Abstract
A receptor that mediates osteoprotegerin ligand (OPGL)-induced osteoclast differentiation and activation has been identified via genomic analysis of a primary osteoclast precursor cell cDNA library and is identical to the tumor necrosis factor receptor (TNFR) family member RANK. The RANK mRNA was highly expressed by isolated bone marrow-derived osteoclast progenitors and by mature osteoclasts in vivo. Recombinant OPGL binds specifically to RANK expressed by transfected cell lines and purified osteoclast progenitors. Transgenic mice expressing a soluble RANK-Fc fusion protein have severe osteopetrosis because of a reduction in osteoclasts, similar to OPG transgenic mice. Recombinant RANK-Fc binds with high affinity to OPGL in vitro and blocks osteoclast differentiation and activation in vitro and in vivo. Furthermore, polyclonal Ab against the RANK extracellular domain promotes osteoclastogenesis in bone marrow cultures suggesting that RANK activation mediates the effects of OPGL on the osteoclast pathway. These data indicate that OPGL-induced osteoclastogenesis is directly mediated through RANK on osteoclast precursor cells.
Bone morphogenesis, remodeling, and resorption are controlled in part by osteoclasts. These cells arise from hematopoietic precursors by physiologically controlled processes that involve growth factors, cytokines, peptide, and steroid hormone interactions with their receptors (1, 2). Osteoclast differentiation and activation is now known to be positively and negatively controlled by members of the tumor necrosis factor (TNF) and TNF receptor (TNFR) superfamilies of proteins. Osteoprotegerin (OPG), a secreted TNFR-related protein, negatively regulates osteoclast differentiation in vitro and in vivo (3, 4), is required for the maintenance of bone density (5, 6), and blocks pathophysiological induction of bone resorption in rodent disease models (7). OPG exerts its effects by binding to, and sequestering, OPG ligand (OPGL), a potent inducer of osteoclast differentiation and activation (8–10). OPGL binds to hematopoietic precursors that correspond to osteoclast progenitor cells and induces changes in patterns of preosteoclast gene expression that manifests osteoclast differentiation and culminates in the production of mature, bone resorbing osteoclasts. In mice, the formation of mature osteoclasts is absolutely dependent on OPGL (11), indicating that it, in addition to colony-stimulating factor 1 (CSF-1)/macrophage colony-stimulating factor (12), is a critical differentiation factor that specifies the osteoclast maturation program, and hence induction of bone resorption.
The precise mechanism of OPGL activity is still unclear but is presumably caused by binding a cell surface receptor(s) that initiates a signal transduction cascade. In appropriate precursors, this cascade culminates in osteoclast differentiation and/or activation (8, 9). OPGL has also been described as the ligand for the TNFR-related protein receptor activator of NFκB (RANK) (13). RANK(TNFRSF11B) was identified as a dendritic cell protein implicated in immune responses (13). Its role in OPGL-mediated osteoclastogenesis remains to be determined.
We took the genomic approach to examine genes expressed in murine osteoclast precursors. In this report, we describe the identification and characterization of the osteoclast differentiation and activation receptor that is present on normal mouse osteoclast progenitors and which mediates OPGL-induced osteoclast differentiation and activation. The identified receptor is indeed identical to the previously reported TNFR family member RANK. Like several known TNFR family members, the signaling pathway of RANK involves the interaction with cytoplasmic TNFR-associated factor (TRAF) proteins. Cumulatively, our findings reveal that OPGL–RANK–OPG comprise key regulatory proteins that govern osteoclast development, and implicate TRAF family members and/or Jun N-terminal kinase (JNK) as potential osteoclastogenic signal transducers.
EXPERIMENTAL PROCEDURES
Recombinant Protein and Ab Generation.
The production of recombinant murine OPGL(158–316) and derivation of a FITC conjugate (FITC-OPGL) has been previously described (8). The PCR product encoding the entire RANK extracellular domain was spliced in-frame to the human IgG-γ1 heavy chain Fc region sequence, and the RANK-Fc fusion protein product was expressed in human 293 Epstein–Barr virus nuclear antigen fibroblasts as described (4). Purified RANK-Fc fusion protein was used as antigen to raise polyclonal anti-RANK antiserum in rabbits (Babco, Berkeley, CA). A PCR fragment encoding RANK extracellular domain (amino acid 31–211), preceded with an artificial methionine, was subcloned for expression in bacteria. The Escherichia coli-produced RANK(31–211) product was purified from the soluble fraction by using anion and cation-exchange chromatography, then conjugated to CNBr-activated Sepharose according to manufacturer’s suggestion (Pharmacia Biotech). The RANK(31–211)-conjugated Sepharose column was then used to affinity purify anti-RANK-specific Abs from rabbit immune serum.
Generation and Pathological Analysis of RANK Transgenic Mice.
A DNA fragment coding for the RANK-Fc fusion protein was excised from the RANK-Fc/pCEP4 vector and cloned into an expression vector under the control of the human ApoE promoter and liver-specific enhancer element (4). RANK transgenic mice were generated, and analyzed as described (4).
Analysis of Recombinant RANK-Fc Protein.
Young rapidly growing male BDF1 mice aged 3–4 weeks received varying doses of RANK-Fc fusion protein by single, daily subcutaneous injections in carrier [PBS containing 0.1% BSA (wt/vol)] for 4 days. Administration of carrier alone served as the control. After sacrifice, the right tibia of treated mice were removed and fixed in 70% ethanol, and the bone density in the proximal tibial metaphysis was analyzed by radiography or peripheral quantitative computerized tomography (PQCT) (Stratec, Pforzheim, Germany). Total bone mineral density was defined as the average of the bone mineral content (mg/bone area cm3) for two cross-sectional images of the tibia at 1.5 mm and 2.0 mm distal to the proximal articular surface. These data were analyzed by ANOVA JMP. In vitro osteoclast-forming assay was performed as described (4, 8).
Transfection, Immunoprecipitation and Cross-Linking.
NF-κB reporter assay and coimmunoprecipitation assay were performed as described (21). For the JNK kinase assay, HA-JNK or endogenous JNK was first immunoprecipitated with anti-HA (Babco) or anti-JNK mAb (PharMingen). The kinase activity was then determined by using 2 μg of GST-JUN as substrate according to the manufacturer’s recommendations (Stratagene). For cross-linking experiment, ≈4 × 106 cells obtained from the FITC-OPGL sorting were incubated with 10 nM 125I-labeled OPGL on ice for 1 hr. Cells were then washed with 10 ml PBS twice and resuspended in 500 μl PBS supplemented with 1 mM disuccinimidyl tartrate. After a 30-min incubation in ice, cross-linking reactions were stopped by addition of Tris⋅HCl to a final concentration of 20 mM. After washing with PBS, cells were lysed with 500 μl RIPA buffer, and subsequent immunoprecipitation was performed as described (21).
RESULTS
RANK Mediates OPGL-Induced Osteoclastogenesis.
We have previously shown that OPGL binds to the surface of the osteoclast precursor population from mouse bone marrow, and that the positively sorted cells readily differentiated into osteoclasts (8). To search for the OPGL receptor on osteoclast precursor cells, the nonadherent fraction of mouse bone marrow cells cultured in the presence of CSF-1 and OPGL for 24 hr were stained and sorted with FITC-OPGL. About 10% of the nonadherent population from these conditions were labeled with FITC-OPGL. About 1 × 108 FITC-OPGL binding cells were used to construct a cDNA library for random expressed sequence tag (EST) analysis (4). Among the first 400 EST sequences obtained, one appeared to encode the amino terminus of RANK, a TNFR-related protein involved in activation of NF-κB (13). The RANK DNA fragment was used as probe in the Northern blot analysis of RNA from the positive and negative FITC-OPGL cells. RANK mRNA was highly expressed in the FITC-OPGL binding cells, but not the negative binding population (Fig. 1A). This expression pattern suggested that RANK might represent a candidate osteoclast differentiation and activation receptor.
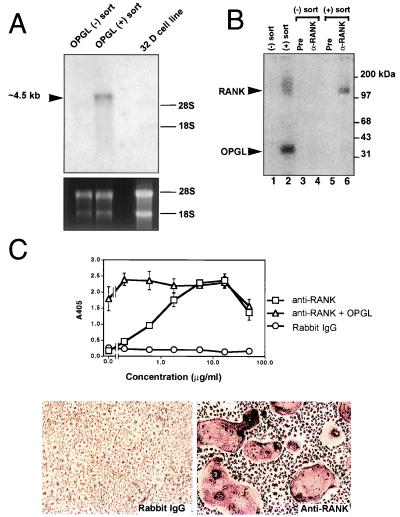
Figure 1.
RANK mediates OPGL-induced osteoclastogenesis. (A) Expression of RANK in osteoclast precursor cells. A RANK-specific probe was used in the Northern blot analysis of 4 μg of total RNA prepared from the negative OPGL-binding population, positive OPGL-binding population, or 32-D cells. (B) Cross-linking of OPGL–RANK complexes on the surface of osteoclast precursors. OPGL positive and negative binding cells were prepared by FACS, incubated with 125I-labeled OPGL, and then subjected to cross-linking with 1 mM disuccinimidyl tartrate. One-tenth of the cell lysates were directly fractionated by SDS/PAGE (lanes 1 and 2). The remainder of the cell lysates were immunoprecipitated with either pre-immune serum (lanes 3 and 5) or anti-RANK Ab (lanes 4 and 6). (C) Anti-RANK Abs stimulate osteoclastogenesis. Mouse bone marrow cells were cultured with CSF-1 (30 ng/ml) with and without 5 ng/ml murine OPGL(158–316) in the presence of various concentrations of either anti-RANK Ab or rabbit IgG. Osteoclastogenesis was quantified by TRAP solution assay. A405 represents absorbance at 405 nm and is presented as the mean ± SD (n = 3). TRAP cytochemical staining was shown at the bottom. (Bar = 200 μm.)
To determine whether OPGL binds RANK present on osteoclast progenitor cells, cross-linking studies were performed with 125I-labeled OPGL and FITC-OPGL positive and negative cells. Analysis of the cell lysates by SDS/PAGE indicates that OPGL binds to a protein from the FITC-OPGL(+) cell pool and migrates in a complex with a molecular weight of ≈150 kDa (Fig. 1B). This protein was not detected in the FITC-OPGL(−) pool, nor were any other labeled proteins identified. Immunoprecipitates of the 125I-labeled OPGL cross-linked cells using anti-RANK Abs showed the same labeled complex from the FITC-OPGL(+) sorted cells. These data indicate that OPGL recognizes RANK expressed on osteoclast precursors. Thus, RANK is the likely candidate for mediating OPGL signaling in osteoclast progenitors.
Definitive proof that RANK is an essential mediator of OPGL in the induction of osteoclastogenesis came from experiments where the effects of Abs specific for the RANK extracellular domain were examined on the in vitro osteoclast formation assay. Various concentrations of affinity-purified polyclonal anti-RANK Abs were added to mouse bone marrow cultures containing CSF-1 with and without OPGL (5 ng/ml), and following 5 days the cultures were evaluated for tartrate-resistant acid phosphatase (TRAP) expression by using both the TRAP solution assay and cytochemistry. The anti-RANK Abs alone stimulated osteoclast differentiation in a dose-dependent manner with an EC50 of ≈1 μg/ml (Fig. 1C). No induction was observed with the control rabbit IgG. The combination of both OPGL and anti-RANK did not result in an increased level of osteoclastogenesis, suggesting that both proteins acted as agonists that were maximally stimulating the same pathway. The agonist activity of RANK Ab provides direct evidence that OPGL exerts its activity on osteoclast precursor cells via RANK.
RANK Expression by Osteoclasts.
In situ hybridization of embryonic and adult mouse bone was performed by using a 33P-labeled RANK riboprobe. In the long bones of the developing embryo, RANK-expressing cells were located in the diaphysis at the interface of the embryonic marrow cavity and developing growth plate (Fig. 2 A and B). The grain density is highest over multinucleated cells, with a morphology and location (on bone and cartilage in the primary spongiosa) consistent with osteoclasts (Fig. 2 C–E). In the adult, RANK-expressing cells with an osteoclast morphology (see arrows in Fig. 2 H and I) were located along both the metaphyseal side of the growth plate in the primary spongiosa (Fig. 2H) and the periosteal surface of the metaphysis (Fig. 2I). These are areas of intense resorptive activity by osteoclasts and the region where the in situ signal for OPGL was most obvious (8). In addition, it appears that a somewhat lower level of RANK message is expressed by hypertrophic chondrocytes (see arrowheads, Fig. 2H). In situ hybridization by using riboprobes for cathepsin K, TRAP, and β3 demonstrate a similar expression pattern, except for β3, which is also detectable in megakaryocytes (data not shown). These data indicate that RANK expression in bone is highest in osteoclasts within regions of active bone resorption, with some expression by maturing chondrocytes. The latter finding suggests a potential role of RANK and OPGL in growth plate physiology in addition to their role in bone modeling and remodeling.
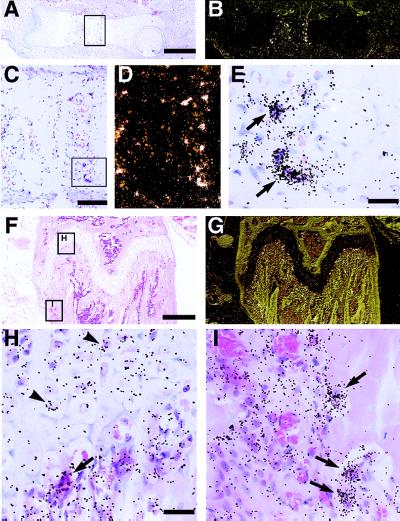
Figure 2.
Localization of RANK mRNA expression in developing and adult bone by in situ hybridization. Sections of embryonic and adult long bones were hybridized with 33P-labeled antisense riboprobes to RANK, processed and stained with hematoxylin/eosin. Panels A–E are from fetal (E15.5) femur, with B and D being darkfield images of A and C, respectively. Panels F–I are from adult (6-week) femur, with G being a darkfield image of F. RANK is expressed by multinucleated cells attached to matrix, consistent with osteoclasts in areas of active bone modeling in the fetus and adult. Hypertrophic chondrocytes in the adult growth plate also appear to express RANK transcripts. Arrows mark the position of osteoclasts, whereas arrowheads mark the position of hypertrophic chondrocytes. [Bars = 400 μm (A and F), 100 μm (C), and 25 μm (E and H).]
Soluble RANK Transgenic Mice Present an Osteopetrotic Phenotype.
To gain further insights into the biological function for RANK, transgenic mice were generated that expressed entire extracellular domain of RANK fused with human IgG-γ1 driven by the apolipoprotein E gene promoter (hepatic expression), a model system that initially revealed the skeletal activity of OPG (4). Founder mice harboring the RANK-Fc transgene were identified by PCR analysis of genomic DNA samples (data not shown). The relative level of RANK-Fc fusion protein expression in liver extracts from mice 10 weeks of age was determined by Western blot analysis with anti-RANK Ab. The high and low RANK-Fc expressors along with their normal littermates were subjected to systemic pathology analysis.
The RANK-Fc-expressing transgenic mice were of normal size and activity with erupted incisors. As with the OPG transgenics, the RANK–Fc expressing animals had increased radiodensity of all bones including the long bones, pelvic girdle, and vertebrae compared with control littermates (Fig. 3A, Left). The bones of the expressors were also of normal size and shape. Histologically, the bone marrow of the expressors had increased amounts of nonresorbed primary spongiosa, consisting of both bone and cartilage (Fig. 3A, Middle). The amount of space for bone marrow was markedly reduced in these animals leading to a compensatory splenomegaly because of extramedullary hematopoiesis. Histochemical stains for the osteoclast enzyme TRAP revealed a decreased number of osteoclasts in the transgenic mice. In the high expressors, osteoclasts were also reduced in size and detached from the trabecular bone surface (Fig. 3A, Right). No changes were observed in other organs or organ systems, including lymph node, thymus, and intestine. In sum, the osteopetrotic phenotype of the RANK-Fc transgenic mice highly resembled those of the OPG transgenic mice (4).
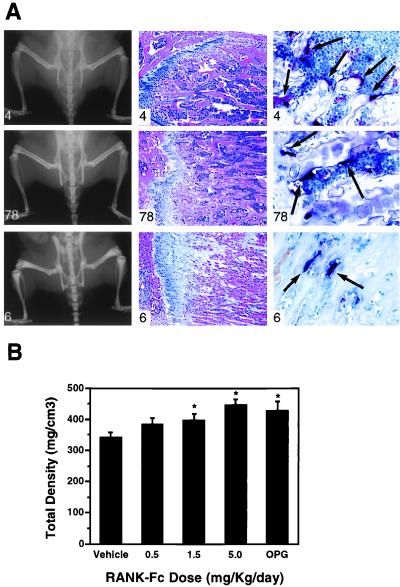
Figure 3.
Soluble RANK-Fc increases bone density. (A) Increased bone density in RANK-Fc transgenic mice. A high RANK-Fc expressing mouse (animal 6), medium expressing mouse (animal 78), and control littermate (animal 4) were subjected to radiographic analysis (Left) and histologic analysis (Middle and Right). Increased radiodensity of the long bones, pelvic bones, and the vertebrae were detected in both the high and low expressors (Left) with no changes in bone shape. The femoral diaphyses of the RANK-Fc transgenic mice contained nonresorbed bone and cartilage with the highest expressor (animal 6) having the greatest accumulation (Middle). TRAP histochemical staining of the bones showed decreased osteoclast numbers in the RANK-Fc expressors (Right). (B) PQCT analysis of mice treated with RANK-Fc recombinant protein. Male BDF1 mice aged 3 weeks received the indicated doses of RANK-Fc fusion protein by single daily subcutaneous injection for 4 days. The bone density in the proximal tibial metaphysis was measured by peripheral quantitative computerized tomography. Total density was presented as the mean ± SD (n = 4). Treatment with RANK-Fc induced a dose-dependent increase of total bone density (P < 0.001).
The osteopetrotic transgenic phenotype was confirmed by direct administration of soluble recombinant RANK-Fc fusion protein. Young, rapidly growing male BDF1 mice were treated by subcutaneous injections with doses of soluble recombinant RANK-Fc fusion protein ranging from 0.5 to 5 mg/kg for 4 days. Radiographs of the proximal tibia demonstrated an obvious accumulation of radiodense material beneath the growth plate (data not shown). The density of the proximal tibial metaphysis was measured by PQCT. The results indicate that RANK-Fc treatments resulted in a dose-dependent increase in total bone density as compared with the control group (Fig. 3B) with significant effects seen at 1.5 and 5 mg/kg. The effects of these two highest doses were comparable to the effects of OPG (5 mg/kg), which served as a positive control. These data clearly support a major role for RANK in mediating osteoclast differentiation and activation signaling.
RANK Signals Through TRAF Proteins.
OPGL is presumed to act as a potent cytokine that induces osteoclast differentiation and activation by inducing receptor-mediated intracellular signal transduction. Darnay et al. (14) and Wong et al. (22) have recently reported that, like some other TNFR family members, the intracellular domain of RANK is able to bind directly with TRAF2, TRAF5, and TRAF6. To identify authentic signaling molecules that RANK utilizes to initiate osteoclastogenesis, a yeast two-hybrid library was generated from primary mouse osteoclast precursor cells purified by FITC-OPGL sorting. The whole intracellular domain of RANK was used as bait in the yeast two-hybrid screen. From ≈20 million transformants, 200 positive clones were recovered as determined by activation of the his reporter gene. Of these clones, the majority encoded TRAF2, and the rest of the positives encoded TRAF5 or TRAF6. Apart from the TRAF proteins, we failed to identify other proteins that interacted specifically with RANK in this two-hybrid screen.
To localize the TRAF interaction region within RANK, a series of deletion mutants within the RANK intracellular domain were fused to the GAL4 DNA-binding domain and tested for their interaction with TRAF proteins in the yeast two-hybrid binding assays. Our deletion analysis demonstrated that the TRAF6-binding domain of RANK is located in a region that is physically separated from the TRAF2/TRAF5-binding domain (Fig. 4A). The TRAF6-binding domain of RANK resides between amino acid residues 336 and 454, whereas the TRAF2/TRAF5-binding domain locate at the C terminus of RANK (Fig. 4A).
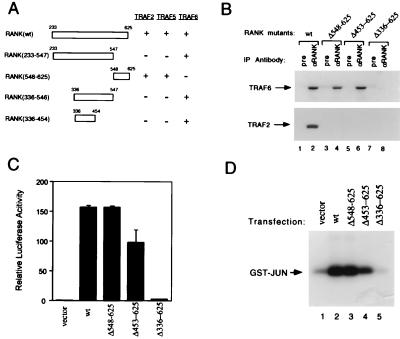
Figure 4.
RANK interactions with TRAF proteins and activation of NF-κB and JNK pathways. (A) Mapping of TRAF-binding domains of RANK. Expression vectors encoding full-length or deletion mutants of RANK intracellular domain fused to the GAL4 DNA-binding domain were cotransformed into HF7C yeast strain with vectors expressing the GAL4 activation domain fused with TRAF2, TRAF5, or TRAF6 as described (21). +, Growth after 1 week on the selection plates. (B) Coimmunoprecipitation of RANK and TRAF proteins. After 24 hr of transfection, 293 cell lysates were immunoprecipitated with pre-immune serum or Ab against RANK extracellular domain. Coprecipitating TRAF2 or TRAF6 were detected by immunoblot analysis with goat anti-TRAF2 or anti-TRAF6 Ab (Santa Cruz Biotechnology). (C) Activation of NF-κB in RANK transfected 293 cells. Luciferase activities were measured after 24 hr and normalized on the basis of β-galactosidase expression levels. The values (means ± SD) represent luciferase activities relative to vector transfection for representative experiments performed in duplicate. (D) Activation of JNK in RANK-transfected 293 cells. After 24-hr transfection, 293 cell lysates were immunoprecipitated with monoclonal anti-HA epitope Ab. Immunoprecipitates were assayed for kinase activity by using 2 μg GST-JUN (Stratagene) as substrate, and resolved by SDS/PAGE.
The yeast two-hybrid mapping of RANK–TRAF interactions was confirmed by using coimmunoprecipitation assays in a mammalian cell system. Expression plasmids that direct the synthesis of full-length or C-terminal deletion mutants of RANK were cotransfected with TRAF2 or TRAF6 expression vectors into human embryonic kidney 293 cells. Both TRAF2 and TRAF6 were coimmunoprecipitated with wild-type RANK by anti-RANK polyclonal Ab but not by pre-immune serum. Anti-RANK immunoprecipitates of C-terminal RANK deletion mutants (Δ548–625 and Δ453–625) contained TRAF6 but not TRAF2 (Fig. 4B). Further C-terminal deletion (Δ336–625) abolished RANK binding with TRAF6 (Fig. 4B). Thus, the associations between RANK and TRAF proteins in mammalian cells directly correlate with our yeast two-hybrid interaction mapping results described above.
Like other TRAF-interacting TNFR family members, RANK may also mediate NF-κB and JNK activation. To test this possibility, we examined whether RANK overexpression in 293 cells could induce these two pathways. An NF-κB-dependent luciferase reporter was transfected into 293 cells. Cotransfection with the RANK expression vector potently induced luciferase activity (Fig. 4C). The RANK C-terminal deletion mutants were also examined for their NF-κB activation activities. The TRAF6-binding mutants (Δ548–625 and Δ453–625) were active, whereas the noninteracting mutant (Δ336–625) was inactive (Fig. 4C), suggesting a correlation between TRAF6-interaction and NF-κB activation. To examine JNK activation, a HA-tagged JNK expression plasmid was cotransfected with vector or RANK expression plasmid into 293 cells. The kinase activity of JNK from cell lysates was determined by immunoprecipitation with mAb against the HA epitope followed by an in vitro kinase assay. RANK overexpression strongly activated JNK kinase activity (Fig. 4D). Similarly, the JNK activation of RANK C-terminal deletion mutants also correlated with their TRAF6-binding activities (Fig. 4D). Considering the available genetic evidence indicating a role for both NF-κB and Fos/AP-1 (via JNK) during osteoclast development in mouse, these data implicate a role for TRAF6 binding to the cytoplasmic region RANK (residues 336–454) during the induction of osteoclastogenesis. The importance of the TRAF2 and TRAF5 signaling pathways during osteoclastogenesis are unknown.
Induction of Osteoclast-Specific Gene Expression in RANK-Expressing Cell Line.
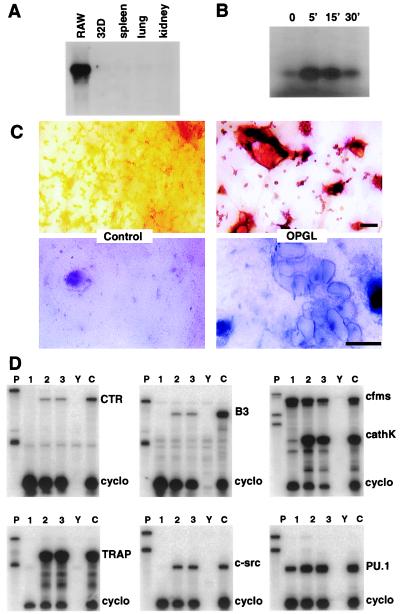
The in vitro study of osteoclastogenesis has historically relied on the use of nontransformed hematopoietic progenitors, usually from the spleen, bone marrow, or peripheral blood. Because RANK expression identifies osteoclast progenitors in bone marrow (Fig. 1) (8), we screened a number of transformed myeloid cell lines for RANK mRNA expression to identify cell lines with osteoclastogenic potential. As shown in Fig. 5A, the RAW 264.7 cell line was found to express high levels of RANK mRNA. Treatment of RAW cells with murine OPGL readily stimulated cell differentiation into osteoclast-like TRAP positive cells (Fig. 5C, Upper Right). Functionally, the osteoclasts readily formed resorption lacunae on bone slices, a critical test for evaluating the osteoclast phenotype of large multinucleated, TRAP positive cells (Fig. 5C, Lower Right). RAW cells that were differentiated into osteoclasts in the presence of OPGL express genes typical of the osteoclast lineage including TRAP, cathepsin K, β3, c-src, and calcitonin receptor (Fig. 5D). To determine whether RANK mediates osteoclastogenesis in RAW cells as it does in primary osteoclast precursors, cultures were also treated with anti-RANK agonistic Abs. Anti-RANK treatment of RAW cell cultures results in the induction of the same pattern of osteoclast-related genes seen following OPGL treatment (5C, lane 2 vs. lane 3). These data further support the role of RANK signaling during osteoclastogenesis, and they reveal the osteoclastogenic potential of this murine myelomonocytic cell line.
Figure 5.
OPGL and anti-RANK agonistic Abs stimulate osteoclast differentiation of the murine myeloid RAW 264.7 cell line. (A) Northern blots of RNA from various sources hybridized with an RANK probe. (B) Approximately 106 RAW cells were exposed to 100 ng/ml OPGL for the indicated length of time. The cell lysates were immunoprecipitated with monoclonal anti-JNK Ab. Immunoprecipitates were assayed for kinase activity by using GST-JUN as substrate and resolved by SDS/PAGE. (C) RAW cells were plated on bone slices and treated for 7 days with OPGL. The Upper and Lower panels show the results of TRAP cytochemistry and toluidine blue staining, respectively. Note that OPGL induces the formation of TRAP positive cells (Upper) that form resorption lacunae (Lower). (Bars = 50 μm.) (D) Total cell RNA was extracted from RAW cells treated with media alone (lane 1) or media supplemented with either 100 ng/ml OPGL (lane 2) or 1,750 ng/ml anti-RANK antibodies (lane 3). RNA from OPGL plus CSF-1-treated bone marrow cultures (lane C) served as the positive control. RNase protection assays were performed by using probes for the calcitonin receptor (CTR), β3 integrin (B3), c-fms, cathepsin k (cathK), TRAP, c-src, PU.1, and cyclophilin (cyclo). Lanes P and Y represent the undigested probe and yeast hybridization controls, respectively.
We also examined whether OPGL induced JNK and/or NF-κB activation in RAW cells. RAW cells were induced with OPGL for various lengths of time. Endogenous JNK was immunoprecipitated with an JNK-specific mAb, and its kinase activity was determined by immune complex kinase assay. Activation of JNK was readily detectable after 5 min of OPGL exposure, and rapidly decreased after 30-min treatment (Fig. 5B). NF-κB activation was not detectable in OPGL-treated RAW cells, as determined by gel shift analysis (data not shown). These data, taken together, strongly suggested Jun kinase as potential important osteoclastogenic signal transducers.
DISCUSSION
The approach used to identify an osteoclast receptor for OPGL described here took into consideration what was known about the biology of OPG (3, 4) and OPGL (8, 9) and focused on the discovery of proteins expressed by osteoclast precursors that were implicated in regulating osteoclastogenesis. We purified a population of primary mouse hematopoietic precursors based on their ability to bind a fluorescent version of OPGL, and prepared cDNA libraries for random expressed sequence tag sequencing and bio-informatic analysis. One of the proteins readily identified by this process was a TNFR-related protein and was identical to the previously reported dendritic cell protein RANK (13).
RANK mRNA was found to be exclusively expressed in osteoclast progenitors within the bone marrow cells sorted by OPGL binding, consistent with the idea that it is a candidate OGPL receptor. In vitro binding assays using soluble murine OPGL(158–316) and soluble RANK-Fc indicate that they interact with a Kd of ≈3 × 10−9 M (M.J.K. and D. Powers, unpublished data). In addition, radiolabeled OPGL binds only to OPGL-sorted osteoclast precursors in bone marrow, and analysis of the protein it binds reveals that it is the RANK polypeptide. Furthermore, soluble RANK blocks osteoclast differentiation and activation in vitro (data not shown) and in vivo, indicating that a major physiological pathway antagonized by soluble RANK is mediated by the osteoclast. Finally, Ab specific for the RANK extracellular domain has agonistic properties, and they induce osteoclast differentiation and activation of osteoclast-specific gene expression. These data indicate that ligation of RANK on osteoclast precursors is sufficient for induction of osteoclastogenesis and for initiation of bone resorption.
Darnay et al. (14) reported the mapping of RANK and TRAF protein interaction to the extreme cytoplasmic tail of RANK. Recently, Wong et al. (22) reported the presence of additional TRAF6-binding sites within RANK intracellular domain. We found that the RANK intracellular domain contains two distinct TRAF binding domains, each of which recognize different TRAF proteins specifically. The C-terminal region of RANK, between amino acid residues 547 and 581, interacts with TRAF2 and TRAF5 but not with TRAF6. The TRAF6-binding domain resides in the middle of the RANK intracellular region, between amino acid residues 336 and 453. In addition, the NF-κB and JNK activation induced by overexpression of RANK C-terminal deletion mutants in 293 cells correlated with TRAF6-interacting activities. A role for the involvement of TRAF proteins in osteoclast signaling is currently under study, and recent results indicate that the TRAF6 knockout mice exhibit an osteopetrotic phenotype because of a defect in bone resorption (M. Lomaga and T. Mak, personal communication). The TRAF2 and TRAF3 knockout mice have been analyzed and no bone phenotypes detected (15, 16), although specific examination of the skeleton was not performed. The role of TRAF5 in osteoclastogenesis remains undefined. These findings suggest that TRAF6 may play an important role in osteoclastogenesis mediated through RANK. Both c-Fos knockout mice and p50/p52 double knockout mice present with impaired osteoclast maturation and a severe osteopetrosis (17–19). Like other TRAF-binding TNFR family members, overexpression of ODAR in 293 cells induced both NF-κB activation and JNK activation. However, OPGL readily activated JNK but not NF-κB in RAW cells, a murine myelomonocytic cell line that differentiates to osteoclasts on OPGL treatment. Although the role of NF-κB in OPGL-induced osteoclastogenesis remains to be determined, our data strongly support JNK as an important early signaling molecule along ODAR signal transduction pathway.
It is now clear that in the mouse, the development and activation of osteoclasts are governed by OPGL—RANK–OPG. Based on both the loss and gain of OPG function previously shown in mouse (4, 5), we conclude that the level of OPG expression in mouse directly correlates to the level of bone mass. Recently, it has been shown that expression of the OPG gene is regulated by estrogen, a critical hormone that controls diverse physiological processes including the maintenance of bone mass (20). In addition, OPGL gene expression has been shown to be induced by parathyroid hormone and vitamin D3, hormones that are known to induce osteoclast function and increase bone resorption (9). Thus, regulation of both OPG and OPGL levels are directly implicated as the end effectors of circulating hormones and cytokines involved in the physiological and pathological regulation of bone density via the osteoclast. At the level of the osteoclast, the diverse humoral signaling regulating bone density ultimately regulates the activation of RANK on the osteoclast/osteoclast precursor cell surface, and the precise signal transduction pathways that emanate from this receptor will define the key intracellular processes that control osteoclastogenesis.
Acknowledgments
We thank all members of the Amgen Genome Program for their collective contributions to this work, and to Ms. Judy from Kansas for her pioneering support of OPG clinical studies.
ABBREVIATIONS
- TNFR
tumor necrosis factor receptor
- OPG
osteoprotegerin
- OPGL
OPG ligand
- CSF-1
colony-stimulating factor 1
- JNK
Jun N-terminal kinase
- PQCT
peripheral quantitative computerized tomography
- TRAP
tartrate-resistant acid phosphatase
- TRAF
TNFR-associated factor
- RANK
receptor activator of NFκB
References
- 1.Suda T, Takahashi N, Martin T J. Endocr Rev. 1992;13:66–80. doi: 10.1210/edrv-13-1-66. [DOI] [PubMed] [Google Scholar]
- 2.Roodman G D. Endocr Rev. 1996;17:308–332. doi: 10.1210/edrv-17-4-308. [DOI] [PubMed] [Google Scholar]
- 3.Yasuda H, Shima N, Nakagawa N, Mochizuki S I, Yano K, Fujise N, Sato Y, Goo M, Yamaguchi K, Kuriyama M, et al. Endocrinology. 1998;139:1329–1337. doi: 10.1210/endo.139.3.5837. [DOI] [PubMed] [Google Scholar]
- 4.Simonet W S, Lacey D L, Kelley M, Chang M S, Luthy R, Nguyen H, Wooden S, Bennett L, Dunstan C, Boone T, et al. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 5.Bucay N, Sarosi I, Dunstan C R, Morony S, Tarpley J, Capparelli C, Scully S, Tan H-L, Xu W, Lacey D L, et al. Genes Dev. 1998;12:1260–1268. doi: 10.1101/gad.12.9.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizuno A, Amizuka N, Irie K, Murakami A, Fujise N, Kanno T, Sato U, Nakagawa N, Yasuda H, Mochizuki S, et al. Biochem Biophys Res Commun. 1998;247:610–615. doi: 10.1006/bbrc.1998.8697. [DOI] [PubMed] [Google Scholar]
- 7.Yamaoto M, Murakami T, Nishikawa M, Tsuda E, Mochizuki S-L, Higashio K, Akatsu T, Motoyoshi K, Nagata N. Endocrinology. 1998;139:4012–4015. doi: 10.1210/endo.139.9.6290. [DOI] [PubMed] [Google Scholar]
- 8.Lacey D L, Timms E, Tan H-L, Kelley M J, Dunstan C R, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, et al. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 9.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S-I, Tomoyasu A, Yano K, Goto M, Murakami A, et al. Proc Natl Acad Sci USA. 1998b;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller K, Wong B, Fox S, Choi Y, Chambers T J. J Exp Med. 1998;188:997–1001. doi: 10.1084/jem.188.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong Y-Y, Yoshida H, Sarosi I, Tan H-L, Timms E, Itie A, Capparelli C, Morony S, Oliveira dos Santos A J, Van G, et al. Nature (London) 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Mishikawa S, Okumura H, Sudo T, Shultz L D, Nishikawa S-I. Nature (London) 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 13.Anderson D M, Maraskovsky E, Billingsley W, Dougall W C, Tometsko M, Roux E R, Teepe M C, DuBose R F, Cosman D, Galiber L. Nature (London) 1997;390:175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 14.Darnay B G, Haridas V, Ni J, Moore P A, Aggarwal B B. J Biol Chem. 1998;273:20552–20555. doi: 10.1074/jbc.273.32.20551. [DOI] [PubMed] [Google Scholar]
- 15.Yeh W C, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, Pompa J L, Ferrick D, Hum B, Iscove N, et al. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Cheng G, Baltimore D. Immunity. 1996;5:407–415. doi: 10.1016/s1074-7613(00)80497-5. [DOI] [PubMed] [Google Scholar]
- 17.Franzoso G, Carlson L, Xing L, Poljak L, Shores E E, Brown K, Leonardi A, Tran T, Boyce B F, Siebenlist U. Genes Dev. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grigorizdis A E, Wang Z-Q, Cecchini M G, Hofstetter W, Felix F, Fleisch H, Wagner E F. Science. 1994;266:443–447. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 19.Iotsova V, Caamano J, Loy J, Yang Y, Lewin A, Bravo R. Nat Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 20.Hofbauer, L. C., Dunstan, C. R. & Spelsberg, T. C., Riggs, B. L. & Khosla, S. (1999) Biochem. Biophys. Res. Commun., in press. [DOI] [PubMed]
- 21.Hsu H, Solovyev I, Colombero A, Elliott R, Kelley M, Boyle W J. J Biol Chem. 1997;272:13471–13474. doi: 10.1074/jbc.272.21.13471. [DOI] [PubMed] [Google Scholar]
- 22.Wong B, Josien R, Lee S Y, Vologodskaia M, Steinman R M, Choi Y. J Biol Chem. 1998;273:28355–28359. doi: 10.1074/jbc.273.43.28355. [DOI] [PubMed] [Google Scholar]