors critical for orofacial development. Prx1-/-/Prx2-/- neonates have mandibular hypoplasia and malformed mandibular incisors. Although the mandibular incisor phenotype has been briefly described (ten Berge et al., 1998, 2001; Lu et al., 1999), very little is known about the role of Prx proteins during tooth morphogenesis. Since the posterior mandibular region was relatively normal, we examined molar tooth development in Prx1-/-/Prx2-/- embryos to determine whether the tooth malformation is primary to the loss of Prx protein or secondary to defects in surrounding tissues. Three-dimensional (3D) morphological reconstructions demonstrated that Prx1-/-/Prx2-/- embryos had molar malformations, including cuspal changes and ectopic epithelial projections. Although we demonstrate that Prx1 protein is expressed only mesenchymally, 3D reconstructions showed important morphological defects in epithelial tissues at the cap and bell stages. Analysis of these data suggests that the Prx homeoproteins are critical for mesenchymal-epithelial signaling during tooth morphogenesis.
Keywords: homeobox, tooth development, molars, patterning
INTRODUCTION
Reciprocal signaling between mesenchyme and dental epithelium are required for normal tooth morphogenesis (Thesleff et al., 1995). Homeodomain-type transcription factors have been implicated in these signaling processes (Weiss et al., 1995; Chen et al., 1996; Tucker et al., 1998; Zhang et al., 1999; Zhao et al., 2000). Paired-related homeobox genes, Prx1 (a.k.a. mHox, K-2, Pmx, Prrx1) and Prx2 (a.k.a. S8 and Prrx2), encode DNA-binding transcription factors (Opstelten et al., 1991; Cserjesi et al., 1992; Kern et al., 1992; ten Berge et al., 1998; Lu et al., 1999). Based on mRNA expression studies, both Prx1 and Prx2 are mesenchymally expressed in tissues that develop into cartilage, bone, and tooth structures (Opstelten et al., 1991; Cserjesi et al., 1992; Kern et al., 1992; de Jong and Meijlink, 1993; Nohno et al., 1993; Kuratani et al., 1994; Leussink et al., 1995; ten Berge et al., 1998; Chesterman et al., 2001). Preceding tooth morphogenesis, Prx1 and Prx2 transcripts are widely expressed throughout the developing maxilla and mandible in undifferentiated mesenchyme. Prx1 mRNA is present in condensed mesenchyme at the bud through the cap stages, but is down-regulated once differentiation occurs at the bell stage (Karg et al., 1997). Although Prx1 RNA expression has been well-studied, we have previously showed that, due to post-transcriptional regulation of this gene, mRNA and protein expressions do not always correlate (Chesterman et al., 2001). Furthermore, an analysis of the spatial and temporal expression patterns for Prx1 protein during tooth development has not been conducted to date.
Prx1 and Prx2 genes are critical for the development of craniofacial and limb structures. The initial characterization of the phenotype in Prx1-/-/Prx2-/- (MUT) mice described severe craniofacial defects with mandibular hypoplasia (ten Berge et al., 1998, 2001; Lu et al., 1999). Anterior/midline regions of the mandible, where incisors develop, are extremely malformed and so hypoplastic that they appear to be nearly missing. As a result, mandibular incisors are either missing or fused at the midline, and the Prx genes were defined as critical regulators of both mandible formation and incisor positioning (ten Berge et al., 1998, 2001). Mandibular incisors were also reported to be arrested in the bud stage, but it was not determined if the Prx proteins were expressed in lower incisor tissues (Lu et al., 1999). Therefore, it was unclear whether the Prx genes played a direct role in tooth development or simply altered the anterior region of the mandible, which, in turn, had an adverse impact on incisor development. The anterior mandible in Prx1-/-/Prx2-/- embryos has altered expression of Shh, Pax9, Dlx2, and Alx3, which have been shown to affect tooth morphogenesis. In contrast, the posterior region of the mandible, where the molars form, is relatively normal and does not show these gene expression alterations or severe jaw malformations (ten Berge et al., 1998, 2001; Lu et al., 1999). We reasoned that analyzing molar development would eliminate the complications of wholesale gene circuitry changes, as well as severe malformations, and thereby provide a better tool to evaluate the role of Prx proteins in tooth development.
When studying protein expression in this study, we focused primarily on the localization and role of the Prx1 protein in molar morphogenesis, and not Prx2, for several reasons. First, the Prx1 homozygous null embryos had malformed and missing craniofacial tissues, while there were no morphological alterations in Prx2 null mice (Martin et al., 1995; ten Berge et al., 1998; Lu et al., 1999). Second, Prx1 is more potent at the biochemical level (Norris and Kern, 2001a,b). Third, the Prx1 primary transcript is alternatively spliced to generate 2 proteins, Prx1a and Prx1b, which have antagonistic effects on target genes and limb chondrogenesis (Norris and Kern, 2001a; Peterson et al., 2005). Therefore, it was more interesting to study the role of Prx1 during molar morphogenesis. Nevertheless, there is significant redundancy between these two loci, and the full spectrum of Prx1's impact on tooth morphogenesis cannot be evaluated unless Prx2 is also deleted. Therefore, tooth morphogenesis was examined in Prx1 and Prx2 double-null embryos.
The primary goal of this study was to examine both mandibular and maxillary molar tooth development in Prx1-/-/Prx2-/- mice. We utilized 3D modeling, since it is a powerful tool that can discern subtle alterations in morphology and facilitate a clearer understanding of tooth phenotypes in mutant mice (Peterkova et al., 2002a). We aimed to elucidate the role of Prx proteins during molar tooth development and determine whether these proteins directly regulate tooth development, or if the incisor defect observed in Prx1-/-/Prx2-/- embryos is a consequence of extreme malformations in adjacent jaw tissue.
MATERIALS & METHODS
Mice
Prx1 and Prx2 null alleles were maintained on a 129 SV/J × C57Bl/6 background. For 3D reconstructions, mice were mated and checked after 2 hrs for evidence of a vaginal plug (E0). Pregnant mice were euthanized at embryonic days (E) 14, E15, E16, and E18. We obtained Prx1-/-/Prx2-/- (MUT) embryos by crossing Prx1+/- mice on a Prx2-/- background and genotyped them as previously described (Lu et al., 1999). MUT mice die shortly after birth, limiting our analysis to gestational timepoints. All animals were treated in accordance with MUSC Institutional Review Board requirements.
Western Blot Analysis
Tissues were isolated from anterior and posterior regions of the mandible, where incisor and molar teeth, respectively, develop. Protein lysates were analyzed via Western Blot analysis as previously described (Chesterman et al., 2001).
Immunohistochemistry
Molar and incisor sections were immunostained with an anti-Prx1 antibody and a fluorescently labeled secondary antibody as previously described (Chesterman et al., 2001). Images were captured via a Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems, Inc., Bannockburn, IL, USA).
3D Reconstructions
We used Amira® 3.1 software (Mercury Computer Systems, San Diego, CA, USA) to generate 3D reconstructions of mandibular and maxillary molars from a series of sequential histological sections. At least 3 reconstructions were generated for both wild-type (WT) and MUT at E14, E15, E16, and E18. Details of the 3D modeling process are in the online APPENDIX.
RESULTS
Expression of the Prx1 Isoforms in Molars and Incisors
Prx1 isoforms have antagonistic effects on the transcription of downstream targets (Norris and Kern, 2001a; Peterson et al., 2005). No reagents are available to discriminate these isoforms by immunohistochemistry or in situ hybridization. However, the different electrophoretic mobilities of Prx1a and Prx1b identify the 2 isoforms, since the anti-Prx1 antibody recognizes the amino region of the protein, which is common to both (Fig. 1A). Prx1a and Prx1b isoforms are both expressed during murine molar and incisor development from the earliest timepoint examined (E13.5) through E16.5. No Prx1 protein was detectable in either incisor or molar tissues at E18.5. During this timecourse, the ratio of the 2 isoforms changes slightly in molars, with Prx1b predominating at E15.5. In contrast, the ratio of the 2 isoforms in developing incisors changes very little, with Prx1a predominating at all times analyzed.
Figure 1.
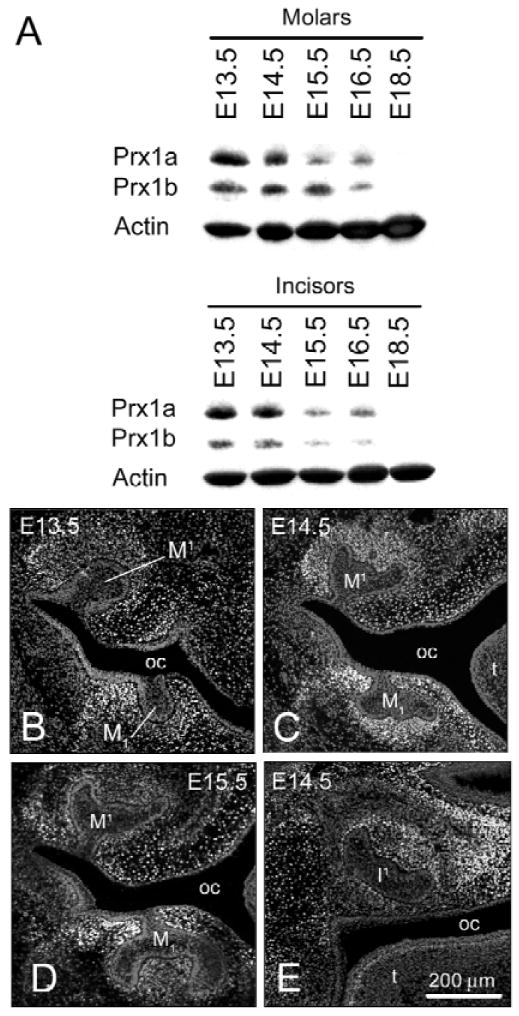
Expression of Prx1 protein during molar and incisor tooth development in WT mice. (A color version of this Fig. is included in the APPENDIX.) (A) Western analysis of Prx1 expression in tissue lysates derived from the anterior and posterior halves of the mandible, containing incisor and molar primordia, respectively. The data illustrate temporal changes in absolute levels of the 2 Prx1 isoforms (Prx1a and Prx1b) and a change in their ratio. (B-D) Immunohistochemistry of Prx1 protein expression (green) in frontal sections during molar and incisor tooth development (the 2 Prx1 isoforms cannot be distinguished by this method). Tissues were counterstained with propidium iodide (red). (B) At E13.5, high levels of Prx1 protein expression are present in mesenchyme surrounding maxillary (M1) and mandibular (M1) molar tooth buds; Prx1 protein is also localized in condensed mesenchyme adjacent to Meckel's cartilage (data not shown). There is no expression in oral or dental epithelium. (C) By E14.5, Prx1 expression is more refined and is strong in condensed mesenchyme surrounding the maxillary and mandibular molars. Prx1 expression is also highly localized in the mesenchyme of the developing oral vestibule. (D) At E15.5, there is reduced Prx1 protein expression in the dental papilla, but expression remains constant in mesenchymal tissues lateral to M1 and M1. (E) In a frontal section at E14.5, Prx1 protein is detected in the dental papilla of the developing maxillary incisor tooth germ (I1). Key: oral cavity (oc), tongue (t).
Prx1 Protein Expression Pattern during Molar and Incisor Development
The localization of Prx1 proteins in WT molars was assessed by immunohistochemistry. At E11.5, total Prx1 (Prx1a and Prx1b) protein is expressed in undifferentiated mesenchyme of the mandibular and maxillary arches, but not in epithelial cells (data not shown). This mesenchymal restriction of Prx1 was maintained throughout tooth development. By E13.5 (Fig. 1B), Prx1 protein was highly expressed in condensed mesenchyme surrounding the dental epithelium and was particularly intense lateral to the mandibular molar tooth bud extending to the vestibular lamina. Prx1 protein was also expressed adjacent to Meckel's cartilage and was localized along its medial aspect (data not shown). Although there was still some residual expression in undifferentiated mesenchyme, by E14.5, expression was further refined and Prx1 was localized predominantly to condensed mesenchyme around molars, lower vestibular lamina (Fig. 1C), and Meckel's cartilage. By E15.5 (Fig. 1D), Prx1 protein was present at low levels within the dental papilla of the bell-staged molar, but there was still pronounced expression along the lateral surface of the molar germ and adjacent to the developing oral vestibule. This pattern is likely repeated for second and third molars, since Prx1 was highly expressed in condensed mesenchyme surrounding the bud-staged second molar (data not shown). A similar pattern of expression was detected for Prx1 proteins during incisor development (Fig. 1E; data not shown).
Prx1-/-/Prx2-/- Phenotype Analyzed via 3D Orofacial Reconstructions
Previously, the orofacial phenotype of Prx1-/-/Prx2-/- embryos was characterized by two-dimensional (2D) sections, as well as by whole-mount Alcian blue and alizarin red histochemistry (Lu et al., 1999). 3D orofacial reconstructions at E14.5 (Appendix Fig. 1) facilitated the reanalysis and identification of novel features of the Prx1-/-/Prx2-/- phenotype, including tongue and molar tooth defects. At E14.5, MUT molar epithelia lack enamel grooves and are hypoplastic compared with WT (Appendix Figs. 1E, 1F).
Prx1-/-/Prx2-/- Molars Show Developmental Defects
For further analysis of tooth defects, 3D molar reconstructions were generated. At E14 (Appendix Fig. 2), there were subtle morphological differences, including a region of ectopic epithelia on the lateral aspect of the mutant mandibular molar (MUT-M1). Lateral to this tooth, the vestibular lamina was absent. By E15 (Appendix Fig. 3), morphological alterations were more severe, especially for MUT-M1. Ectopic epithelium, first evident at E14, was very pronounced and extended anteriorly-posteriorly along the lateral surface of MUT-M1. Both MUT-M1 and MUT-M1 had severe cervical loop hypoplasia. By E16 (Fig. 2), WT maxillary second molar (M2) and mandibular second molar (M2) buds were forming posterior to WT-M1 and WT-M1. Similar to E15, the epithelial cap and dental papilla were not very pronounced in MUT-M1 (Fig. 2L). Compared with WT, MUT-M1 epithelium was considerably hypoplastic and smaller in every dimension, while MUT-M1 was shortened along the anterior-posterior axis (Fig. 2E), but was enlarged medio-laterally (Fig. 2H). As a result, MUT-M1 epithelium was much larger than MUT-M1. Unlike E15, small cervical loops had formed in the MUT, but with a miniscule dental papilla region. The flange of ectopic epithelium was still present on the lateral surface and extended the full length of MUT-M1 (Figs. 2E, 2H), while the vestibular lamina was absent lateral to MUT-M1. MUT-M2 was substantially reduced in size (Fig. 2F) compared with WT (Fig. 2B), while the MUT-M2 was completely absent (Fig. 2G). Based on immunohistochemical analysis, Prx1 protein was highly expressed in condensed mesenchyme of WT-M2 and WT-M2 (data not shown). Therefore, based on this expression and morphological defects evident in the MUT, Prx genes are also involved in the morphogenesis of M2 and M2 tissues.
Figure 2.
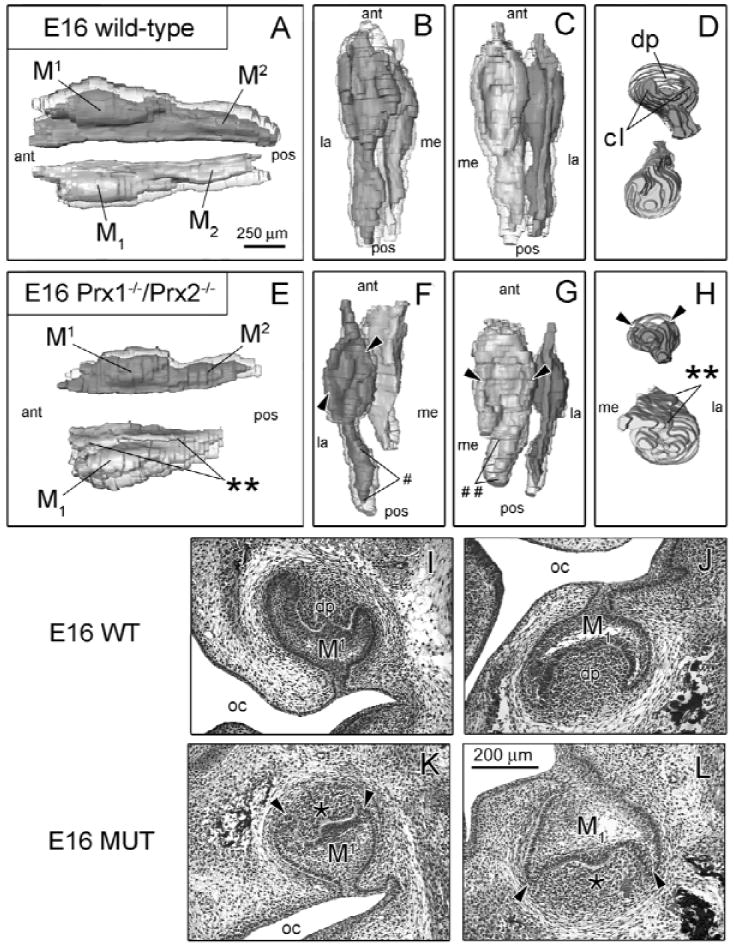
Prx1-/-/Prx2-/- molars show patterning defects at E16. (A color version of this Fig. is included in the APPENDIX.) WT (n = 3) and MUT (n = 3) 3D reconstructions (A-H) are presented from various viewpoints: lateral (A,E), superior (B,F), inferior (C,G), and frontal (D,H). Histological images of frontal sections are displayed for WT mice (I,J) and MUT (K,L). Morphological differences between WT and MUT include hypoplastic cervical loops (cl) (arrowheads), and dental papilla (dp) in the MUT-M1 and MUT-M1 are hypoplastic. In addition, MUT-M1 epithelium is more malformed and larger than the corresponding M1. WT-M1 is still inclined medially, while MUT-M1 lacks this positional displacement. WT maxillary (M2) and mandibular (M2) second molars are developing posterior to bell-stage WT-M1 and WT-M1, while MUT-M2 is hypoplastic (#) and MUT-M2 is absent (##). Key: oral cavity (oc), is anterior (ant), posterior (pos), medial (me), lateral (la).
Volumetric Analysis of 3D Reconstructions
For quantitative assessment of molar development in MUT embryos, volumetric measurements were extracted from all 3D reconstructions (E14-E16). At E16 (Appendix Fig. 4), volumetric measurements confirmed the epithelial hypoplasia of MUT-M1 detected by 3D reconstructions. This analysis demonstrated that MUT-M1 epithelia remained similar in size to WT, while both MUT-M1 and MUT-M1 mesenchyme were hypoplastic. MUT-M1 patterning defects (i.e., lacking a well-defined dental papilla region and exhibiting a flange of ectopic epithelium along the buccal surface of the enamel organ) reinforced the specific nature of the change while retaining the general size.
E18 Defects in Prx1-/-/Prx2-/- Molars Include Cuspal Alterations
By E18 (Fig. 3), alterations in MUT molar morphology evident at earlier stages were still manifested and were generally more severe for MUT-M1. Additionally, M2 development was so hypoplastic that a distinct MUT-M2 could not be detected (Fig. 3G). Ectopic epithelial tissue in MUT-M1 (Figs. 3E, 3H, 3J) still extended along the lateral surface of the molar epithelium from M1 to the ill-defined M2 region, while the lower vestibular lamina was absent lateral to molars.
Figure 3.
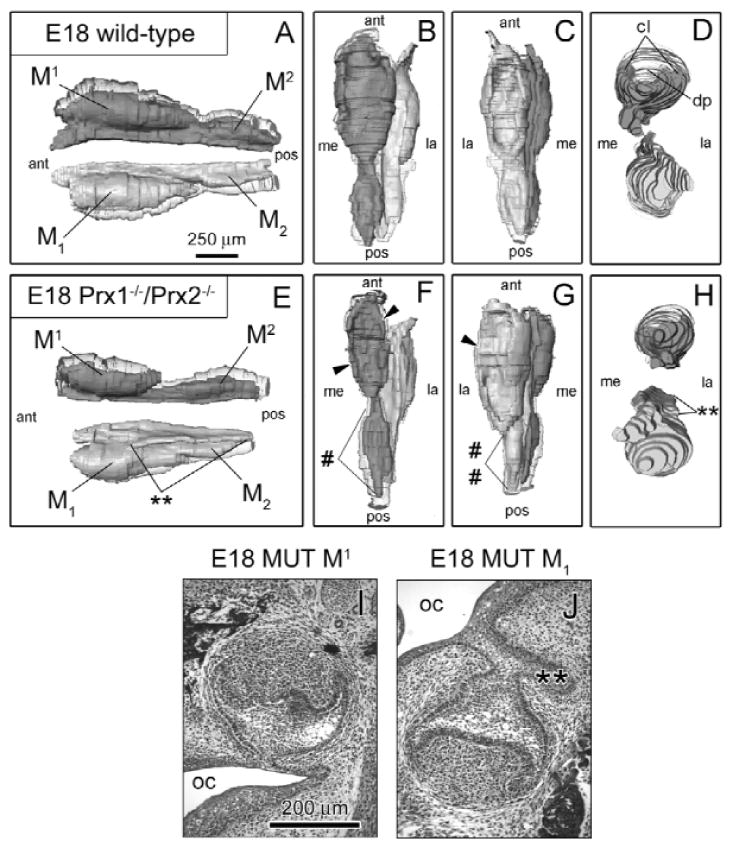
Altered morphology in MUT-M2 and MUT-M2 was even more evident at E18 (A-H). (A color version of this Fig. is included in the APPENDIX.) Histological images of frontal sections are also displayed for MUT-M1 (I) and MUT-M1 (J). By E18, WT epithelia of first (M1 and M1) and second (M2 and M2) molars had undergone morphological refinement to form the bell and early cap stages, respectively. Compared with preceding timepoints, the cervical loops (cl) of MUT-M1 were more developed, resulting in a more substantive region of dental papilla (dp), yet lateral cervical loops were underdeveloped (arrowhead) compared with WT (G). Epithelium posterior to MUT-M1 that normally gives rise to M2 was severely hypoplastic (##), and a distinct MUT-M2 was not readily apparent (G). MUT-M1 ectopic epithelia (double stars), first seen at E14, was larger and extended along the entire lateral surface (E,H,J). As demonstrated at E15 and E16, MUT-M1 was enlarged in every dimension compared with its corresponding M1. Although MUT-M1 and MUT-M2 epithelia had the general outline of WT, these tissues were severely hypoplastic, including the cervical loops (arrowhead) and the epithelial connection between MUT-M1 and MUT-M2. Key: oral cavity (oc), anterior (ant), posterior (pos), medial (me), lateral (la).
At this resolution (Fig. 3F), the general shapes of MUT-M1 and MUT-M2 epithelial tissue did not appear to be as affected as MUT-M1. To determine whether maxillary molars also had subtle malformations of tooth patterning, we examined cuspal development. To facilitate this analysis, we generated additional 3D reconstructions from a higher density of histological sections through M1 at E18 (Fig. 4). Though putative cuspal regions were evident in the MUT, patterning of these sites was drastically altered compared with WT (Figs. 4B, 4D), including shallower cusps and deeper pits. In addition, the aboral surface of the dental epithelium was irregular, and the medial cervical loop was very hypoplastic (Fig. 4D). Therefore, both MUT-M1 and MUT-M1 had specific developmental defects, supporting a direct role for Prx genes in tooth morphogenesis.
Figure 4.
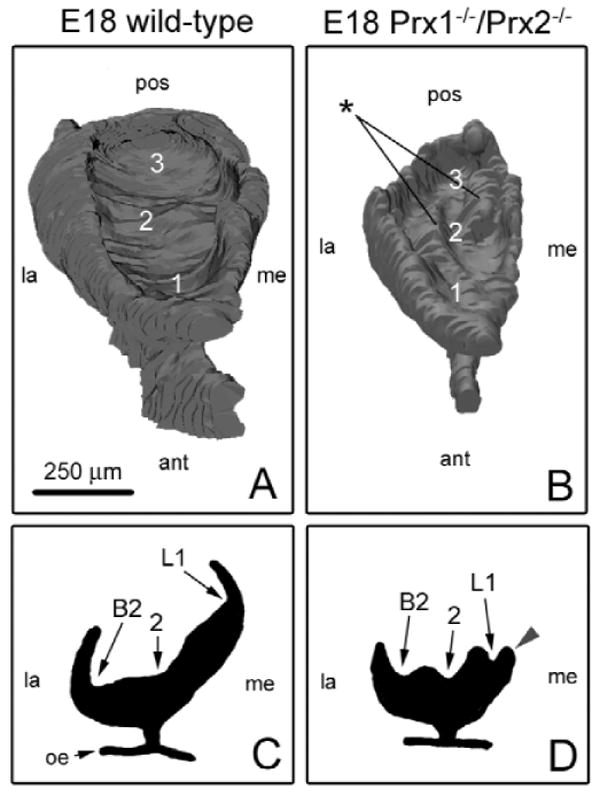
Cuspal patterning is altered in MUT-M1 at E18. (A color version of this Fig. is included in the APPENDIX.) M1 cuspal surfaces for WT (A) and MUT (B) are displayed from an elevated, anterior viewpoint; representative frontal sections through the widest portion of the same molar models are also depicted (C,D). 3D reconstructions of M1 were generated with 80 and 110 serial frontal sections for MUT and WT, respectively. Although MUT-M1 was overall hypoplastic and had hypoplastic cervical loops compared with WT, it also had altered morphology. Eight cuspal regions lie along the aboral surface of WT-M1 epithelium; 3 primary cuspal regions are labeled in panel A. In contrast, the MUT-M1 epithelial surface was very irregular, with the primary cuspal regions having a very different shape compared with WT, as well as containing unique and prominent mounds (stars), which together dramatically altered cuspal patterning (blue arrowhead). Key: oral epithelium (oe), anterior (ant), posterior (pos), medial (me), lateral (la).
DISCUSSION
The combination of Prx1 protein localization and detailed 3D reconstructions of Prx mutants facilitated a greater understanding of the role of Prx1 in molar morphogenesis. Analysis of our data demonstrated that although Prx1 protein is mesenchymally expressed at the bud and cap stages, the defects in the Prx null embryos were more pronounced later and predominantly in the epithelium. Epithelial malformations included ectopic epithelial projections, cervical loop hypoplasia, and cuspal patterning defects. This supports a model that Prx1 proteins are integrally associated with signaling pathways critical for normal tooth development. Furthermore, analysis of these data suggests that Prx genes have a direct impact on molar development.
Sonic hedgehog (Shh) is one member of a growing list of signaling molecules that are critical for normal tooth morphogenesis. It is expressed in the epithelium from the early stage of bud initiation (Hardcastle et al., 1999) to its later expression in the enamel knot, which is a critical signaling center that regulates tooth shape and cuspal patterning (Jernvall and Thesleff, 2000). Shh expression levels in the mandibular epithelium of Prx1-/-/Prx2-/- embryos were shown to be decreased compared with those in WT (ten Berge et al., 2001). Alterations in the complex pattern or levels of Shh expression during tooth development could elicit the molar alterations in Prx1-/-/Prx2-/- embryos, including abnormal cuspal patterning, cervical loop hypoplasia, and an ectopic flange of epithelium along the buccal surface of MUT-M1 (Hardcastle et al., 1998). Future experiments should evaluate Shh expression in the epithelium of the MUT molar tooth germ compared with that in WT.
In normal mice, Prx1 is strongly expressed lateral to M1, including the lower vestibular lamina (Figs. 1B-1D). This suggests that Prx genes can also play a role during differentiation of the lower oral vestibule. The presence of the ectopic flange of epithelium along the buccal surface of MUT-M1 was associated with an absence of the vestibular lamina lateral to the tooth germ. We propose that the ectopic flange of epithelium might correspond to the vestibular lamina fused with MUT-M1 as a consequence of failed differentiation of the oral vestibule in the lower jaw of MUT mice. In the upper jaw, fusion between vestibular and dental lamina occurs physiologically in mice (Peterkova et al., 2002b) and humans (Hovorakova et al., 2005).
Another important consideration and an area of potential future work is the specific role of the Prx1 isoforms, Prx1a and Prx1b, in tooth development. Based on the antagonistic roles of Prx1a and Prx1b on target gene transcription and chondrogenesis (Norris and Kern, 2001a; Peterson et al., 2005), coupled with our morphological and expression data, it is logical to predict that differential expression of the 2 isoforms could regulate tooth morphogenesis. For example, Prx1a may be expressed near the cervical loops stimulating their proliferation. Conversely, Prx1b may be expressed along the dental sac, thereby limiting epithelial proliferation and outgrowth. However, an analysis of Prx1a and Prx1b expression patterns awaits the development of a new set of reagents, either isoform-specific in situ probes or antibodies, both of which are technically difficult, due to the limited differences in the 2 isoforms.
Supplementary Material
A supplemental appendix to this article, and color images of Figures 1-4, are published electronically only at http://www.dentalresearch.org.
APPENDIX
Acknowledgments
The authors thank Mary Ann Baybo for assistance with the mice; Dr. Thomas Trusk and Josh Spruill for assistance with Amira® 3.1; Sue Tjepkema-Burroughs for assistance with figures; and Jamie Lee and Richard Peterson for invaluable discussions. This work was supported by NIH grants DE007337, RR16434, RR017677, RR016461, and DE16776; by the MUSC College of Dental Medicine, the Furman University Summer Research Program, and the Grant Agency of the Czech Republic (Grant 304/05/2665).
References
- Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- Chesterman ES, Gainey GD, Varn AC, Peterson RE, Jr, Kern MJ. Investigation of Prx1 protein expression provides evidence for conservation of cardiac-specific posttranscriptional regulation in vertebrates. Dev Dyn. 2001;222:459–470. doi: 10.1002/dvdy.1198. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Lilly B, Bryson L, Wang Y, Sassoon DA, Olson EN. MHox: a mesodermally restricted homeodomain protein that binds an essential site in the muscle creatine kinase enhancer. Development. 1992;115:1087–1101. doi: 10.1242/dev.115.4.1087. [DOI] [PubMed] [Google Scholar]
- de Jong R, Meijlink F. The homeobox gene S8: mesoderm-specific expression in presomite embryos and in cells cultured in vitro and modulation in differentiating pluripotent cells. Dev Biol. 1993;157:133–146. doi: 10.1006/dbio.1993.1118. [DOI] [PubMed] [Google Scholar]
- Hardcastle Z, Mo R, Hui CC, Sharpe PT. The Shh signalling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development. 1998;125:2803–2811. doi: 10.1242/dev.125.15.2803. [DOI] [PubMed] [Google Scholar]
- Hardcastle Z, Hui CC, Sharpe PT. The Shh signalling pathway in early tooth development. Cell Mol Biol (Noisy-le-grand) 1999;45:567–578. [PubMed] [Google Scholar]
- Hovorakova M, Lesot H, Peterka M, Peterkova R. The developmental relationship between the deciduous dentition and the oral vestibule in human embryos. Anat Embryol (Berl) 2005;209:303–313. doi: 10.1007/s00429-004-0441-y. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- Karg H, Burger EH, Lyaruu DM, Bronckers AL, Woltgens JH. Spatiotemporal expression of the homeobox gene S8 during mouse tooth development. Arch Oral Biol. 1997;42:625–631. doi: 10.1016/s0003-9969(97)00057-5. [DOI] [PubMed] [Google Scholar]
- Kern MJ, Witte DP, Valerius MT, Aronow BJ, Potter SS. A novel murine homeobox gene isolated by a tissue specific PCR cloning strategy. Nucleic Acids Res. 1992;20:5189–5195. doi: 10.1093/nar/20.19.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratani S, Martin JF, Wawersik S, Lilly B, Eichele G, Olson EN. The expression pattern of the chick homeobox gene gMHox suggests a role in patterning of the limbs and face and in compartmentalization of somites. Dev Biol. 1994;161:357–369. doi: 10.1006/dbio.1994.1037. [DOI] [PubMed] [Google Scholar]
- Leussink B, Brouwer A, el Khattabi M, Poelmann RE, Gittenberger-de Groot AC, Meijlink F. Expression patterns of the paired-related homeobox genes MHox/Prx1 and S8/Prx2 suggest roles in development of the heart and the forebrain. Mech Dev. 1995;52:51–64. doi: 10.1016/0925-4773(95)00389-i. [DOI] [PubMed] [Google Scholar]
- Lu MF, Cheng HT, Kern MJ, Potter SS, Tran B, Diekwisch TG, et al. prx-1 functions cooperatively with another paired-related homeobox gene, prx-2, to maintain cell fates within the craniofacial mesenchyme. Development. 1999;126:495–504. doi: 10.1242/dev.126.3.495. [DOI] [PubMed] [Google Scholar]
- Martin JF, Bradley A, Olson EN. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- Nohno T, Koyama E, Myokai F, Taniguchi S, Ohuchi H, Saito T, et al. A chicken homeobox gene related to Drosophila paired is predominantly expressed in the developing limb. Dev Biol. 1993;158:254–264. doi: 10.1006/dbio.1993.1184. [DOI] [PubMed] [Google Scholar]
- Norris RA, Kern MJ. The identification of Prx1 transcription regulatory domains provides a mechanism for unequal compensation by the Prx1 and Prx2 loci. J Biol Chem. 2001a;276:26829–26837. doi: 10.1074/jbc.M100239200. [DOI] [PubMed] [Google Scholar]
- Norris RA, Kern MJ. Identification of domains mediating transcription activation, repression, and inhibition in the paired-related homeobox protein, Prx2 (S8) DNA Cell Biol. 2001b;20:89–99. doi: 10.1089/104454901750070292. [DOI] [PubMed] [Google Scholar]
- Opstelten DJ, Vogels R, Robert B, Kalkhoven E, Zwartkruis F, de Laaf L, et al. The mouse homeobox gene, S8, is expressed during embryogenesis predominantly in mesenchyme. Mech Dev. 1991;34:29–41. doi: 10.1016/0925-4773(91)90089-o. [DOI] [PubMed] [Google Scholar]
- Peterkova R, Kristenova P, Lesot H, Lisi S, Vonesch JL, Gendrault JL, et al. Different morphotypes of the tabby (EDA) dentition in the mouse mandible result from a defect in the mesio-distal segmentation of dental epithelium. Orthod Craniofac Res. 2002a;5:215–226. doi: 10.1034/j.1600-0544.2002.02226.x. [DOI] [PubMed] [Google Scholar]
- Peterkova R, Peterka M, Viriot L, Lesot H. Development of the vestigial tooth primordia as part of mouse odontogenesis. Connect Tissue Res. 2002b;43:120–128. doi: 10.1080/03008200290000745. [DOI] [PubMed] [Google Scholar]
- Peterson RE, Hoffman S, Kern MJ. Opposing roles of two isoforms of the Prx1 homeobox gene in chondrogenesis. Dev Dyn. 2005;233:811–821. doi: 10.1002/dvdy.20412. [DOI] [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, Korving J, Martin JF, Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, Korving J, Reijnen MJ, van Raaij EJ, Verbeek F, et al. Prx1 and Prx2 are upstream regulators of sonic hedgehog and control cell proliferation during mandibular arch morphogenesis. Development. 2001;128:2929–2938. doi: 10.1242/dev.128.15.2929. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Vaahtokari A, Kettunen P, Åberg T. Epithelial-mesenchymal signaling during tooth development. Connect Tissue Res. 1995;32:9–15. doi: 10.3109/03008209509013700. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- Weiss KM, Ruddle FH, Bollekens J. Dlx and other homeobox genes in the morphological development of the dentition. Connect Tissue Res. 1995;32:35–40. doi: 10.3109/03008209509013703. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao X, Hu Y, Amand T, Zhang M, Ramamurthy R, et al. Msx1 is required for the induction of Patched by Sonic hedgehog in the mammalian tooth germ. Dev Dyn. 1999;215:45–53. doi: 10.1002/(SICI)1097-0177(199905)215:1<45::AID-DVDY5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Stock D, Buchanan A, Weiss K. Expression of Dlx genes during the development of the murine dentition. Dev Genes Evol. 2000;210:270–275. doi: 10.1007/s004270050314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A supplemental appendix to this article, and color images of Figures 1-4, are published electronically only at http://www.dentalresearch.org.
APPENDIX


