Abstract
PriA, a 3′ → 5′ DNA helicase, directs assembly of a primosome on some bacteriophage and plasmid DNAs. Primosomes are multienzyme replication machines that contribute both the DNA-unwinding and Okazaki fragment-priming functions at the replication fork. The role of PriA in chromosomal replication is unclear. The phenotypes of priA null mutations suggest that the protein participates in replication restart at recombination intermediates. We show here that PriA promotes replication fork assembly at a D loop, an intermediate formed during initiation of homologous recombination. We also show that DnaC810, encoded by a naturally arising intergenic suppressor allele of the priA2∷kan mutation, bypasses the need for PriA during replication fork assembly at D loops in vitro. These findings underscore the essentiality of replication fork restart at recombination intermediates under normal growth conditions in bacteria.
Primosome assembly on bacteriophage φX174 DNA requires seven proteins: PriA, PriB, PriC, DnaT, DnaB, DnaC, and DnaG (1). Only three of these proteins, DnaB, the replication fork helicase (2), DnaC, and DnaG, the replication fork primase, (3) were known to be required for cellular replication from the chromosomal origin, oriC (4). However, strains carrying priA null mutations are constitutively induced for the SOS response (5). We suggested that this observation indicated that replication forks that formed at oriC did not complete replication of the chromosome and had to be rescued by a PriA-dependent process (6). Subsequent genetic analyses demonstrated that priA null strains acquired suppressor mutations in dnaC very rapidly (7, 8) and were defective in homologous recombination (7, 8), repair of UV damage (7, 8), double-strand break-repair (8), and both induced and constitutive stable DNA replication (9). These results led to the proposal that the absence of PriA-catalyzed replication fork assembly at recombination intermediates could account for all of these phenotypes (7, 10). This idea was supported by the demonstration that PriA could specifically bind D loop DNA (11). In this report we have investigated PriA-catalyzed replication fork assembly at D loops by using defined templates and purified DNA replication proteins.
MATERIALS AND METHODS
DNA Replication Proteins.
A dnaC810 ORF was constructed by means of splicing by overlap extension PCR and cloned into the NdeI site of the pET11C overexpression plasmid (Novagen). Overexpression and purification of DnaC810 was as for the wild-type protein. PriA, PriB, PriC, DnaT, DnaB, and DnaC were purified as described by Marians (12). Single-stranded DNA-binding protein (SSB) was purified according to Minden and Marians (13). The DNA polymerase III holoenzyme was reconstituted either from Pol III* and β subunit as described by Wu et al. (14) or from purified subunits as described by Marians et al. (15).
Preparation of D Loop Template DNA.
A 100-nt-long oligonucleotide having the sequence 5′-ACATACATAAAGGTGGCAACGCCATTCGAAATGAGCTCCATATGCTAGCTAGGGAGGCCCCCGTCACAATCAATAGAAAATTCATATGGTTTACCAGCGC-3′ was annealed to f1R408 viral DNA (16). The central 42 nt of this oligonucleotide are nonhomologous with the template. The heteroduplex was converted to a nicked form II DNA with a 42-nt-long bubble region by incubation with the DNA polymerase III holoenzyme in the presence of SSB. During the last 2 min of this incubation, the 2′,3′-dideoxynucleotides ddTTP and ddATP at concentrations 20-fold higher than dTTP and dATP were introduced to the reaction to ensure that the complementary strand synthesized could not be extended further. After extraction with phenol and precipitation with ethanol, the DNA products were purified by electrophoresis through nondenaturing agarose gels. Complete form II bubble DNA was recovered from the gel and a 5′-32P-labeled minus-strand oligonucleotide with the sequence 5′-ATATAAAAGAAACGCAAAGACACCACGGAATAGTTTATTTT-3′ was then annealed to form the D loop form II template. The template was then gel filtered through Bio-Gel A5M to remove both unannealed oligonucleotide and unincorporated [γ-32P]ATP.
DNA Replication Reaction.
Reaction mixtures (12 μl) containing 50 mM Hepes–KOH (pH 8.0), 10 mM Mg(OAc)2, 10 mM DTT, 80 mM KCl, 200 μg/ml BSA, 2 mM ATP, 40 μM dNTPs, 0.42 nM 32P-labeled form II D loop DNA template, 0.5 μM SSB, 225 nM DnaC, and 30 nM PriA, PriB, PriC, DnaT, DnaB, and DNA polymerase III holoenzyme were incubated at 37°C for 10 min. Reactions were terminated by the addition of EDTA to 25 mM and NaOH to 50 mM and analyzed by electrophoresis at 2 V/cm for 20 h at room temperature through horizontal 0.7% alkaline agarose gels using 30 mM NaOH and 2 mM EDTA as the electrophoresis buffer. Gels were neutralized, dried, and autoradiographed. When used, PriAK230R, PriAK230A, PriAK230D, and DnaC810 were present at the same concentrations as their wild-type counterparts.
RESULTS
PriA-Directed Replication Fork Assembly at D Loops.
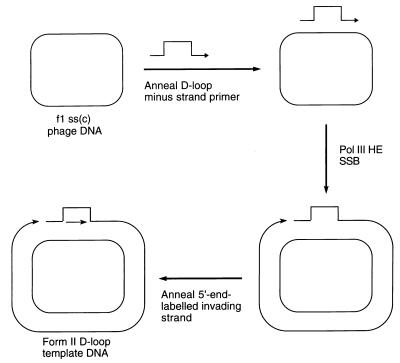
The scheme for preparing a double-stranded circular template carrying a D loop with a 3′-invading strand is given in Fig. 1. A replication fork assembled on such a template could use the 3′-OH of the invading strand as the primer for leading-strand synthesis. To focus on this event, the only radioactive label present in our replication reactions was at the 5′ end of the invading strand oligonucleotide. Extension of the 3′ end of the minus strand of the template was inhibited because of the presence of a terminal dideoxy-nucleotide.
Figure 1.
Scheme for preparation of the form II D loop template DNA. Pol III HE, DNA polymerase III holoenzyme; ss(c), single-stranded circular.
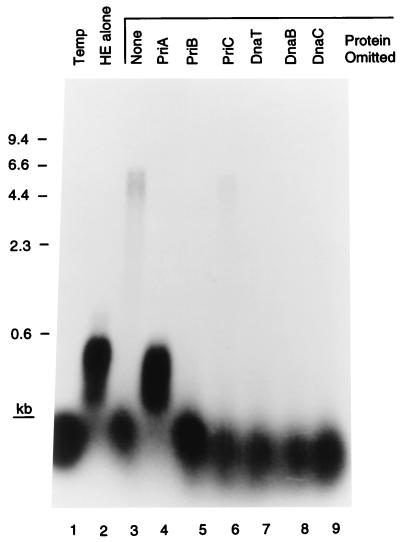
Incubation of the D loop template, the seven primosomal proteins, SSB, and the DNA polymerase III holoenzyme, the ten-subunit replicase of Escherichia coli (1), resulted in extension of the invading strand oligonucleotide (42 nt) to full-length template size (6.4 kb). This activity could be seen quite clearly when the reaction products were analyzed on a denaturing alkaline agarose gel (Fig. 2). The efficiency of the reaction varied, but generally 15–30% (as determined by PhosphorImager analysis) of the invading strand could be elongated to full length in a 10-min incubation. The reaction exhibited an absolute requirement for all of the primosomal proteins except PriC. Omission of this protein resulted in a decrease in DNA synthesis to one-third that of the complete reaction. This result is similar to what we have observed in the past with replication on different templates (17). Some extension of the invading strand by the holoenzyme alone could be observed, but this was suppressed by the presence of PriA. If the invading strand was omitted from the reaction and [α-32P]dATP was included, no DNA replication could be observed (data not shown). These observations suggested that PriA was directing primosome assembly at the D loop.
Figure 2.
Primosomal protein requirements for D loop-dependent DNA replication. The indicated proteins were omitted from standard replication reaction mixtures. Temp, template only; HE alone, reaction mixture contained only the DNA polymerase III holoenzyme and SSB. The markers were a HindIII digest of bacteriophage λ DNA.
In the experiment shown in Fig. 2, extension of the invading strand could result from one of two processes: (i) either assembly of a bona fide replication fork at the D loop followed by elongation of the leading strand coupled with unwinding of the duplex DNA template or (ii) uncoupled unwinding of the template DNA leaving an oligonucleotide annealed to the viral single-stranded DNA that could be elongated in a primer-extension reaction by the polymerase. We showed previously that coupled replication fork action requires a protein–protein interaction between DnaB and the τ subunit of the holoenzyme (18). In the presence of this interaction replication forks could move rapidly at nearly 1,000 nt/sec, whereas in its absence, the polymerase becomes stuck behind a slow-moving helicase and replication fork progression proceeds at only about 30 nt/sec.
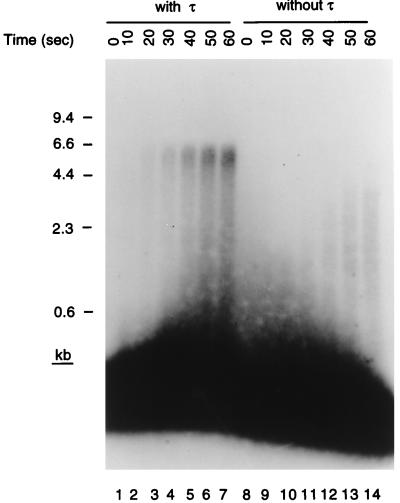
Accordingly, we assessed the speed of elongation of the invading strand in the presence and absence of τ (Fig. 3). For these experiments we used holoenzyme reconstituted from individually purified subunits. Ten-second time points were taken from the start of the reaction, and the elongated products were examined on denaturing gels. Full-length material could be observed in the presence of τ after 10 sec, whereas even after 60 sec no full-length material was observed in its absence. This gives a rate of replication fork progression in the presence of τ of 600–700 nt/sec, similar to what we have measured in the past in other replication systems (19). Thus, we conclude that bona fide replication fork assembly was occurring at the D loop on the template in the presence of the primosomal proteins, SSB, and the holoenzyme.
Figure 3.
Bona fide replication forks are formed on the D loop template. Standard replication reaction mixtures were increased in size 8-fold and contained holoenzyme reconstituted with purified subunits in either the presence or the absence of τ. Aliquots (12 μl) were removed before the start of the incubation (0 sec) and at 10-sec intervals thereafter, and the reaction products were analyzed as described in the text. The overexposed region at the bottom of the gel represents unelongated invading strand and arises because less than 1% of the primer is extended during the first minute of the reaction.
Replication Fork Assembly Does Not Require the PriA Helicase Activity.
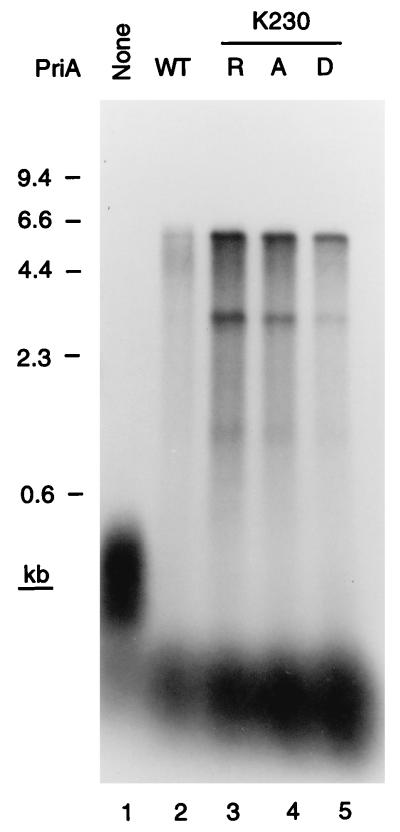
Although PriA is a DNA helicase (20, 21), all of the phenotypes of priA null mutations can be suppressed by mutated priA alleles that encode PriA proteins that are no longer ATPases or DNA helicases, but still catalyze primosome assembly (22). These mutations are substitutions of the invariant Lys in the Walker A box nucleotide-binding motif. If the PriA-dependent replication fork assembly described here were relevant to what happened in the cell, we would expect these mutant proteins to substitute fully for wild-type PriA in the replication reaction. This proved to be the case. Three mutant proteins, having the K230R, K230A, and K230D substitutions, were tested. All three supported replication on the D loop template to a greater extent than the wild-type protein (Fig. 4). We have observed this improved activity of the mutant proteins in other systems (22) and believe it arises because they remain bound to the site of DNA binding, providing a better target than the wild-type protein, which can move off the site because of its DNA helicase activity.
Figure 4.
Only the primosome assembly function of PriA is required for D loop-dependent DNA replication. Replication reaction mixtures contained either the wild-type PriA (WT) or the K230R, K230A, or K230D mutant PriA protein.
DnaC810 Bypasses the Requirement for PriA, PriB, PriC, and DnaT During Replication Fork Assembly.
Strains carrying priA null mutations are very difficult to grow. They are rich media-sensitive, form huge filaments, and have a viability roughly 1/100th that of the wild type (5, 7, 9). Suppressor mutations that restore viability, as well as ablate constitutive induction of the SOS response and the defects in homologous recombination and repair of UV-damaged DNA, arise overnight after transduction of the priA2∷kan allele into fresh recipient cells (7, 9). These mutations map to dnaC (7). DnaC forms a complex with DnaB in solution (23) and is required for the efficient transfer of DnaB to DNA in the presence of other replication proteins (1). To assess the biochemical properties of these altered DnaC proteins, one such suppressor allele, dnaC810, was molecularly cloned into an overexpression plasmid and the mutant protein was purified.
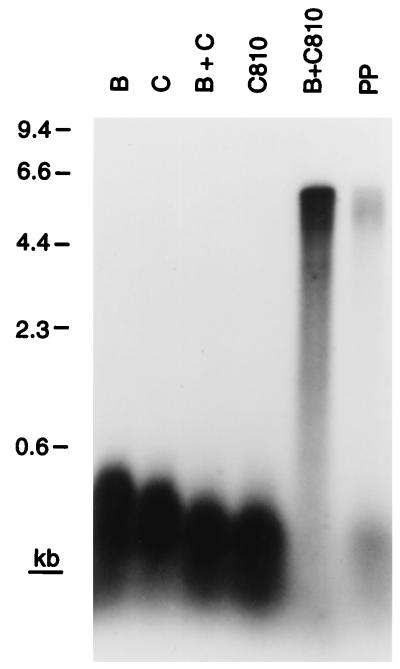
Strains carrying dnaC810 no longer require PriA for viability. This finding suggests that if the essential role for PriA in cellular metabolism is to catalyze assembly of replication forks at recombination intermediates, DnaC810 must be able to bypass the requirement for PriA to recognize the D loop and nucleate the assembly of a primosome. Accordingly, we tested whether DnaC810 alone could direct transfer of DnaB to the D loop template DNA (Fig. 5).
Figure 5.
DnaC810 bypasses the requirement for PriA, PriB, PriC, and DnaT during D loop-dependent DNA replication. Replication reactions contained the indicated proteins. PP, this reaction mixture contained all the primosomal proteins.
In the presence of SSB and the holoenzyme, the combination of wild-type DnaC and DnaB did not support elongation of the invading strand in the D loop (Fig. 5). This result was expected, both from the data presented in Fig. 2 and because SSB binding to DNA inhibits DnaB loading in the absence of either DnaA at oriC or PriA, PriB, PriC, and DnaT on bacteriophage φX174 DNA (1). On the other hand, DnaC810 was clearly able to load DnaB to the D loop on the template in the absence of the other primosomal proteins, as evinced by the elongation of the invading strand to full length (Fig. 5). Thus, the E176G substitution in DnaC810 represents a true gain-of-function mutation that allows bypass of the DnaB loading pathway that involves PriA, PriB, PriC, and DnaT.
Interestingly, the relative efficiencies of the replication reactions catalyzed in the presence of DnaC810 and DnaB varied compared with the reaction catalyzed by the complete set of primosomal proteins. At 80 mM KCl in the experiment shown, the DnaC810 reaction was 5- to 10-fold more efficient. However, at 600 mM potassium glutamate (data not shown), the reaction catalyzed by the complete set of proteins was more efficient by a factor of 2 (the efficiency of the DnaC810 reaction decreases, whereas that of the primosomal reaction increases). We assume that this difference reflects the relative stability of intermediate complexes that are formed during the loading of DnaB onto the DNA.
DISCUSSION
The E. coli replication fork possesses an extraordinary processivity that has been measured during rolling circle DNA replication in vitro at in excess of 1 megabase (Mb) (14, 19). Thus it has been the natural assumption that the two replication forks formed at oriC during chromosomal DNA replication have a sufficient inherent processivity to each synthesize roughly half the genome, about 2.2 Mb. Congruence of genetic and biochemical data now suggests that this may not be what happens in the cell.
Strains carrying priA null mutations are constitutively induced for the SOS response, that is, no exogenous DNA-damaging agent is required. Of the known activities of PriA, only the primosome assembly function is necessary to complement this defect, and we have shown here that the likely target for primosome assembly in the cell is a D loop. This is strongly reinforced by our demonstration that naturally arising suppressor mutations in dnaC encode a protein that has gained the ability to bypass the PriA-directed D loop primosome assembly pathway and load DnaB directly to the D loop for subsequent replication fork assembly. D loops represent the only known target for PriA in the cell. Although searched for extensively, primosome assembly sites similar to the sequence present in φX174 viral DNA have never been identified (24). SOS induction in priA null strains presumably relates to stalling of replication forks. Essentially all cells in a culture of a priA null mutant are filamented, and the filamentation can be suppressed by introduction of the sulA mutation (5). Thus, under normal growth conditions it seems unlikely that the replication forks that form at oriC complete synthesis of the genome. Instead, they must be subverted, most likely by encountering endogenous DNA damage or perhaps by colliding with protein roadblocks on the DNA (frozen topoisomerases? stalled RNA polymerases?). Replication restart would then become crucial to survival.
Formation of the D loop required for replication fork assembly for restart of DNA synthesis depends on the recombination proteins. Strains carrying priA null mutations are also severely defective in homologous recombination (7, 8). Both the dnaC810 and priAK230R alleles suppress that defect as well. On the basis of the data presented here, this observation suggests that the homologous recombination defect relates to the lack of replication fork assembly at recombination intermediates. Two interesting possible interpretations of this are as follows: Either the frequency of recombination in the cell is such that almost all recombination intermediates that form tend to block the advance of a replication fork, generating a requirement for replication restart on the downstream side of the intermediate, or resolution of most recombination intermediates includes an obligate DNA replication step.
Acknowledgments
Sincerest thanks are given to A. J. Clark, in whose lab S.J.S. started his genetic studies of priA. This work was supported by National Institutes of Health Grant GM34557 to K.J.M.
ABBREVIATION
- SSB
single-stranded DNA-binding protein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Marians K J. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 2.LeBowitz J H, McMacken R. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 3.Bouche J-P, Zechel K, Kornberg A. J Biol Chem. 1975;250:5995–6001. [PubMed] [Google Scholar]
- 4.Kaguni J M, Kornberg A. Cell. 1984;38:183–190. doi: 10.1016/0092-8674(84)90539-7. [DOI] [PubMed] [Google Scholar]
- 5.Nurse P, Zavitz K H, Marians K J. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zavitz K H, Marians K J. Mol Microbiol. 1991;5:2869–2873. doi: 10.1111/j.1365-2958.1991.tb01846.x. [DOI] [PubMed] [Google Scholar]
- 7.Sandler S J, Sawra H S, Clark A J. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kogoma T, Cadwell G W, Barnard K G, Asai T. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masai H, Asai T, Kubota Y, Arai K-I, Kogoma T. EMBO J. 1994;13:5338–5345. doi: 10.1002/j.1460-2075.1994.tb06868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kogoma T. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 11.McGlynn P, Al-Deib A A, Liu J, Marians K J, Lloyd R G. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 12.Marians K J. Methods Enzymol. 1995;262:507–521. doi: 10.1016/0076-6879(95)62042-7. [DOI] [PubMed] [Google Scholar]
- 13.Minden J S, Marians K J. J Biol Chem. 1985;260:9316–9325. [PubMed] [Google Scholar]
- 14.Wu C A, Zechner E L, Marians K J. J Biol Chem. 1992;267:4030–4044. [PubMed] [Google Scholar]
- 15.Marians K J, Hiasa H, Kim D R, McHenry C S. J Biol Chem. 1998;273:2452–2457. doi: 10.1074/jbc.273.4.2452. [DOI] [PubMed] [Google Scholar]
- 16.Russell M, Kidd S, Kelley M R. Gene. 1986;45:333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- 17.Ng J Y, Marians K J. J Biol Chem. 1996;271:15642–15648. doi: 10.1074/jbc.271.26.15642. [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Dallmann H G, McHenry C, Marians K J. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 19.Mok M, Marians K J. J Biol Chem. 1987;262:16644–16654. [PubMed] [Google Scholar]
- 20.Lee M S, Marians K J. Proc Natl Acad Sci USA. 1987;84:8345–8349. doi: 10.1073/pnas.84.23.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lasken R S, Kornberg A. J Biol Chem. 1988;263:5512–5518. [PubMed] [Google Scholar]
- 22.Zavitz K H, Marians K J. J Biol Chem. 1992;267:6933–6940. [PubMed] [Google Scholar]
- 23.Wickner S, Hurwitz J. Proc Natl Acad Sci USA. 1975;72:921–925. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marians K J. CRC Crit Rev Biochem. 1984;17:153–215. doi: 10.3109/10409238409113604. [DOI] [PubMed] [Google Scholar]