Abstract
We have developed a combinatorial approach, using incremental truncation libraries of overlapping N- and C-terminal gene fragments, that examines all possible bisection points within a given region of an enzyme that will allow the conversion of a monomeric enzyme into its functional heterodimer. This general method for enzyme bisection will have broad applications in the engineering of new catalytic functions through domain swapping and chemical synthesis of modified peptide fragments and in the study of enzyme evolution and protein folding. We have tested this methodology on Escherichia coli glycinamide ribonucleotide formyltransferase (PurN) and, by genetic selection, identified PurN heterodimers capable of glycinamide ribonucleotide transformylation. Two were chosen for physical characterization and were found to be comparable to the wild-type PurN monomer in terms of stability to denaturation, activity, and binding of substrate and cofactor. Sequence analysis of 18 randomly chosen, active PurN heterodimers revealed that the breakpoints primarily clustered in loops near the surface of the enzyme, that the breaks could result in the deletion of highly conserved residues and, most surprisingly, that the active site could be bisected.
For a variety of proteins in which the backbone polypeptide chain has been broken by chemical, proteolytic, or genetic means, interchain packing interactions have been sufficient to maintain function. The combination of two protein fragments to restore activity has been termed protein fragment complementation. Such complementation, which results from conversion of a monomer to a heterodimer, is the reverse of evolutionary processes in which functional structures or domains are recruited and then become fused at the genetic level. Building on classic examples of protein fragment complementation, such as ribonuclease S (1) and β-galactosidase (2), researchers have used protein fragment complementation to examine theories of protein evolution (3), protein folding (4), macromolecular assembly (5), structure-function relationships (6), and for mapping contacts in membrane-embedded proteins (7). Protein fragment complementation has proved useful for the development of several enzyme-based two-hybrid systems in Escherichia coli (8–11).
To date, however, there has been little systematic exploration of the scope of protein fragment complementation. Isoleucyl-tRNA synthetase was found to tolerate bisection at 11 of 18 locations chosen to sample a variety of different cut-point strategies (6). Active pairs revealed that cleavage of the amide backbone need not occur at well-defined junctions of structural units/domains and that the two fragments did not require overlapping sequences. Active heterodimers were found with cut-points within conserved and nonconserved regions as well as within secondary structures such as α-helixes. Combined with results stemming from the systematic exploration of the circular permutation of E. coli aspartate transcarbamoylase (12), one concludes that enzymes tolerate bisection in a variety of locations along the amino acid backbone.
An exhaustive evaluation of all possible cleavage points in an average size protein would require the individual construction of hundreds of plasmids. Instead, we developed a method by which a library of all possible combinations of lengths of N-terminal and C-terminal protein fragments can be constructed. We have applied this methodology to explore the structural foundation for the catalytic activity of E. coli glycinamide ribonucleotide formyltransferase (PurN). Formylation of glycinamide ribonucleotide (GAR) is an early step in the de novo synthesis of purines. E. coli has two GAR transformylases: the product of the purT gene catalyzes the production of formyl GAR (fGAR) from formate, ATP, and GAR (13, 14); that of purN catalyzes the production of fGAR from 10-formyltetrahydrofolate and GAR (15). High-resolution crystal structures (16–18) and site-directed mutagenesis studies (19, 20) on the 212-aa enzyme product of the purN gene have identified three key active site residues important for catalysis: N106, H108, and D144.
MATERIALS AND METHODS
Plasmids.
Phagemids pDIM-N2 and pDIM-C6 were constructed by a series of oligo replacements into vectors pMOpelB.H and pMOpelB.L designed for creating very large Fab antibody libraries (M.O. and S.J.B., unpublished work). These antibody vectors were derived from pBP107 (21) and pTC01 (22), respectively. The pertinent features of these vectors are described in Figs. 1 and 2.
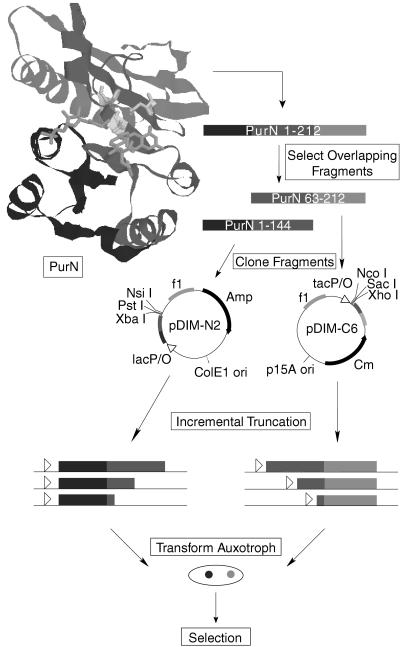
Figure 1.
Experimental overview. Based on the structure of PurN (16), the purN gene was divided into nonactive overlapping fragments. The N-terminus fragment (purN[1–144]) consists of the DNA coding for residues 1–144 and the C-terminus fragment (purN[63–212]) consists of the DNA coding for residues 63–212. By experimental design, the cut-point search is limited to the region of overlap between the two fragments (63–144). The two fragments were cloned into vectors pDIM-N2 and pDIM-C6 designed for incremental truncation. Libraries of incremental truncations from the C terminus of the purN[1–144] gene fragment and from the N terminus of the purN[63–212] gene fragment were created by using Exo III digestion at low temperatures. These incremental truncation libraries then were transformed together into an auxotrophic E. coli strain unable to grow on minimal media lacking purines unless GAR transformylase activity is supplied in trans. Active PurN heterodimers then were selected by plating the crossed library onto selective minimal plates. Colonies that grew contained an active PurN enzyme.
Figure 2.
Vectors for incremental truncation. The •••••• sequence corresponds to the sequence of the gene fragment to be truncated. The start and stop codons for the incremental truncation libraries are shown. The stop codon for pDIM-C6 is at the 3′ end of the gene fragment to be truncated.
Creation of Incremental Truncation Libraries.
Two micrograms of PstI/XbaI-digested pDIM-N2 or SacI/XhoI-digested pDIM-C6 was equilibrated at 12°C in 60 μl of 66 mM Tris, pH 8.0/0.66 mM MgCl2. At time zero, 200 units of exonuclease III (Exo III) were added. One-microliter samples were removed every 30 sec thereafter for 30 min and added to a tube incubating at 4°C containing 180 μl of S1 nuclease buffer (41 mM K-acetate, pH 4.6/365 mM NaCl/1.4 mM ZnSO4/6.8% glycerol), and 25 units of S1 nuclease. After all samples were collected, the tube was incubated at room temperature for 30 min. Subsequently, 24 μl of S1 stop buffer (0.3 M Tris/50 mM EDTA) was added, and the tube was incubated at 72°C for 20 min to fully inactivate S1 nuclease as well as Exo III. After an ethanol precipitation with ammonium acetate, the DNA was resuspended in 88 μl of water and digested with either NsiI (pDIM-N2) or NcoI (pDIM-C6). After a second ethanol precipitation, the pDIM-C6 DNA was incubated with 2.5 units of Klenow (in 2 mM Tris, pH 8.0/10 mM MgCl2 containing 0.125 mM each dATP, dCTP, dGTP, and dTTP) for 5 min at 37°C. For pDIM-N2, the DNA was first incubated in the same buffer without the dNTPs for 3 min at 37°C to use Klenow’s 3′ to 5′ exonuclease activity to blunt the 3′ overhang left by NsiI digestion. Subsequently pDIM-N2 DNA was incubated with the dNTPs as above. After heat inactivation of Klenow by incubation at 75°C for 20 min, 400 μl of ligase mix (50 mM Tris⋅HCl, pH 7.6/10 mM MgCl2/1 mM ATP/5% PEG-800/1 mM DTT) containing 15 units of DNA ligase was added for unimolecular blunt end ligation overnight at room temperature. The DNA was concentrated by ethanol precipitation into 30 μl of water and was electroporated into JS5 cells (Bio-Rad) by six electroporations of 5 μl of DNA each. After recovery at 37°C for 1 hr in 6 ml of SOB media (2% Bacto-Tryptone/0.5% Bacto-Yeast extract/10 mM NaCl/2.5 mM KCl/10 mM MgSO4) containing 2% glucose, the cells were plated onto a 243 × 243-mm TY media plate (0.8% Bacto-Tryptone/0.5% Bacto-Yeast extract/0.5% NaCl/1.5% agar) plate containing 2% glucose and either ampicillin (100 μg/ml) or chloramphenicol (50 μg/ml). After growth overnight at 37°C, the library was recovered from the plate into 20 ml of 2× TY/2% glucose/15% glycerol, concentrated by centrifugation, and frozen in small aliquots.
Selection of PurN Heterodimers.
The N-terminal and C-terminal truncation libraries were packaged into phage particles with the use of helper phage and infected into a 10-ml culture of exponentially growing E. coli strain TX680F′ (constructed by mating TX680 with XL-1 blue) at a titer such that approximately 1–5% of the cells became infected with both plasmids. Infection proceeded for 30 min at 37°C without shaking. The cells then were centrifuged, washed once with 10 ml of selective media (M9 salts, 0.2% glucose, 0.06% caseine, 2 μg/ml of thiamine, 40 μg/ml of kanamycin), and resuspended in 2 ml of selective media. The culture was shaken at 37°C for 2 hr before plating dilutions on selective plates with 0.3 mM isopropyl β-d-thiogalactoside. Plates were incubated at 37°C for up to 48 hr. Randomly chosen colonies that appeared within 28 hr were restreaked on selective plates to affirm complementation and ensure isolation of a single positive. From these plates, positives were restreaked onto rich plates (TY) containing ampicillin and chloramphenicol. Colonies from the rich plates were tested for PurN recombination by a PCR screen by using primers for the beginning and end of the purN gene. The plasmid DNA from those positives that were not recombinants was isolated and transformed at very dilute concentrations into E. coli strain DH5α so that the two plasmids could be isolated and sequenced. The plasmid DNA from these DH5α transformants was retransformed back into the auxotroph (both separately and together) to confirm complementation resulted from PurN heterodimers. After complementation was confirmed, sequencing of the truncated genes was performed by the Nucleic Acid Facility at The Pennsylvania State University.
Growth Rate.
Cells from overnight cultures in LB media (supplemented with 2% glucose, 100 μg/ml of ampicillin, 50 μg/ml of chloramphenicol, and 12.5 μg/ml of tetracyline) were washed once in 5 vol of minimal growth media [M9 salts, 0.2% glucose, 2 μg/ml of thiamine, all 20 amino acids at recommended levels (23), 40 μg/ml of kanamycin, 12.5 μg/ml of tetracylcine, and 0.3 mM isopropyl β-d-thiogalactoside] and diluted 1,000-fold into 50 ml of minimal growth media in 250-ml shake flasks. Cultures were shaken at 200 rpm at 37°C, and growth was monitored by removing 1-ml samples at various times and measuring the OD at 600 nm. Doubling time was calculated during early exponential phase (OD600 = 0.02–0.10). Because the lag times for auxotrophic cells expressing either wild-type monomer PurN or the heterodimers were essentially identical (approximately 2.5 hr), the growth rates measured cannot have been a result of a recombination event.
RESULTS AND DISCUSSION
Two overlapping purN gene fragments were cloned into compatible vectors pDIM-N2 and pDIM-C6 designed specifically for creating incremental truncation libraries (Figs. 1 and 2). The length of the overlap was set by selecting long N- and C-terminal fragments that were catalytically inactive. Both vectors can be maintained in the same cell because the vectors’ replicons belong to different compatibility groups and the vectors code for different antibiotic resistances. The plasmids also have f1 origins so that they can be packaged into filamentous phage particles and thus can be used for efficient transformation of E. coli.
Libraries of incremental truncations from the C terminus of the purN[1–144] gene fragment and from the N terminus of the purN[63–212] gene fragment were created by using Exo III. The amino acids of PurN coded for by the gene fragments are indicated by the numbers in brackets. Digestion with Exo III at 37°C has been used previously in the shortening of DNA fragments in the construction of large deletions (24) and for sequencing purposes (25). The rate of Exo III digestion at 37°C (≈500 bp/min, ref. 26) is too fast for purposes of incremental truncation where every one codon deletion is desired. Although it has been reported that the digestion rate of Exo III at 4°C is 25 bp/min (27), we found Exo III to digest at a rate of ≈2 bp/min at 4°C and 8–9 bp/min at 12°C.
For incremental truncation, the supercoiled plasmid first was digested with two restriction enzymes near the end of the gene to be truncated. The restriction enzyme site nearest the gene leaves a 3′ recessed end whereas the restriction site further away leaves a 5′ recessed end. The 5′ recessed ends are resistant to Exo III digestion whereas the 3′ recessed ends are not. The linear plasmid then was digested with Exo III at 12°C, and small samples were removed every 30 sec for N/9 min (where N is the maximum number of bases to be deleted). Samples all were removed to a single tube containing a low pH, zinc buffer that inhibited further digestion by Exo III. The buffer also contained S1 nuclease that removed the remaining single-stranded tails. After heat inactivation of S1 nuclease, digestion with a third restriction enzyme and treatment with Klenow polymerase to create blunt ends, unimolecular ligation resulted in the fusion of the truncated gene to either a series of stop codons in all three frames (for pDIM-N2) or to a start codon (for pDIM-C6). Some members of the N-terminal gene fragment library will have additional amino acids on the C terminus depending on which stop codon is in-frame with the gene. Likewise, two-thirds of the C-terminal gene fragment library will be out of frame with the start codon. However, one-third of each library will be in-frame and will not code for any amino acids that are not in the original protein except for the methionine start in the C-terminal library.
Libraries of 105–106 transformants typically were obtained by this methodology, well exceeding the target number of possible library members. For example, the target number of possible library members for an 81 codon truncation is 243. Exceeding the number of possible library member by three orders of magnitude ensured that all possible members were present despite any small biases inherent in the library construction. After the two libraries are crossed into the auxotoroph, the target number of possible heterodimers is 59,049. This number can be easily exceeded by standard transformation techniques or by phagemid infection.
Truncation size diversity was confirmed in each library by picking 10 random library members and examining the size of the truncated gene by restriction enzyme or PCR methods. The fraction of library members having genes within the desired size range was always found to be 80% or greater. Although truncations encompassed the full size range expected, there was some bias toward smaller truncations.
To search for active PurN heterodimers, we used a genetic selection strategy by using TX680, an auxotrophic strain of E. coli that lacks functional purT and purN genes. In TX680, the purT gene was inactivated by a spontaneous point mutation that introduces a stop codon in the middle of the gene (13). The purN gene was inactivated by the introduction of a lacZ/KanR cassette in between residues 89 and 90 to create a purN[1–89]-lacZ fusion (15). This auxotroph is unable to grow on minimal media in the absence of purines unless an active GAR transformylase is supplied in trans.
The individual N-terminal and C-terminal truncation libraries were packaged into phage particles and coinfected into strain TX680F′. The cells were plated on minimal plates and those colonies that grew (positives) were examined further. Several steps were taken to ensure that the positives selected were not the result of recombination or reversion. A PCR screen was used to identify any positive that contained an intact purN gene (a recombinant). Furthermore, all plasmids from positives were isolated and retransformed, both individually and together, back into the auxotroph to confirm that complementation only resulted when both plasmids were present.
Although the frequency of recombination was high, the frequency of functional heterodimers was found to be higher. For library A, generated from crossing truncated fragments derived from purN[1–144] and purN[63–212], the frequency of functional heterodimers and recombination were 0.67% and <0.04%, respectively. Recombination was found to occur, ostensibly because the auxotroph is recA+ and because of the large overlap of homogenous sequences (up to 243 bases) if a cell happens to receive a long N-terminal and C-terminal purN gene fragment. Several recombinants were examined and found to contain an intact purN gene, apparently resulting either from recombination between the two plasmids or between the purN[1–89] gene fragment on the chromosome and the C-terminal gene fragment on the pDIM-C6 plasmid.
Sequencing of a limited fraction of the hundreds of positives in library A indicated that C-terminal gene fragments coding for amino acids 63–212, in some cases with extra N-terminal amino acids from the BamHI cloning site, predominated. Although these positives did not have the purN[63–212] gene truncated, this finding did not reflect any problem with the incremental truncation methodology. By design, both vectors have an approximately 25-bp spacer between the gene to be truncated and the start point for incremental truncation (Fig. 2). Thus, all selected C-terminal fragments were derived from phagemids with digested DNA, the digested DNA being the 25-bp spacer. The starting C-terminal fragment (purN[63–212]) turned out to be an excellent place to functionally bisect PurN as this fragment was very promiscuous in the N-terminal pieces it could complement.
To investigate whether heterodimers with a C-terminal fragment smaller than the 63–212 sequence would be functional without sequencing hundreds of positives in library A to find them, a second library (B) was created as above starting from fragments purN[1–144] and purN[79–212]. For library B the frequency of functional heterodimers and recombination was approximately the same (0.03%). The frequency of functional heterodimers was lower in library B because library B is a subset of library A and most of the functional heterodimers of library A lie outside the range of library B. Functional heterodimers in library B were found with C-terminal fragments commencing at amino acid 107, illustrating that the required C-terminal fragment is not limited to initiation at position 63.
The amino acid sequences of randomly selected functional PurN heterodimers are shown in Table 1. Cut-points generally were found near the surface of the protein (Fig. 3) and tended to cluster within loops (Fig. 4). Indeed, every positive examined had at least one fragment either ending or starting within a loop. Cut-points were found to occur in α-helixes and β-sheets in one fragment (not both), but only if the α-helix or β-sheet was intact in the other half of the heterodimer. Thus, no heterodimers were discovered that result from a complete break in an α-helix or β-sheet. The B series of heterodimers bisects the active site with N106 and H108 on the N-terminal fragment and D144 on the C-terminal fragment.
Table 1.
Sequences of active PurN heterodimers
| Number | N-terminal fragment | C-terminal fragment | Relative doubling time* |
|---|---|---|---|
| A46 | 1–72NQ | MLGS63–212 | 1.2 ± 0.1 |
| A36 | 1–65 | MLGS63–212 | 1.3 |
| A37 | 1–69VTS | MGS63–212 | 1.3 |
| A43 | 1–62 | MGS63–212 | 1.3 |
| A47 | 1–80GVAR | M63–212 | 1.3 |
| A42 | 1–62 | MGS63–212 | 1.4 ± 0.2 |
| A35 | 1–74 | M63–212 | 1.4 |
| A38 | 1–66LTS | MLGF63–212 | 1.4 |
| A39 | 1–72QRS | MLGS63–212 | 1.5 |
| A44 | 1–61 | MLGS63–212 | 1.5 |
| A34 | 1–63LTS | M63–212 | 1.5 |
| B31 | 1–113 | M112–112 | 1.2 |
| B15 | 1–114LTS | M114–212 | 1.2 |
| B30 | 1–112LTS | M109–212 | 1.3 |
| B13 | 1–111 | M114–212 | 1.4 ± 0.3 |
| B18 | 1–111 | M107–212 | 1.6 |
| B28 | 1–118TS | M112–212 | 2.7 |
| B27 | 1–112A | M112–212 | 5.7 |
| B17 | 1–112RS | M114–212 | 6.1 |
| WT control | 1–212 | 0.98 ± 0.06 | |
| WT control | 1–212 | 1.0 ± 0.02 |
The amino acid sequences were determined from the DNA sequence of the purN gene fragments. Because proteolytic processing of the fragments may occur (e.g. to shorten overlapping sequences), the exact amino acid sequence of active PurN heterodimers may differ.
The growth rate in minimal media of auxotrophic strain TX680F′ expressing the heterodimers was determined (see Materials and Methods). The doubling time during exponential growth is expressed relative to that of TX680F′ cells expressing the wild-type monomer from phagemid pDIM-C6. TX680F′ bearing control plasmids without any purN gene or with only one gene fragment showed no growth.
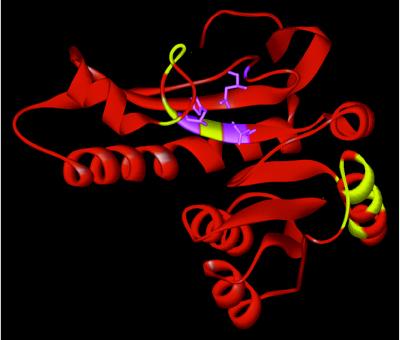
Figure 3.
Cut-points mapped onto the crystal structure of PurN (16). The new N and C termini for all the functional PurN heterodimers are indicated in yellow. Active site residues N106, H108, and D144 are indicated in violet. The original C terminus of the wild-type monomer is at the top.
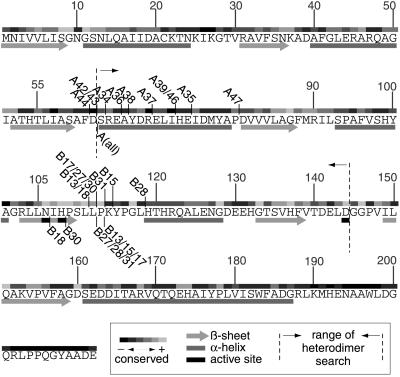
Figure 4.
Cut-points relative to secondary structure and conserved residues. The cut-points of all positives sequenced are shown relative to the amino acid sequence of PurN. If the number of the positive is shown above the sequence, it indicates the end of an N-terminal fragment. If the number of the positive is shown below the sequence, it indicates the beginning of the C-terminal fragment. For the A series of positives, all C-terminal fragments start at the same residue indicated by A(all). The degree of conservation of each amino acid is indicated above the sequence and was determined by alignment of 16 GAR transformylases from E. coli to human by using megalign (DNASTAR, Madison, WI). Secondary structure (16) is indicated below the sequence.
Heterodimers were found with cut-points within nonconserved and also, surprisingly, within conserved regions of the protein (Fig. 4). The motif NIHPSLLP (in PurN, residues 106–113) is highly conserved not only among GAR transformylases but also in other enzymes that use 10-formyltetrahydrofolate as a cofactor, including formyltetrahydrofolate hydrolases (28) and formyl-methionyl tRNA transferases (29). It is unexpected that heterodimers resulting from cleavage within such a highly conserved region would tolerate the corresponding disruptions, which include overlapping sequences, foreign amino acids, and break-points right next to active site residues (e.g., B18 contains the active site residue H108 in both fragments). Even more surprisingly, a heterodimer (B13) not containing residues L112 or P113 in either fragment was found to retain activity. This finding strongly suggests that this motif, despite its conservation, provides a nonessential linkage, but one whose conformation is satisfied only by a unique amino acid sequence. This finding illustrates that our combinatorial approach, unbiased by the current wisdom of enzyme structure and function, can identify heterodimers with cut-points at unexpected locations. It is difficult to see how B13, which assembles from two fragments derived from a cut within a highly conserved region and which lacks two absolutely conserved residues only four amino acids away from an active site histidine essential for activity, could have been expected or predicted by rational methods.
To examine the association and activity of these heterodimers, A42, A46, and B13 were chosen for further characterization and their protein fragments were expressed in the auxotroph TX680F′ from the lac and tac promoters. The specific activity of clarified cell lysates from the three heterodimers varied greatly (B13/A46/A42 = 500:6:1), but this activity difference was found to result primarily from differences in protein production levels and not activity (see below). Clarified cell lysates were fractionated by a G-75 gel filtration column. Maximum GAR transformylase activity for all three heterodimers was found in the same fraction as the wild-type monomeric enzyme, in accord with the strong association of the two fragments. Indeed, the activity of A46 was as stable as the monomer up to about 1 M urea, at which concentration the activity of A46 decreased faster than the monomer. In 2 M urea, A46 had lost virtually all of its activity, whereas the monomer still retained 25% of its activity measured in the absence of urea. Western analysis of the active gel filtration fraction resolved by SDS/PAGE revealed two bands reacting with anti-PurN antisera of approximately the expected size of the two protein fragments and no band at the size of a monomer PurN protein.
Initial velocity studies were initiated to determine the kinetic parameters of A46 and B13. Both were found to have kinetic constants near that of monomeric PurN (Table 2). For both heterodimers, the Kms for substrate GAR were increased by a factor of less than two. The effect of heterodimerization on the Kms for cofactor 10-formyl-5,8-dideazafolate (fDDF) was more pronounced, but were within a factor of 10. The greater effect on Km (fDDF) may result from the proximity of the cut-points to the binding site for fDDF. Based on kcat/Km (GAR), the heterodimers A46 and B13 have 5–10% of the activity of wild-type monomer PurN.
Table 2.
Kinetic constants of purN heterodimers
| Enzyme | Km (GAR), μM | Km (fDDF), μM | kcat, s−1 | kcat/Km (GAR), μM−1⋅s−1 |
|---|---|---|---|---|
| A46 | 159 ± 5 | 31.4 ± 3.9 | ∼8 | ∼0.05 |
| B13 | 196 ± 34 | 120 ± 16 | ∼12 | ∼0.06 |
| WT | 118 ± 3 | 12.3 ± 1.3 | 90 ± 2 | 0.76 |
GAR and fDDF were obtained and kinetic measurements were performed as described (30). Wild-type E. coli PurN was prepared as described by using a pMSW2 vector in MW12 cells (19). Heterodimer PurN was prepared by the same method using the vectors isolated from the positives (pDIM-N2 and pDIM-C6) and TX680F′ cells. Heterodimer concentration was estimated by densitometry of SDS/PAGE separation of the most active gel filtration fraction.
Although the two heterodimers selected for physical characterization were found to be near wild type in activity, heterodimers with much lower activity and/or expression levels can be identified by our selection strategy. Auxotrophic cells expressing heterodimers B13 and A42 had identical doubling times in minimal media, but 500-fold different levels of total activity. This finding strongly suggests that our screen, at a minimum, will select for heterodimers with expression levels the same as B13, but with 500-fold lower activity and for heterodimers as active as B13, but with 500-fold lower expression levels. Also, previous work using mutants of PurN (19) has shown that the cut off for successful complementation of the auxotroph appears to be a difference in kcat/Km (fDDF) of somewhere between 2,000- and 12,000-fold.
We have created a general method using incremental truncation libraries to search for bisection points to convert a monomeric enzyme into its functional heterodimer. This combinatorial method rapidly generates a wealth of information on loci for protein fragment complementation and is not limited by the availability of structural information or biased by our limited understanding of proteins. Applying this method to E. coli PurN GAR transformylase, we found that functional bisection points primarily clustered in loops. Active heterodimers often included short, overlapping segments that presumably point out into solution, because most cut-points were located near the surface. Cut-points occurred both within conserved and nonconserved regions of the enzyme. Active heterodimers were found with deletions of the primary amino acid sequence and with active site residues divided between the two fragments. Although we used a selection system, the frequency of functional bisection-points in PurN suggests that screening is a feasible alternative.
We envision that this general method for converting a monomer into a series of heterodimers will have applications in the study of enzyme evolution and protein folding. In addition, this methodology potentially subverts an impediment in the chemical synthesis of proteins: proteins too large to be chemically synthesized as a monomer can be synthesized as fragments, which allows the introduction of unique side-chain functions.
Our methodology could prove to be a powerful strategy for protein engineering in general by delineating domains and structures and defining their roles in catalysis. Furthermore, assigning function to protein motifs should prove useful in the design of domain-swapped enzymes (31) by identifying potential fusion points for exchange of domains within enzyme superfamilies. This strategy, which mimics evolutionary processes of domain swapping, should complement existing strategies of directed evolution (32, 33).
Acknowledgments
We thank Steven C. Tizio for constructing control vectors for the growth rate experiments. This work was supported in part by National Institutes of Health Grant GM24129 (S.J.B.) and National Institutes of Health Postdoctoral Fellowship GM18560 (M.O.).
ABBREVIATIONS
- PurN
E. coli glycinamide ribonucleotide formyltransferase
- GAR
glycinamide ribonucleotide
- Exo III
exonuclease III
- fDDF
10-formyl-5,8-dideazafolate
References
- 1.Kato I, Anfinsen C B. J Biol Chem. 1969;244:1004–1007. [PubMed] [Google Scholar]
- 2.Ullmann A, Jacob F, Monod J. J Mol Biol. 1967;24:339–343. doi: 10.1016/0022-2836(67)90341-5. [DOI] [PubMed] [Google Scholar]
- 3.Bertolaet B L, Knowles J R. Biochemistry. 1995;34:5736–5743. doi: 10.1021/bi00017a005. [DOI] [PubMed] [Google Scholar]
- 4.Ladurner A G, Itzhaki L S, Gray G P, Fersht A R. J Mol Biol. 1997;273:317–329. doi: 10.1006/jmbi.1997.1303. [DOI] [PubMed] [Google Scholar]
- 5.Tasayco M L, Carey J. Science. 1992;255:594–597. doi: 10.1126/science.1736361. [DOI] [PubMed] [Google Scholar]
- 6.Shiba K, Schimmel P. Proc Natl Acad Sci USA. 1992;89:1880–1884. doi: 10.1073/pnas.89.5.1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, Kono M, McKee T D, Oprian D D. Biochemistry. 1995;34:14963–14969. doi: 10.1021/bi00046a002. [DOI] [PubMed] [Google Scholar]
- 8.Johnsson N, Varshavsky A. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossi F, Charlton C A, Blau H M. Proc Natl Acad Sci USA. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karimova G, Pidous J, Ullmann A, Ladant D. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelletier J N, Campbell-Valois F-X, Michnick S W. Proc Natl Acad Sci USA. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graf R, Schachman H K. Proc Natl Acad Sci USA. 1996;93:11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nygaard P, Smith J M. J Bacteriol. 1993;175:3591–3597. doi: 10.1128/jb.175.11.3591-3597.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marolewski A, Smith J M, Benkovic S J. Biochemistry. 1994;33:2531–2537. doi: 10.1021/bi00175a023. [DOI] [PubMed] [Google Scholar]
- 15.Smith J M, Daum H A. J Biol Chem. 1987;262:10565–10569. [PubMed] [Google Scholar]
- 16.Almassy R J, Janson C A, Kan C-C, Hostomska Z. Proc Natl Acad Sci USA. 1992;89:6114–6118. doi: 10.1073/pnas.89.13.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P, Schulze-Gahmen U, Stura E A, Inglese J, Johnson D L, Marolewski A, Benkovic S J, Wilson I A. J Mol Biol. 1992;227:283–292. doi: 10.1016/0022-2836(92)90698-j. [DOI] [PubMed] [Google Scholar]
- 18.Klein C, Chen P, Arevalo J H, Stura E A, Marolewski A, Warren M S, Benkovic S J, Wilson I A. J Mol Biol. 1995;249:153–175. doi: 10.1006/jmbi.1995.0286. [DOI] [PubMed] [Google Scholar]
- 19.Warren M S, Marolewski A E, Benkovic S J. Biochemistry. 1996;35:8855–8862. doi: 10.1021/bi9528715. [DOI] [PubMed] [Google Scholar]
- 20.Warren M S, Benkovic S J. Protein Eng. 1997;10:63–68. doi: 10.1093/protein/10.1.63. [DOI] [PubMed] [Google Scholar]
- 21.Posner B, Lee I, Itoh T, Pyrati J, Graff R, Thorton G B, La Polla R, Benkovic S J. Gene. 1993;128:111–117. doi: 10.1016/0378-1119(93)90161-u. [DOI] [PubMed] [Google Scholar]
- 22.Collet T A, Roben P, O’Kennedy R, Barbas C F, Burton D R, Lerner R A. Proc Natl Acad Sci USA. 1992;89:10026–10030. doi: 10.1073/pnas.89.21.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerhardt P, Murray R G E, Wood W A, Krieg N R. Methods for General and Molecular Bacteriology. Washington, DC: Am. Soc. Microbiol.; 1994. [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 25.Henikoff S. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 26.Henikoff S. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- 27.Promega. Erase-a-Base System Technical Manual, #TM006. Madison, WI: Promega; 1995. [Google Scholar]
- 28.Nagy P L, McCorkle G M, Zalkin H. J Bacteriol. 1993;175:7066–7073. doi: 10.1128/jb.175.21.7066-7073.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guillon J M, Mechulam Y, Schmitter J M, Blanquet S, Fayat G. J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shim J H, Benkovic S J. Biochemistry. 1998;37:8776–8782. doi: 10.1021/bi980244k. [DOI] [PubMed] [Google Scholar]
- 31.Nixon A E, Warren M S, Benkovic S J. Proc Natl Acad Sci USA. 1997;94:1069–1073. doi: 10.1073/pnas.94.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuchner O, Arnold F H. Trends Biotechnol. 1997;15:523–530. doi: 10.1016/S0167-7799(97)01138-4. [DOI] [PubMed] [Google Scholar]
- 33.Crameri A, Raillard S, Bermudez E, Stemmer W P. Nature (London) 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]