Abstract
We have designed ribozymes based on a self-splicing group I intron that can trans-splice exon sequences into a chosen RNA target to create a functional chimeric mRNA and provide a highly specific trigger for gene expression. We have targeted ribozymes against the coat protein mRNA of a widespread plant pathogen, cucumber mosaic virus. The ribozymes were designed to trans-splice the coding sequence of the diphtheria toxin A chain in frame with the viral initiation codon of the target sequence. Diphtheria toxin A chain catalyzes the ADP ribosylation of elongation factor 2 and can cause the cessation of protein translation. In a Saccharomyces cerevisiae model system, ribozyme expression was shown to specifically inhibit the growth of cells expressing the virus mRNA. A point mutation at the target splice site alleviated this ribozyme-mediated toxicity. Increasing the extent of base pairing between the ribozyme and target dramatically increased specific expression of the cytotoxin and reduced illegitimate toxicity in vivo. Trans-splicing ribozymes may provide a new class of agents for engineering virus resistance and therapeutic cytotoxins.
Self-splicing group I introns catalyze their excision and the ligation of flanking exon sequences via two sequential transesterification reactions (1). Group I introns from T4 phage (2) and Tetrahymena thermophila (3, 4) have been bisected, and the separated components have been coexpressed in various cell types. Formation of natural base pairings and, presumably, tertiary interactions between the two components in vivo resulted in trans-splicing reactions analogous to the naturally occurring cis-splicing reactions. Demonstration that the Tetrahymena ribozyme catalyzes the ligation of exons in trans has led to speculation that these molecules could be used for therapeutic repair of defective transcripts (3, 5). Alteration of endogenous myotonic dystrophy protein kinase transcripts (6) and repair of sickle β-globin transcripts by trans-splicing ribozymes in mammalian cells have recently been demonstrated (7). However, this type of ribozyme interacts with the intended target mRNA via a base-paired helix (P1) of only 6 bp and shows relatively poor specificity, with products of illegitimate splicing seen in vivo (8).
We recently described the modification of trans-splicing ribozymes for improved activity in vivo (9). These ribozymes are also based on the Tetrahymena intron but contain altered flanking sequences to allow precise alignment of both 5′- and 3′-exon sequences during trans-splicing, via modified P1 and P10 helices. They also contain an additional region of extended complementarity to the chosen target mRNA (Fig. 1). This design proved to be general, and several ribozymes were targeted at sites within different mRNAs. Each of these ribozymes contained a 3′ exon encoding a portion of the lacZ gene. Coexpression of the ribozymes with their intended target mRNAs in Escherichia coli resulted in the production of accurately spliced chimeric mRNAs and readily detectable levels of β-galactosidase activity. Inactivation of the ribozyme or mutation of the target mRNA abolished activity. Furthermore, modification of the ribozyme internal guide sequences (IGSs) and inclusion of an extended antisense domain were shown to be crucial for improved biological activity.
Figure 1.
Ribozyme-mediated trans-splicing. Ribozyme and target RNAs are indicated schematically, with conserved sequences shown explicitly. In the first step, ribozyme and target RNAs base pair to form the P1 and extended antisense helices. Guanosine-mediated transesterification results in cleavage of the target RNA. The distal portion of helix P1 is displaced by sequences from the 3′ exon to form helix P10, which allows the second transesterification to proceed, and results in ligation of the two exons.
These ribozymes might be useful for the selective delivery of new gene activities in virus-infected or malignant cells. For example, trans-splicing ribozymes could provide the basis for a virus resistance strategy in plants. Trans-splicing ribozymes could be designed to recognize a particular virus mRNA and to catalyze the in-frame fusion of sequences encoding a toxic peptide. Virus entry and expression of the target sequence in cells expressing the ribozyme would result in localized cell death and thereby inhibit virus replication and spread. For this approach to be successful, trans-splicing and production of the toxin need to be highly specific to minimize toxicity in uninfected cells, and biologically efficient to minimize spread of the virus. To test this strategy, we selected the coat protein (CP) mRNA of cucumber mosaic virus (CMV) as a target. CMV is an important agricultural pathogen with a host range exceeding 800 species and has been identified as the causal agent of several disease epidemics worldwide (10). In the presence of CMV CP sequences, our trans-splicing ribozymes are designed to deliver the ADP ribosyltransferase activity of diphtheria toxin A chain (DTA). DTA modifies elongation factor 2 in a rapid catalytic fashion and results in the cessation of translation (11). We have established a simple genetic approach for the testing and improvement of cytotoxic ribozymes in S. cerevisiae and have demonstrated that the new ribozymes can be used to efficiently and specifically trigger toxin expression in cells expressing the chosen target mRNA.
MATERIALS AND METHODS
Ribozyme Construction.
All plasmids were constructed by standard techniques (12). A CMV-green fluorescent protein (GFP) target gene and its mutant derivative (9) were isolated as BamHI–SacI fragments and inserted between the ADH1 promoter and terminator sequences of pVT103-L (13), to create pCMV-GFP and pMUT-GFP. The ribozyme genes were constructed in a modular fashion, with antisense, ribozyme and exon regions separated by restriction endonuclease sites for BamHI, XhoI, KpnI, and SacI (Fig. 2). Plasmid pcisRz-DTA (Rz indicates ribozyme) was made by fusion of CMV and P10 sequences to nucleotides 28 to 414 of the Tetrahymena intron (14) by using PCR amplification with the primers CGAATTCAAGCTTGGATCCGAAGGAGATATAACAATGGACAAATCTGACTCGAGTACCTAGGTTTGTAAAAGTTATCAGGCATGCAC and CTAAGGTACCTAACGAGTACTCCAAAACTAATC (BamHI, XhoI, and KpnI sites italic). A deleted form of the intron, corresponding to the ΔP5abc mutation (15), was constructed and used as a template to provide an inactive control. The DTA ORF (16) was PCR amplified with the 5′ and 3′ primers CGGCGGTACCTCGATGATGTTGTTGATTCTTCTAAATC and CGGCGAGCTCTCACAAAGATCGCCTGACACGATTTCC, respectively (KpnI and SacI sites italic). The ribozyme and DTA ORF PCR products were cut and cloned in tandem within the BamHI and SacI sites between the ADH1 promoter and terminator sequences of pVT103-U (13).
Figure 2.
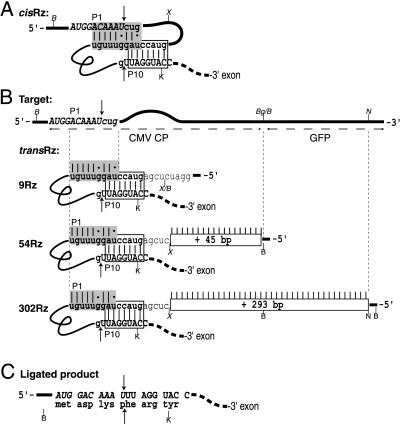
Design of ribozyme and target RNAs. (A) Schematic diagram of the cis-splicing ribozyme in pcisRz-DTA. The 5′-exon sequences are italic and uppercase, the 3′-exon sequences are nonitalic and uppercase, and intron sequences are lowercase. The IGS of the intron is represented in bold. The modified P1 helix sequences are shaded, and the modified P10 helix sequences are boxed. Watson-Crick base paring is indicated by |, and G:U base pairs are represented by dots. Arrows indicate the 5′- and 3′-splice sites. (B) Schematic diagram of the trans-splicing ribozymes. The target transcript is represented by the upper horizontal line with sequences around the splice site shown italicized. Trans-splicing ribozymes are represented below. Potential base pairing between the ribozyme and the target is indicated by |, and the length of the extended antisense sequences are indicated in rectangular boxes. The unpaired bulged region between the P1 helix and the extended antisense sequence is represented by the curved region in the target diagram. (C) Sequence of the expected ligated splice junction (indicated by arrows) and the encoded amino acids. Restriction endonuclease recognition sites: B, BamHI; X, XhoI; K, KpnI; S, SacI; P, PstI; N, NdeI; Bg, BglII. The figures are not drawn to scale.
Trans-splicing ribozymes were created from pcisRz-DTA as follows: p9Rz-DTA was created by digesting pcisRz-DTA with BamHI and XhoI, filling in the 5′ overhangs with the Klenow fragment of E. coli DNA polymerase I, and performing blunt end ligation. Complementary oligonucleotides containing the sequence GATCCACCACGACGCAAACGACGTCGACGGTTACGACCAGCACTGGTCTCGA were synthesized and subcloned into the BamHI and XhoI sites of pcisRz-DTA to create p54Rz-DTA. For the creation of p302Rz-DTA, a 245-nt fragment of mGFP5 was PCR amplified from the target vector by using as a 5′ primer GGCGGATCCATGAGTAAAGGAGAAGAACTTTTCACT and as a 3′ primer CGGCGAAGCTTGGATCCCATATGATCTGGGTATCTTG (BamHI and NdeI site italic). The fragment was digested with BamHI and subcloned into the BamHI site of p54Rz-DTA, and a clone was identified with the fragment in the antisense orientation.
Site-directed mutagenesis (17) was used to introduce the mutations W50A and G52E into the DTA ORF. The mutagenic oligonucleotide had the sequence GGATGATATCTTAAGGAATCGTAGTAGCAG.
Bioassay.
All yeast manipulations were performed by standard protocols (18, 19). Ribozyme-containing plasmids derived from pVT103-U were maintained in yeast strain FY833 (MATa his3Δ200 ura3–52 leuΔ1 lys2Δ202 trp1Δ63) (20) and grown on media lacking uracil. CMV target-containing plasmids derived from pVT103-L were maintained in FY834 (MATα his3Δ200 ura3–52 leuΔ1 lys2Δ202 trp1Δ63) (20), on media lacking leucine. Growth rates were measured by taking OD600 readings of liquid cultures maintained in log phase for at least six generations. Diploids resulting from mating of the two strains were selected on media lacking uracil and leucine. Reverse transcription (RT) of 4 μg of total yeast RNA was performed with a poly(dT)-primer and Super RT (HT Biotechnology) according to the supplier’s protocol. A fifth of the reaction (10 μl) was then used as a template for PCR amplification with a nested 3′ primer (also used for DTA ORF construction, see above), and a 5′ primer complementary to the target, with the sequence TCCAAGGAGATATATAACAATGG. PCR was carried out in 50 μl with Taq DNA polymerase (Promega) for 35 cycles with 60 s at 94°C, 60 s at 55°C, and 60 s at 72°C. Aliquots of the reaction were analyzed on 1.2% agarose gels stained with ethidium bromide. The PCR bands were gel eluted and reamplified with the same primers used in the RT-PCR. Aliquots were directly sequenced with the Thermo Vent cycling kit (New England Biolabs), according to the supplier’s protocol, and with a primer that is homologous to nucleotides 73–94 of the DTA-coding region (GAATGGAATCTACATAACCAGG). Steady-state levels of ribozyme and URA3 RNAs were measured by Northern blot analysis using 32P-radiolabeled probes.
RESULTS
Ribozyme Design.
The trans-splicing ribozymes used in this work are based on the group I intron of the large rRNA subunit of T. thermophila that catalyzes its own excision and ligation of the exon sequences via two sequential transesterification reactions (1). The reaction mechanism of trans-splicing is assumed to be the same as for cis-splicing, with the exception that the 5′-splice site is on a separate molecule, necessitating an intermolecular association via base pairing. To increase the potential association between the trans-acting ribozyme and the target, additional sequences complementary to the target were fused to the 5′ end of the intron in some of the constructs tested (Figs. 1 and 2B). The sole sequence requirement in the target mRNA is a uridine that forms a conserved wobble base pair with a guanosine in the IGS of the intron to define the 5′-splice site (21, 22). Annealing of the target and the ribozyme occurs via the P1 helix and an extended “antisense” region fused to the 5′ end of the ribozyme. Cleavage at the 5′-splice site is initiated by nucleophilic attack from a guanosine cofactor (GOH). In the second step of splicing, the distal portion of helix P1 is presumably displaced by sequences from the 3′ exon to form helix P10 and align the two exons across the IGS. The 3′-hydroxyl group of the cleaved 5′ exon (UOH) then attacks the precisely positioned 3′-splice site, resulting in ligation of the exons, and product release (Fig. 1). The design of a new trans-splicing ribozyme requires the selection of (i) a target mRNA site and (ii) 3′-exon sequence and (iii) the introduction of compensating base changes within the IGS sequence.
The CP mRNA of the CMV was selected as a target, because it is the most highly transcribed of the virus sequences, and the 5′ region is highly conserved between isolates (23). A uridine 10 nt downstream of the CP initiation codon was chosen as the 5′-splice site. Selecting a uridine close to the AUG minimizes the size of the amino-terminal fusion produced after splicing to the 3′-exon ORF. Furthermore, selecting a splice site close to the 5′ terminus of the target transcript may facilitate helix formation between the target and the ribozyme (24). Sequences in the intron IGS were altered from that of the wild-type Tetrahymena intron to allow formation of an artificial P1 duplex with the target while maintaining the essential U:G wobble base pair at the 5′-splice site.
We chose to use the coding sequence of DTA as the 3′ exon. DTA is a potent inhibitor of translation in eukaryotic cells but is not toxic in prokaryotes. This allows easy manipulation of the sequences in bacterial cells. The 3′ exon encoding DTA was fused downstream of the ribozyme and positioned so that splicing would result in an in-frame fusion to the coding sequence of the target mRNA (Fig. 2C). Sequences at the 5′ end of the 3′ exon and in the IGS were adjusted to allow formation of the P10 duplex, which is proposed to align the 3′ exon within the intron core (25). The P10 duplex is dispensable for splicing in vitro, but it prevents the utilization of cryptic splice sites (26, 27), and a potential P10 helix is present in most group I introns sequenced to date (28). The start codon of the DTA ORF was removed to minimize expression before splicing. It was also found necessary to alter a methionine at position 14 of the wild-type DTA protein sequence to reduce spurious expression of the toxin (mutation M14I; J.H., A. H. Brand, N. Perrimon, and H.M.G., unpublished results).
Cis-Splicing Ribozymes.
To test the catalytic and biological activity of the modified ribozyme, we first produced a cis-splicing form. The CMV target sequence was fused to the ribozyme domain and the DTA 3′ exon and cloned behind the constitutive ADH1 promoter in a yeast multicopy plasmid vector (pcisRz-DTA; Fig. 2A). A derivative that is inactive under physiological conditions was produced by deletion of the P5abc region (15) of the ribozyme (pcisΔP5-DTA). Self-splicing should produce a translatable CMV-DTA-fused mRNA and result in toxicity. Transformation of pcisRz-DTA into yeast cells resulted in no colonies, whereas the expected frequency of colony formation was obtained with pcisΔP5-DTA. The DTA ORF within pcisΔP5-DTA-harboring cultures was subcloned into the active cis-splicing construct. Cells transformed with this construct failed to form colonies (data not shown), confirming that the DTA sequence in pcisΔP5-DTA is fully active and that the lack of toxicity was caused by the inactivated ribozyme domain. The toxicity of the cis-splicing construct and sequence analysis of the spliced products (see below) demonstrate that (i) the modifications made to sequences flanking the catalytic core of the intron do not prevent splicing activity, (ii) splicing yields a translatable chimeric mRNA encoding DTA, and (iii) fusion of DTA with the short CMV CP amino-terminal peptide produces an active toxin.
CMV-Specific Trans-Splicing.
The cis-splicing ribozymes were converted to trans-splicing ribozymes by the replacement of sequences upstream of the ribozyme, which were flanked by BamHI and XhoI sites (Fig. 2B). Different lengths of sequence complementary to the CMV CP target were inserted to produce p9Rz-DTA, which was capable of forming 9 bp with the target, which is analogous to the length of the naturally occurring Tetrahymena P1 helix. Two other ribozymes were constructed with additional antisense sequences to give a total of 54 and 302 bp complementary to the target mRNA (p54Rz-DTA, and p302Rz-DTA, respectively). In the latter two constructs, an unpaired bulge region was maintained between the P1 duplex and the antisense region to mimic the unpaired bases in the loop terminating the P1 helix in the Tetrahymena intron. Maintaining an unpaired region at the end of the P1 helix should facilitate formation of the P10 duplex (27) by complementary sequences in the IGS and the 5′ end of the 3′ exon during the second transesterification step. Additional vectors were constructed with the ΔP5abc version of the ribozyme (p9ΔP5-DTA, p54ΔP5-DTA, and p302ΔP5-DTA) as inactive controls. Northern blot analysis showed that the ribozyme transcripts accumulated to similar levels in vivo (data not shown).
A synthetic target was created by fusing sequences encoding the N terminus of CMV CP with the ORF of a modified GFP (mGFP5) (29) (Fig. 2B). A mutated version of the CMV-GFP target was also constructed in which the U residue involved in the essential U:G wobble base pair at the 5′-splice site (position 10 of the CMV CP sequence) was changed to a G (pMut-GFP). Base pairing between the ribozyme and the mutated target should be similar to that formed with the correct target, but the first transesterification reaction should be prevented, prohibiting exon ligation. The target genes were cloned into the multicopy yeast plasmid pVT103-L, and expression of the target genes could be monitored by the production of green fluorescence in vivo.
The various ribozyme and target genes were maintained in separate heterothallic yeast strains. Ribozyme-containing plasmids were transformed into an a mating type yeast strain (FY833), and the target constructs were transformed into an α mating type (FY834). This allowed the maintenance of haploid lines and also allowed different combinations of ribozyme and target to be brought together by mating. The number and size of diploid colonies that grew after mating of the relevant ribozyme and target-containing strains provided a measure of trans-splicing-mediated toxicity. We also used strains with defective forms of the target mRNA, DTA, and ribozyme domain as controls, to confirm the specificity of ribozyme-mediated toxicity.
Fig. 3 shows the result of crossing strains that express various ribozymes with strains expressing wild-type or mutant target genes. Normal frequencies of colony formation were seen after mating of strains containing the parental plasmid (pVT103-U) and the ribozyme lacking the antisense domain (p9Rz-DTA) (Fig. 3 A and B). Introduction of the ribozyme with 54 nt of antisense sequence (p54Rz-DTA) into cells containing the target resulted in a significantly reduced frequency and rate of colony development, relative to control cultures (Fig. 3C). Furthermore, mating of a strain containing the ribozyme with 302 nt of complementarity to the target (p302Rz-DTA) with the strain containing the correct target (pCMV-GFP) resulted in complete failure of colony growth (Fig. 3D). In contrast, expression of the trans-splicing ribozymes in cells containing a mutated target (pMut-GFP) always resulted in normal colony growth. The pMut-GFP differs from pCMV-GFP in only a single nucleotide at the intended splice site. This is expected to confer immunity to trans-splicing and provides a strong indication that the observed cytotoxicity is caused by specific trans-splicing.
Figure 3.
Targeted cell ablation with trans-splicing ribozymes and increased complementarity to the target RNA. Yeast strains expressing various ribozymes, pVT103-U (A), p9Rz-DTA (B), p54Rz-DTA (C), or p302Rz-DTA (D), were mated with strains containing the CMV target gene (pCMV-GFP, left) or a mutated version with a single nucleotide alteration at the 5′-splice site (pMut-GFP, right). The diploid progeny of the cross were plated onto selective media, and colony growth was recorded after 3 days.
RNA Analysis of Spliced Products.
To establish whether the ribozymes were using the expected splice sites within the target, RT-PCR was used to analyze the products of the in vivo splicing reactions. Because the toxicity observed in the above experiments would limit the recovery of trans-spliced mRNA, the DTA-coding sequence was mutated to encode an inactive protein. These mutations were W50A, which reduces ADP ribosyltransferase activity nearly 105-fold (30), and G52E, which was identified as a nontoxic mutation in yeast cells (31) and subsequently was shown to be temperature sensitive (32). Mutated cis- and trans-splicing constructs did not reduce the growth rates of yeast in liquid culture, and Northern blot analysis showed that the mutations did not alter the stability of the RNAs in vivo (data not shown).
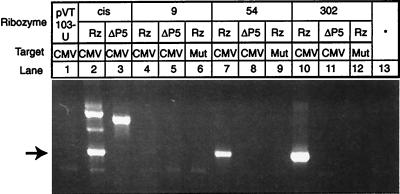
RT-PCR assays were performed on RNA isolated from diploid cultures coexpressing active or inactive ribozymes with the mutated DTA ORF (null) and either pCMV-GFP or pMut-GFP (Fig. 4). Bands corresponding to the expected splice products (632 nt) were obtained with pcisRz-null (lane 2), p54Rz-null (lane 7), and p302Rz-null (lane 10) when coexpressed with pCMV-GFP. A small quantity of RT-PCR product was obtained from cultures coexpressing p9Rz-null and pCMV-GFP (lane 4), indicating a low level of trans-splicing in these cells despite little evidence of impaired growth (Fig. 3B). Additional bands corresponding to the unspliced RNA were amplified from cultures harboring pcisRz-null (1042 nt, lane 2) and pcisΔP5-null (973 nt, lane 3), because of the arrangement of oligonucleotide priming sites. Bands corresponding to spliced products were not obtained when the trans-splicing ribozymes were coexpressed with the mutated target or when any of the ΔP5abc ribozymes were coexpressed with the correct target. RNA isolated from a culture harboring the pCMV-GFP target construct and RNA from a separate culture expressing p302Rz-null were mixed and analyzed by RT-PCR: absence of any RT-PCR products indicates that the observed product bands result from trans-splicing in vivo and not in vitro (Fig. 4, lane 13). The 632-nt bands shown in Fig. 4, lanes 2, 7, and 10, were reamplified, sequenced, and shown to correspond to the expected cis- and trans-spliced products (AUG GAC AAA U/UU AGG UAC; Fig. 2C).
Figure 4.
Amplified products of in vivo cis- and trans-splicing. RT-PCR was used to amplify the RNA products of splicing from yeast cells expressing the indicated ribozyme and target genes. Bands migrating at a rate expected for DNA fragments 632 nt in length (indicated by an arrow) correspond to spliced products, and the major slower migrating bands in lanes 2 and 3 correspond to unspliced cis-acting ribozyme and inactive ΔP5abc derivative, respectively. A faint band corresponding to 632 nt was seen in lane 4.
DTA Provides a Sensitive Assay for Target-Independent Expression.
The above experiments demonstrate that trans-splicing ribozymes can be engineered to deliver a toxin to eukaryotic cells depending on the presence of a specific RNA target. However, the trans-splicing reaction must also be highly specific to prevent undesirable “leaky” expression of the new gene activity in the absence of the target. This is particularly critical when the new gene activity is a toxin. ADP ribosylation of elongation factor 2 by DTA is diffusion limited (33), and very small quantities of DTA are sufficient to block translation when microinjected into mouse cells (34). We reasoned that cytotoxicity could be used as a sensitive assay for assessing target-independent effects of the ribozymes.
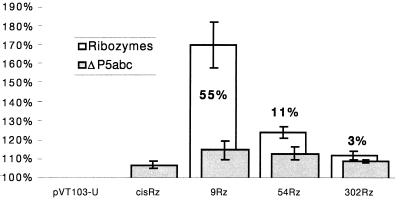
Haploid cultures harboring the cis- and trans-acting ribozyme-DTA fusions were tested for evidence of toxicity by measuring growth rates in liquid culture, which is more sensitive than the qualitative plating assay described above. These experiments were performed in the absence of CMV target sequences. Doubling time was measured relative to the ΔP5abc control ribozymes and a parental plasmid without an Rz-DTA insert (pVT103-U). Cultures harboring p9Rz-DTA, p54Rz-DTA, and p302Rz-DTA doubled 55%, 11%, and 3% slower than the respective ΔP5abc controls (the difference in growth rates between strains containing p302Rz-DTA and p302ΔP5-DTA is within the bounds of experimental error and may not be significant). These figures correspond to 70%, 24%, and 12% slower, respectively, than the pVT103-U control (Fig. 5). Cells expressing certain active ribozymes have a slower growth rate than those expressing the corresponding ΔP5abc derivative, and this indicates that toxicity may result from ribozyme-dependent, target-independent events, such as illegitimate splicing. This is most clearly seen with ribozymes containing the smallest extent of sequence complementarity with the target, suggesting that increasing the length of sequence fused to the 5′ end of the ribozyme increases specificity in vivo. A modest increase in doubling time (7–15%) was observed in cultures expressing the ΔP5abc derivatives relative to cultures harboring pVT103-U. Because these ribozymes are splicing-deficient (i.e., RT-PCR results for pcisΔP5-null; Fig. 4, lane 3), the reduced growth rates are most likely attributable to spurious expression of the DTA ORF. However, these nonspecific effects on growth are minor and have no obvious effect on rates of colony formation.
Figure 5.
Increasing the length of the antisense sequence reduces leaky expression of the toxin. The growth of haploid yeast cultures harboring indicated ribozyme was measured and normalized with respect to cultures containing the parental plasmid, pVT103-U (100%). The relative doubling times of each culture are represented by bars, white for active ribozymes and shaded for inactive ΔP5abc derivatives. The increased doubling times of cultures with active ribozymes relative to the corresponding ΔP5abc controls are shown as percentages on the diagram. Error bars correspond to the SEM from three experiments.
DISCUSSION
The first ribozymes to be designed with specificity for a particular mRNA substrate were based on small “hammerhead” ribozyme domains and possessed endoribonuclease activities (35). These work well in vitro and show clear potential as agents for inactivation of gene expression. However, they have not found routine use in vivo because of the difficulty of delivering these artificial RNAs within cells and efficiently cleaving and eliminating a continually replenished pool of target mRNAs. In contrast, trans-splicing ribozymes catalyze the formation of a novel hybrid mRNA. This has two major benefits. (i) The products of trans-splicing are more readily assayed. Even inefficient catalysis can give rise to a fused RNA product that is unique, easily detected and may give rise to an assayable gene product. This is a considerable advantage compared with the problems of assaying endoribonucleolytic ribozymes, with which in vivo catalysis merely results in target RNA depletion and generation of unstable and inactive products. (ii) Trans-splicing ribozymes may be useful for triggering gene expression solely in cells that contain a chosen mRNA species. Thus, trans-splicing ribozymes could be used to genetically mark or kill cells that are virus infected or of a particular differentiated or malignant type.
In this study, new trans-splicing ribozymes were used to deliver the ADP ribosyltransferase activity of DTA in yeast cells “infected” by CMV sequences. In a multicellular organism, ribozyme-triggered expression of DTA in virus-infected cells could confer resistance not by destroying the viral RNA but by blocking translation in the infected cells and thereby inhibiting virus replication and spread to healthy tissues. For this approach to be successful, it is crucial that the ribozymes produce sufficient quantities of toxin to inhibit cell growth and that cytotoxicity only results from specific trans-splicing with the intended target mRNA. In our yeast model system, cytotoxicity was easily measured by simple assays for cell growth, and we used this to systematically test modified ribozymes for improved selectivity and activity.
Ribozyme-mediated expression of DTA in diploid cells expressing a CMV-GFP target transcript was analyzed by a plating assay. Expression of a ribozyme without an antisense domain (p9Rz-DTA) did not overtly reduce colony formation in the presence of the CMV target mRNA. However, the addition of antisense sequences to the ribozyme caused increased activity. With 54 nt of complementary sequence, p54Rz-DTA caused reduced colony formation and slower growth, whereas a ribozyme with 302 nt of complementary sequence (p302Rz-DTA) prevented colony formation. The toxic effect of these ribozymes was highly specific. Mutation of the intended splice site within the target mRNA allowed normal colony growth. We also found that increased complementarity between the ribozyme and the target RNAs results in improved biological activity in E. coli (9), and it appears to provide a general benefit for trans-splicing in vivo. The antisense sequences are not expected to contribute directly to the ribozyme-catalyzed reaction and are likely to aid the association of ribozyme and target RNAs in vivo.
Specificity is an important aspect of ribozyme utility and is particularly pertinent when delivering a toxin. Cytoxicity provides a sensitive measure for aberrant expression of the toxin. This could result from illegitimate cis- or trans-splicing, direct translation of exon sequences, or some other indirect effect, and we have included a number of controls to test this in yeast. Early experiments in Drosophila showed that it was necessary to mutate an in-frame methionine to avoid nonspecific expression of DTA exon sequences (M14I; J.H., A. H. Brand, N. Perrimon, and H.M.G., unpublished results), probably because of translation of unspliced exon sequences, and the mutation has been incorporated in our constructions. We have also produced yeast strains containing ribozymes with defective catalytic domains (ΔP5abc) and inactive forms of the DTA exon and have expressed ribozymes alone or with mutant target mRNAs. The mating of control strains and assay of plating efficiency show no obvious indication of illegitimate toxin expression. However, the measurement of log phase growth rates in liquid culture provides a more sensitive indicator for minor differences between the ribozymes. Yeast cells harboring the DTA exon exhibited slightly slower growth rates in comparison to a control containing the parental plasmid (pVT103-U). These included cells expressing cis- and trans-splicing ribozymes with deleted catalytic domains (ΔP5abc) that are inactive under physiological conditions. The observed 7–15% slower doubling times are independent of the ribozyme and may be caused by noncanonical initiation or frame-shift translation of the DTA exon (there is no in-frame AUG). If so, this leaky expression might be eliminated by the incorporation of additional stop codons within the ribozyme, upstream of the 3′ exon.
Different extents of nonspecific toxicity were seen after expression of the active ribozymes in cells that did not contain the intended target mRNA. Active ribozymes with 9, 54, and 302 nt of target sequence complementarity gave rise to 55%, 11%, and 3% slower doubling times relative to the ΔP5abc controls, respectively (Fig. 5). Although it is possible that the extra sequences present within the active ribozyme domain may promote spurious translation of the DTA exon, this would need to be modulated by differences within the antisense domains, and it is more likely that leaky expression of DTA resulted from nonspecific catalysis, probably illegitimate splicing. The nonspecific activity is highest for p9Rz-DTA, which lacks the antisense domain. This is consistent with the report of illegitimate trans-spliced products in mammalian cells after the expression of a ribozyme with only six nucleotides of sequence complementary to the target (4). In contrast, increasing the length of the antisense region clearly reduces nonspecific ribozyme-dependent toxicity. For example, strains containing p302Rz-DTA or p302ΔP5-DTA showed almost identical growth rates, which were similar to the control pVT103-U. The addition of a region of extended complementarity confers a crucial improvement in the biological activity and specificity of these trans-splicing ribozymes.
This observation appears to contradict the prediction that an increased length of complementarity would promote illegitimate interactions, rather than enhance ribozyme specificity (36). However, a plausible explanation for our results is that fortuitous secondary or tertiary interactions between the antisense domain and the IGS might exclude nonspecific mRNAs from entering the active site, even those that might contain a suitable target site. In contrast, interaction of the intended target with any accessible portion of the antisense domain would initiate formation of an extended duplex, which would then unmask the IGS and allow formation of helix P1. If this explanation is correct, it should be feasible to engineer or to select for shorter antisense domains that act in a similar way to confer improved ribozyme specificity, and we are currently testing this. Similar “shielding” of complementary sequences has been noted in natural antisense RNAs (37).
In summary, we have demonstrated the prototype of a new class of cytotoxins based on trans-splicing ribozymes. These ribozymes can be targeted at a particular site within a chosen mRNA species and contain modified sequences for substrate recognition, a 3′ exon encoding a toxin and an antisense domain to confer affinity for the target mRNA. It was possible to ablate yeast cells containing CMV mRNA, with a high degree of biological efficiency and specificity, by using a ribozyme with 302 nt of sequence complementarity (p302Rz-DTA). Mutation of a single residue at the intended slice site in CMV resulted in “immunity.” We are now testing the ability of trans-splicing ribozymes to confer CMV resistance to higher plants. Trans-splicing ribozymes may be useful for triggering gene expression in other virus-infected, differentiated, or malignant cell types that contain unique mRNA species. If so, they could form the basis of new therapeutic approaches.
The array of genetic tools and ease of manipulation make S. cerevisiae an attractive model for developing RNA-based therapeutics, but the organism has proved recalcitrant to the application of cleaving ribozymes (38). We have found that targeted gene delivery provides a simple means of assaying the efficiency and fidelity of trans-splicing ribozymes in yeast cells. We anticipate that there are further improvements to be made in the design, transcription, cellular localization and association of trans-splicing ribozymes, and we have recently adapted the ribozymes to deliver other gene activities in yeast, including a transcriptional activator, and selectable metabolic gene (B.G.A., U.K., and J.H., unpublished results). Powerful techniques of genetic selection can now be applied directly to the improvement of ribozymes and their integration into eukaryotic RNA metabolism.
Acknowledgments
We thank Roisin McGarry and Andrew Newman for comments concerning the manuscript and Andrea Brand, Andrew Ellington, and Rob Arkowitz for helpful discussions and materials. We thank Ed Ullman and Sheba Mahmoodian for technical assistance. The work was supported by grants from AgrEvo (J.H.) and Hoechst AG (H.M.G.).
ABBREVIATIONS
- CMV
cucumber mosaic virus
- CP
coat protein
- DTA
diphtheria toxin A chain
- IGS
internal guide sequence
- GFP
green fluorescent protein
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Cech T R. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 2.Galloway-Salvo J L, Coetzee T, Belfort M. J Mol Biol. 1990;211:537–549. doi: 10.1016/0022-2836(90)90264-m. [DOI] [PubMed] [Google Scholar]
- 3.Sullenger B A, Cech T R. Nature (London) 1994;371:619–622. doi: 10.1038/371619a0. [DOI] [PubMed] [Google Scholar]
- 4.Jones J T, Lee S W, Sullenger B A. Nat Med. 1996;2:643–648. doi: 10.1038/nm0696-643. [DOI] [PubMed] [Google Scholar]
- 5.Sarver N, Cairns S. Nat Med. 1996;2:641–642. doi: 10.1038/nm0696-641. [DOI] [PubMed] [Google Scholar]
- 6.Phylactou L A, Darrah C, Wood M J A. Nat Genet. 1998;18:378–381. doi: 10.1038/ng0498-378. [DOI] [PubMed] [Google Scholar]
- 7.Lan N, Howrey R P, Lee S W, Smith C A, Sullenger B A. Science. 1998;280:1593–1596. doi: 10.1126/science.280.5369.1593. [DOI] [PubMed] [Google Scholar]
- 8.Jones J T, Sullenger B A. Nat Biotechnol. 1997;15:902–905. doi: 10.1038/nbt0997-902. [DOI] [PubMed] [Google Scholar]
- 9.Köhler U, Ayre B G, Goodman H M, Haseloff J. J Mol Biol. 1999;284:1935–1950. doi: 10.1006/jmbi.1998.2447. [DOI] [PubMed] [Google Scholar]
- 10.Palukaitis P, Roossinck M J, Dietzgen R G, Francki R I B. Adv Virus Res. 1992;41:281–348. doi: 10.1016/s0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- 11.Pappenheimer A M., Jr Annu Rev Biochem. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Vernet T, Dignard D, Thomas D Y. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 14.Green R, Ellington A D, Szostak J W. Nature (London) 1990;347:406–408. doi: 10.1038/347406a0. [DOI] [PubMed] [Google Scholar]
- 15.Joyce G F, van der Horst G, Inoue T. Nucleic Acids Res. 1989;17:7879–7889. doi: 10.1093/nar/17.19.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmiter R D, Behringer R R, Quaife C J, Maxwell F, Maxwell I H, Brinster R L. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 19.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology: Methods in Enzymology. Vol. 194. New York: Academic; 1991. [PubMed] [Google Scholar]
- 20.Winston F, Dollard C, Ricupero-Hovasse S L. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 21.Doudna J A, Cormack B P, Szostak J W. Proc Natl Acad Sci USA. 1989;86:7402–7406. doi: 10.1073/pnas.86.19.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barford E T, Cech T R. Mol Cell Biol. 1989;9:3657–3666. doi: 10.1128/mcb.9.9.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwinghammer M W, Symons R H. Virology. 1977;97:88–108. doi: 10.1016/0042-6822(77)90337-3. [DOI] [PubMed] [Google Scholar]
- 24.Bertrand E, Pictet R, Grange T. Nucleic Acids Res. 1994;22:293–300. doi: 10.1093/nar/22.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies R W, Waring R B, Ray J, Brown T A, Scazzochio C. Nature (London) 1982;300:719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- 26.Price J V, Cech T R. Genes Dev. 1988;2:1439–1447. doi: 10.1101/gad.2.11.1439. [DOI] [PubMed] [Google Scholar]
- 27.Suh E R, Waring R B. Mol Cell Biol. 1990;10:2960–2965. doi: 10.1128/mcb.10.6.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel F, Westhof E. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 29.Siemering K R, Golbik R, Sever R, Haseloff J. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 30.Wilson B A, Blanke S R, Reich K A, Collier R J. J Biol Chem. 1994;269:23296–23301. [PubMed] [Google Scholar]
- 31.Giannini G, Rappouli R, Ratti G. Nucleic Acids Res. 1984;12:4063–4069. doi: 10.1093/nar/12.10.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bellen H J, D’Evelyn D, Harvey M, Elledge S J. Development (Cambridge, UK) 1992;114:787–796. doi: 10.1242/dev.114.3.787. [DOI] [PubMed] [Google Scholar]
- 33.Blanke S R, Huang K, Wilson B A, Papini E, Covacci A, Collier R J. Biochemistry. 1994;33:5155–5161. doi: 10.1021/bi00183a019. [DOI] [PubMed] [Google Scholar]
- 34.Yamaizumi M, Mekada E, Uchida T, Okada Y. Cell. 1978;15:245–250. doi: 10.1016/0092-8674(78)90099-5. [DOI] [PubMed] [Google Scholar]
- 35.Haseloff J, Gerlach W L. Nature (London) 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 36.Herschlag D. Proc Natl Acad Sci USA. 1991;88:6921–6925. doi: 10.1073/pnas.88.16.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simons R W, Kleckner N. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- 38.Arndt G M, Atkins D. In: Nucleic Acids and Molecular Biology. Eckstein F, Lilley D M J, editors. Vol. 10. Berlin: Springer; 1996. [Google Scholar]