Abstract
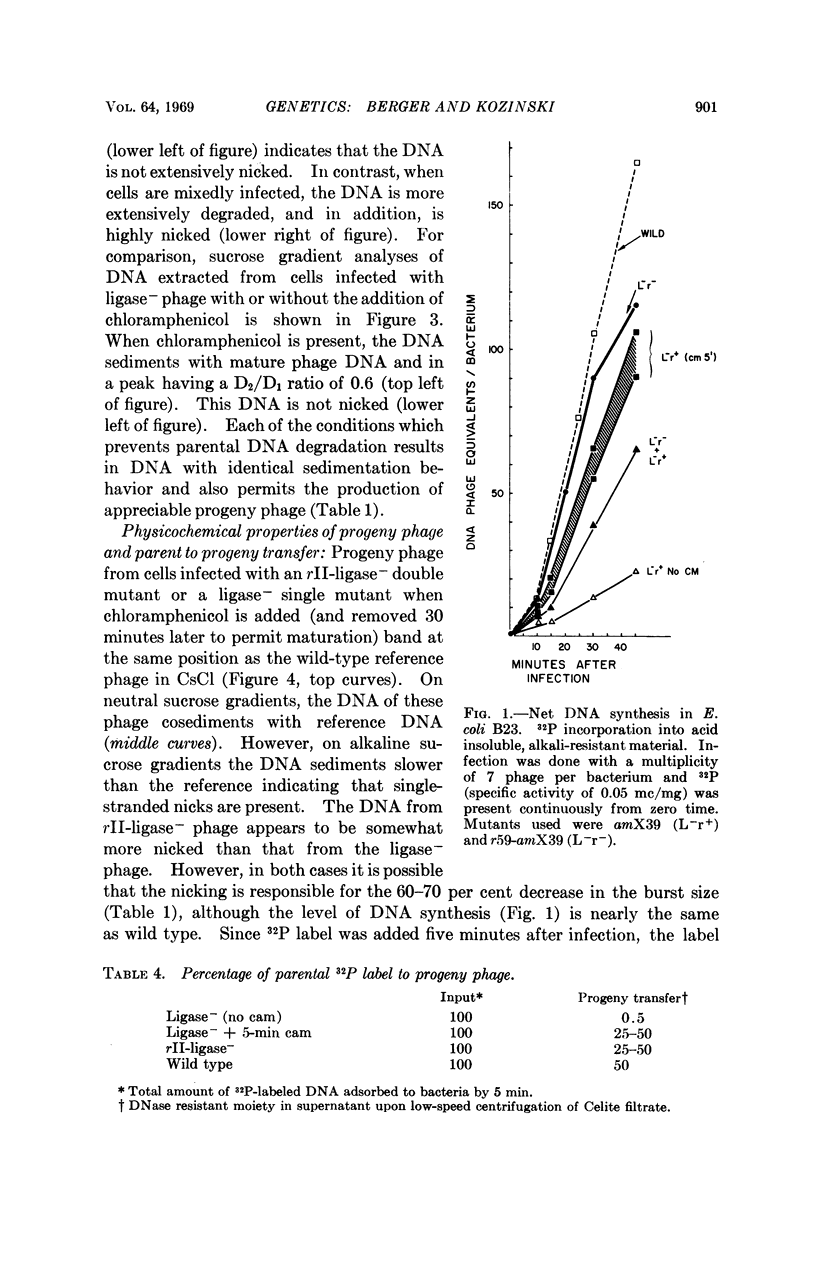
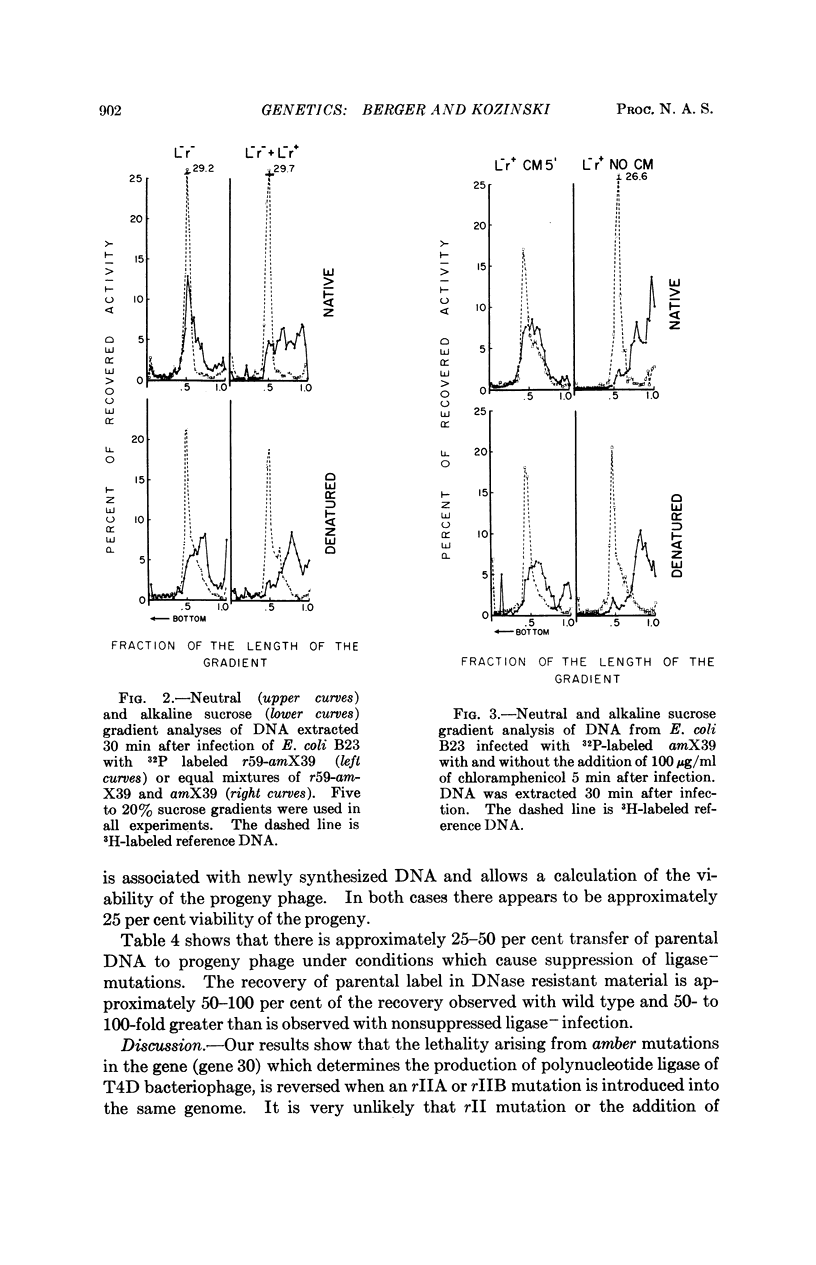
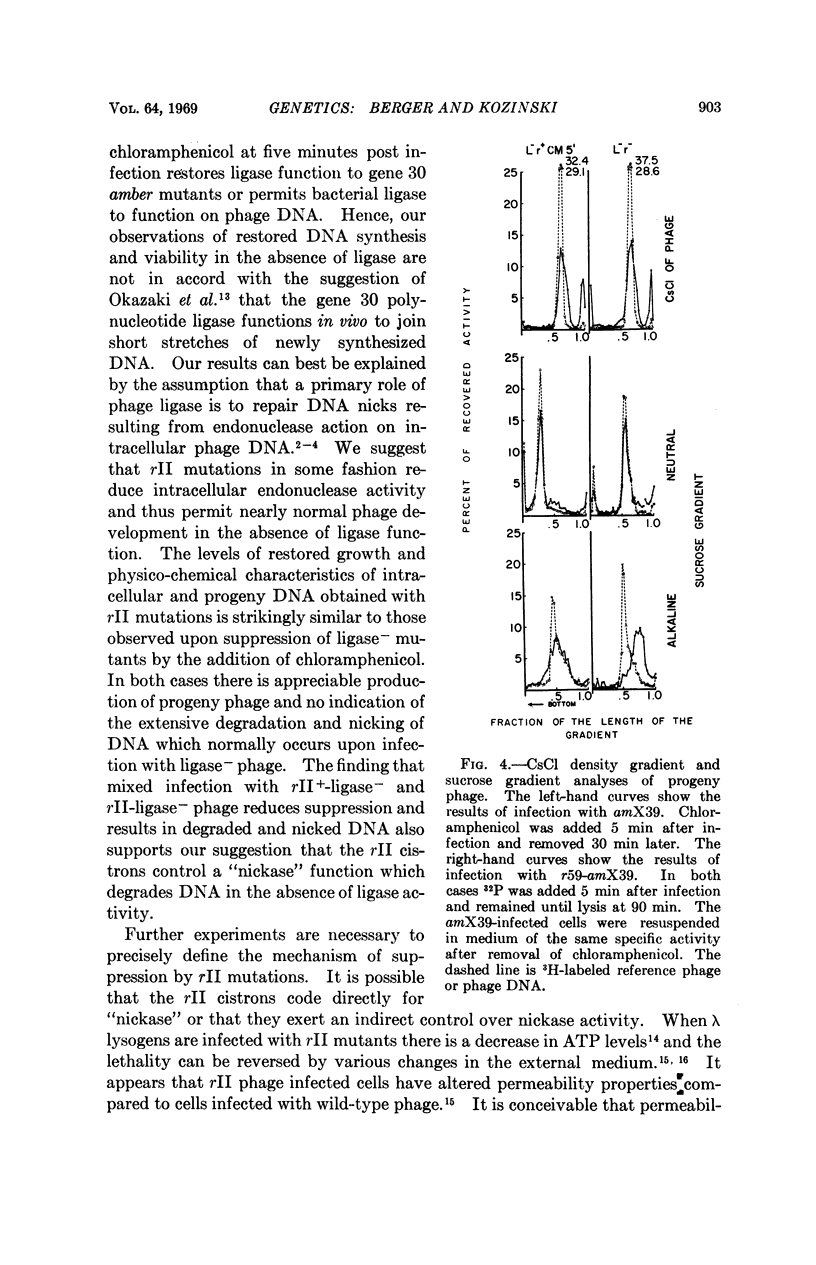
The introduction of rII mutations into polynucleotide ligase amber mutants (gene 30) of T4D bacteriophage results in the restoration of phage growth in the nonpermissive host E. coli B. When cells are mixedly infected with rII+ -ligase- and rII-ligase- phage, growth is reduced, indicating that the rII mutations are recessive to the rII+ alleles. Infection with the rII-ligase- double mutants results in nearly complete restoration of phage DNA synthesis and prevents the extensive degradation of parental phage DNA observed after infection with ligase- single mutants. Similar results are observed when cells are infected with ligase- mutants and chloramphenicol is added five minutes post infection.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buller C. S., Astrachan L. Replication of T4rII bacteriophage in Escherichia coli K-12 (lambda). J Virol. 1968 Apr;2(4):298–307. doi: 10.1128/jvi.2.4.298-307.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doermann A. H., Parma D. H. Recombination in bacteriophage T4. J Cell Physiol. 1967 Oct;70(2 Suppl):147–164. doi: 10.1002/jcp.1040700411. [DOI] [PubMed] [Google Scholar]
- Fareed G. C., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. II. The structural gene for polynucleotide ligase in bacteriophage T4. Proc Natl Acad Sci U S A. 1967 Aug;58(2):665–672. doi: 10.1073/pnas.58.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAREN A. Physiological effects of rII mutations in bacteriophage T4. Virology. 1961 Jun;14:151–163. doi: 10.1016/0042-6822(61)90190-8. [DOI] [PubMed] [Google Scholar]
- Kozinski A. W., Kozinski P. B., James R. Molecular recombination in T4 bacteriophage deoxyribonucleic acid. I. Tertiary structure of early replicative and recombining deoxyribonucleic acid. J Virol. 1967 Aug;1(4):758–770. doi: 10.1128/jvi.1.4.758-770.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozinski A. W. Molecular recombination in the ligase negative T4 amber mutant. Cold Spring Harb Symp Quant Biol. 1968;33:375–391. doi: 10.1101/sqb.1968.033.01.044. [DOI] [PubMed] [Google Scholar]
- Kozinski A. W. Unbiased participation of T4 phage DNA strands in replication. Biochem Biophys Res Commun. 1969 Apr 29;35(2):294–299. doi: 10.1016/0006-291x(69)90281-2. [DOI] [PubMed] [Google Scholar]
- Sekiguchi M. Studies on the physiological defect in rII mutants of bacteriophage T4. J Mol Biol. 1966 Apr;16(2):503–522. doi: 10.1016/s0022-2836(66)80188-2. [DOI] [PubMed] [Google Scholar]
- Weiss B., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid, I. Repair of single-strand breaks in DNA by an enzyme system from Escherichia coli infected with T4 bacteriophage. Proc Natl Acad Sci U S A. 1967 Apr;57(4):1021–1028. doi: 10.1073/pnas.57.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]



