Abstract
Human p32 (also known as SF2-associated p32, p32/TAP, and gC1qR) is a conserved eukaryotic protein that localizes predominantly in the mitochondrial matrix. It is thought to be involved in mitochondrial oxidative phosphorylation and in nucleus–mitochondrion interactions. We report the crystal structure of p32 determined at 2.25 Å resolution. The structure reveals that p32 adopts a novel fold with seven consecutive antiparallel β-strands flanked by one N-terminal and two C-terminal α-helices. Three monomers form a doughnut-shaped quaternary structure with an unusually asymmetric charge distribution on the surface. The implications of the structure on previously proposed functions of p32 are discussed and new specific functional properties are suggested.
Human p32 was originally isolated by copurification with the nuclear pre-mRNA splicing factor SF2/ASF, with which it appeared to be associated, although no evidence of a splicing function was obtained (1). Since the original study, it has become apparent that p32 is an evolutionarily conserved eukaryotic protein. Homologous genes have been identified in a number of eukaryotic species, ranging from fungi to mammals, as a result of both biochemical studies and genome sequencing efforts. In addition to its ability to bind to the splicing factor SF2/ASF in vitro, many additional properties or apparent functions of p32 have been reported. It can interact with many viral proteins, including HIV-1 Tat (2–4) and Rev (5, 6), EBNA-1 of Epstein–Barr virus (7), ORF P of herpes simplex virus (8), and core protein V of adenovirus (9). It has also been reported that p32 interacts with the globular head domain of the plasma complement component C1q, whose hemolytic activity is inhibited as a result (10). Binding of p32 to transcription factor IIB (11), the lamin B receptor (12), high molecular-weight kininogen and factor XII (13), vitronectin (14), and hyaluronic acid (15) has also been reported.
Unexpectedly, immunofluorescence and subcellular fractionation studies have shown that p32 and its yeast homologue localize predominantly in the mitochondrial matrix (9, 16–18). We have confirmed these findings in HeLa cells using multiple anti-p32 monoclonal antibodies, both by immunoblotting of subcellular fractions and by indirect immunofluorescence (I. Watakabe and A.R.K., unpublished results). It is puzzling how p32, a mitochondrial protein, can participate in the aforementioned processes, which take place at various nonmitochondrial locations. There is some evidence that a small fraction of p32 protein localizes in the nucleus (9). Furthermore, the distribution of p32 appears to be altered during adenovirus infection, with p32 migrating to the nucleus together with a viral core protein (9). These observations suggested that p32 plays a bridging role in nucleus–mitochondrion interactions.
p32 has been shown to be important for the maintenance of mitochondrial oxidative phosphorylation (16). However, the exact physiological role of p32 in mitochondria remains unknown. In an effort to understand the structure and function of p32, we have crystallized and determined the structure of the human protein at 2.25 Å resolution. To our knowledge, the structure is the first to be solved among this class of proteins, and it has a new fold.
MATERIALS AND METHODS
Protein Purification and Crystallization.
Recombinant p32 protein (aa 74–282, with Leu74 to Met74 substitution) was produced in Escherichia coli as described (1). p32 was purified by a 65% to 90% ammonium sulfate cut, followed by chromatography on Mono Q (Amersham Pharmacia). Peak p32 fractions eluted from the Mono Q column [3–5 mg/ml, in 20 mM Hepes (pH7.5)/5% glycerol/0.6M NaCl/1 mM EDTA/1 mM EGTA/0.1% β-mercaptoethanol] were readily crystallized in buffers containing acetate. The best diffracting crystals were grown by hanging-drop vapor diffusion at 16°C by using a well solution containing 10% glycerol, 25% 2-methyl-2,4-pentanediol, and 5–10 mM sodium acetate (pH 4.6). The optimal protein concentration used was approximately 3 mg/ml. After mixing equal volumes of protein solution and well solution, crystals began to form between a few minutes to a few days, depending on the acetate concentration. p32 crystallized in spacegroup P21 with cell dimensions of a = 58.63 Å, b = 56.48 Å, c = 93.83 Å, and β = 96°. There are three p32 molecules per asymmetric unit. Crystals used for x-ray diffraction had typical dimensions of 0.1 × 0.1 × 0.3 mm3.
Selenomethionine (SeMet)-substituted p32 was produced in the methionine auxotroph DL41(DE3) in a defined medium that supplies SeMet (19). Purification and crystallization of SeMet p32 was carried out as outlined for the native protein.
Data Collection and Structure Determination.
A complete 2.45-Å resolution native diffraction data set was collected at beamline X26C at the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory, by using an MAR (MAR Research, Hamburg) imaging plate detector. We attempted to solve the structure by the multiple isomorphous replacement method, but an extensive search for heavy-atom derivatives was not successful. We then turned to the multiwavelength anomalous diffraction (MAD) method using SeMet as anomalous scatterers. MAD data were collected at beamline X12C by using a single-cell Brandeis charge-coupled device detector at NSLS. Four data sets at wavelengths λ0 = 1.00 Å, λ1 = 0.9789 Å, λ2 = 0.9786 Å, and λ3 = 0.95 Å were collected from a single crystal at 100 K. All raw diffraction data were processed by using the hkl software package (20). Data processing statistics are shown in Table 1.
Table 1.
Statistics from the crystallographic analysis
| Measurement | Values
|
|||
|---|---|---|---|---|
| Diffraction data | λ0 | λ1 | λ2 | λ3 |
| Resolution, Å | 2.38 | 2.38 | 2.38 | 2.25 |
| Observed reflections | 143,315 | 145,771 | 152,031 | 170,171 |
| Unique reflections | 22,536 | 22,989 | 24.077 | 27,284 |
| Completeness, % | 90.7 | 92.0 | 89.1 | 92.6 |
| Average I/σ | 13.9 | 12.8 | 13.3 | 15.4 |
| Rmerge, %* | 3.5 | 4.1 | 5.2 | 4.5 |
| Phasing | ||||
| Se sites | 9 | |||
| Phasing power† at 2.5 Å | ||||
| Isomorphous | 2.65 | — | 1.53 | 3.24 |
| Anomalous | — | 1.78 | 3.65 | 2.25 |
| Overall figure of merit | 0.67 | |||
| Refinement | ||||
| Resolution range, Å | ∞−2.25 | |||
| R-factor,‡Rfree, % | 17.3 23.4 | |||
| Reflections with |F| > 2σ | 26,812 | |||
| Number of protein atoms | 4,259 | |||
| Number of water molecules | 340 | |||
| Average B-factor, Å2 | 34.2 | |||
| Rms deviations | ||||
| Bond lengths, Å | 0.007 | |||
| Bond angles, ° | 1.18 | |||
| Dihedrals, ° | 21.8 | |||
| Improper, ° | 0.64 | |||
Rmerge = Σ|I − 〈I〉|/Σ〈I〉, where I is the measured intensity and 〈I〉 is the averaged intensity of multiple measurements of the same reflection. The summation is over all the observed unique reflections.
Phasing power = rms (〈FH〉/E), where FH is the calculated structure factor of the heavy atoms and E is the residual lack of closure.
R-factor = Σ∥Fo| − |Fc||/Σ|Fo|, where Fo denotes the observed structure factor amplitude and Fc denotes the structure factor calculated from the model.
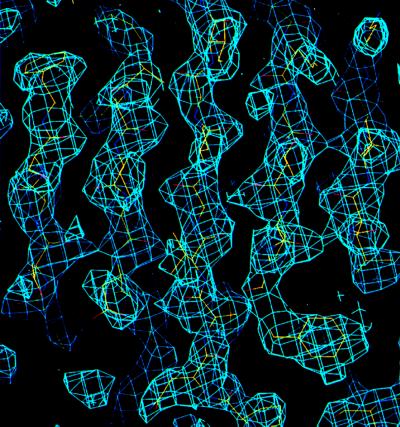
Determination of selenium atom positions, refinement of the selenium atom parameters, and phase calculation were all carried out by using the solve (21) and phases (22) software packages. The quality of the electron density from MAD phasing is excellent (Fig. 1). Solvent flattening and noncrystallographic symmetry averaging were not necessary for chain tracing and amino acid assignments. Almost all of the amino acids present in the refined model were placed into the MAD electron density by using the graphics program o (23).
Figure 1.
MAD-phased electron density map. A portion of the MAD-phased electron density covering the central β-strands is shown with a refined model superimposed onto the map. The electron density was computed to 2.5 Å resolution and contoured at 1.2 σ level.
The structure was refined by using x-plor and cns (24). An initial round of x-plor refinement brought the R-factor from 48.7 to 27.2%, by using the data in the resolution range of 8.0–3.0 Å. Extension to higher resolution and inclusion of low-resolution data with bulk solvent correction were carried out by using cns. The stereochemistry of the refined model was assessed by the procheck program (25).
The programs cath (26), dali (27), and scop (28) were used to search for structural and folding similarities against the Protein Data Bank (29) protein structure database. molscript (30), raster3d (31), and grasp (32) programs were used to generate figures.
RESULTS
We have crystallized and solved the crystal structure of human p32 to 2.25 Å resolution. The electron density map (Fig. 1) produced from SeMet MAD phasing allows unambiguous Cα-chain tracing and side-chain assignments, except for two disordered segments in each molecule within the asymmetric unit. The crystallized protein corresponds to the mature form of p32 (Fig. 2A), which lacks a 73-residue signal sequence at the N terminus. The signal sequence is necessary to target the protein to mitochondria (9, 16–18). The structure has been refined to 2.25 Å resolution with a crystallographic R-factor of 17.3% (Rfree = 23.4%). The refined model has excellent stereochemistry. The Ramachandran plot shows 90.2% of the nonglycine nonproline residues within the most favored region and 9.0% in the additional allowed regions.
Figure 2.
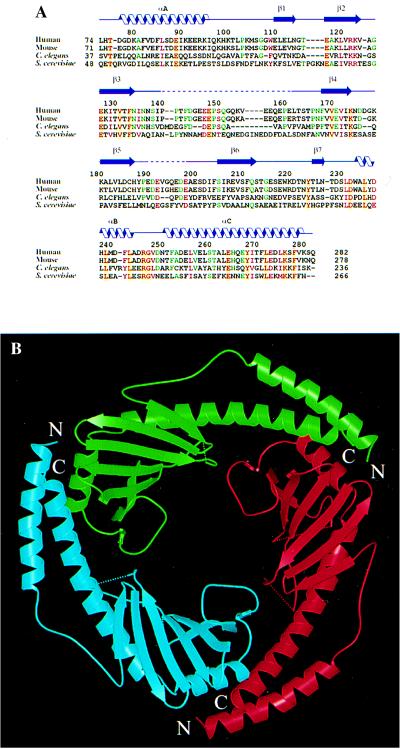
Overall structure of p32. (A) Structure-based alignment of p32 sequences from human (PID g338043), mouse (PID g743485), Caenorhabditis elegans (PID g3334445) and S. cerevisiae (PID g557799). Only the sequences corresponding to the mature human protein are shown. Positions with four identical amino acids are shown in red letters over yellow background; positions with four similar amino acids are shown in plain red letters; positions with three identities are shown in green letters. Related expressed sequence tag fragments or genomic sequences can also be found in Drosophila (NID g3112239), zebrafish (NID g2446858), and Arabidopsis (PID g3334441). PID and NID are protein and nucleotide identification numbers in GenBank. (B) Ribbon representation of the p32 trimer, looking down the noncrystallographic three-fold axis. The dotted lines show disordered segments in the structure. The three monomers that make up the trimer are colored red, green and cyan, and are referred to as subunits A, B, and C, respectively. The same color codes are used in other figures.
Structure Description.
There are three p32 molecules in an asymmetric unit, and they form a tightly bound trimer (Fig. 2B) with a noncrystallographic three-fold symmetry relating the monomers. This oligomeric state is most likely physiological, because it is consistent with solution data obtained by gel-filtration chromatography (18) and by laser-scattering molecular sizing (unpublished data). The doughnut-shaped trimer has an outer diameter of approximately 75 Å, an average inner diameter of approximately 20 Å, and a thickness of approximately 30 Å.
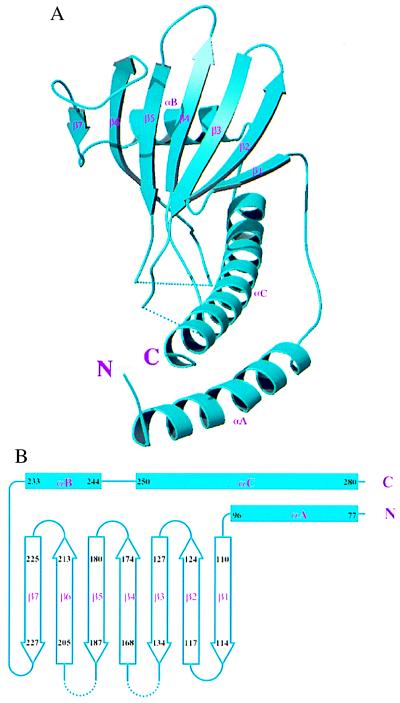
All three subunits have very similar conformations, despite our deliberate effort not to enforce the noncrystallographic symmetry during the process of model building and refinement. The pair-wise rms deviations of Cα positions were 0.89 Å, 0.84 Å, and 0.60 Å, respectively, between subunits A and B, A and C, and B and C. Monomeric p32 does not have any obviously distinct domains. Each monomer consists of seven consecutive β-strands, designated β1 through β7 (Fig. 3A), which form a highly twisted antiparallel β-sheet, with β1 nearly perpendicular to β7. The β-strands are flanked by one N-terminal (αA) and two C-terminal (αB and αC) α-helices. The topological arrangement of secondary structure elements is shown schematically in Fig. 3B. All three α-helices are located on the same side of the β-sheet. Helix αB lies parallel to the β-sheet, with the helix axis perpendicular to the direction of individual strands. Helix αB and the N-terminal portion (four turns) of helix αC make extensive hydrophobic contacts with the β-sheet, which appear to be essential for the stability of the structure. The N-terminal helix αA does not contact the β-sheet within the same monomer, but forms an antiparallel coiled-coil with the C-terminal portion (five turns) of αC. This coiled-coil region is important for protein–protein interactions responsible for homooligomerization. Despite its simplicity, the p32 structure does not belong to any known protein fold. A search for structure similarity against folding and structure databases did not produce any significant matches.
Figure 3.
Folding topology of p32. (A) Ribbon representation of a p32 monomer. The secondary structure elements are labeled. (B) Schematic drawing of the p32 topology.
The Trimer.
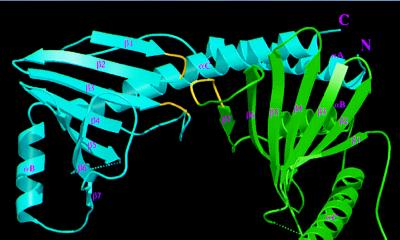
Three p32 molecules form a doughnut-shaped quaternary structure. The channel has a diameter of about 20 Å, but the loops connecting β6 and β7 partially cover the channel (Fig. 2B), reducing the diameter of the opening to about 10 Å. The channel wall is formed by the β-sheets from all three subunits. However, this all-β wall is distinct from other all-β structures (33), such as up-and-down β-barrels, in that the β-sheets from all three p32 monomers are highly twisted and can form only a hyperboloid surface rather than a cylindrical wall as in β-barrels. Because of the high degree of twisting, β-strands from adjacent monomers do not form contiguous β-sheets. Instead, residues located at the tip of β1-turn-β2 and the loop connecting β3 and β4 interact with β7 of the adjacent molecule (Fig. 4). In particular, the main chain carbonyl of Asn-114 and the amide group of Thr-116 form intermolecular hydrogen bonds to Oγ and the carbonyl group of Thr-222, respectively; the side chain atoms of Asn-134 and Asn-136 are involved in several intermolecular hydrogen bonds with main chain atoms of Asp-221, Asn-223, and Thr-225.
Figure 4.
Subunit interface between p32 monomers. The interactions between monomers B (green) and C (cyan) within the p32 trimer are shown, as viewed from the inside of the channel. The portions of the two loops in subunit C and one loop in subunit B that interact with each other (see text) are indicated in yellow. In addition, extensive intermolecular interactions involve contacts between αC and αA of monomer C with the β-sheet and αB of monomer B, respectively, as described in the text.
The coiled-coil region of αA and αC forms extensive intermolecular contacts: αA packs with the antiparallel αB of an adjacent monomer, and the C-terminal region of αC packs against the back of the β-sheet (Fig. 4). Most of these interactions are hydrophobic in nature, except for the intermolecular electrostatic pairing of Arg-246 and Asp-79. Asp-79 is not a conserved residue, but Arg-246 is invariant and is located in a region surrounded by several other invariant residues (see below). In summary, the overall architecture of the trimer can be visualized as if the β-sheets form a hyperboloid-shaped spool with the α-helices wrapping around it.
Surface Characteristics.
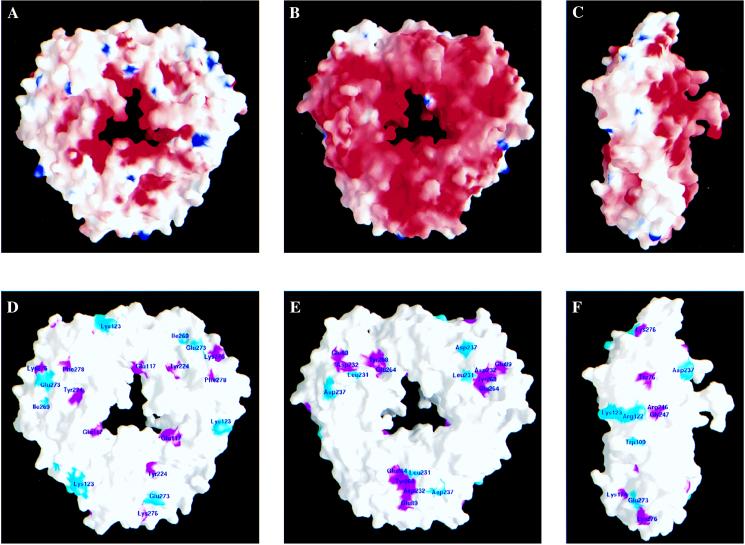
p32 is a very acidic protein, with a calculated pI of 4. However, the charge distribution on the protein surface is highly asymmetric (Fig. 5 A–C). Both the side of the doughnut that includes the loops connecting β3 to β4 and β5 to β6 and the inside of the channel have a high concentration of negatively charged residues (Fig. 5A). Both loops contain disordered segments, which have a combined net charge of −12 units. In contrast, the opposite side of the doughnut is much less negatively charged (Fig. 5B). This polarity in charge distribution clearly suggests asymmetric functional roles for the two sides of the protein.
Figure 5.
Surface distribution of charged and phylogenetically conserved amino acids. (A) Charge distribution on the protein surface, viewed from the same direction as in Fig. 1B. This side is referred to as the positive side. Positively and negatively charged electric potential are indicated in blue and red, respectively. The surface was calculated by using a probe radius of 1.4 Å, and the potential is displayed at the scale of −20 kBT to +20 kBT, where kB is the Boltzmann constant. (B) A significantly higher number of negatively charged residues is present on the opposite side of the protein surface. This side is referred to as the negative side. (C) A side view of the protein surface shows the asymmetry of charge distribution on the two sides of the protein surface. (D) Distribution of conserved amino acids on the positive side of the protein surface. Magenta indicates identical residues, and cyan indicates similar residues. They are shown in Fig. 2A as red letters over yellow background and plain red letters, respectively. (E) Several conserved and spatially clustered residues form a shallow pocket on the negative side of the protein surface. (F) Distribution of conserved amino acids on the outer perimeter of the protein surface.
Sequence comparisons among homologous proteins from different species show that the region spanning αB and αC is highly conserved. This conservation suggests that oligomerization is an inherent feature of this class of proteins. In addition, several conserved residues scattered throughout the p32 sequence cluster together in space, forming conserved regions on the protein surface (Fig. 5 D–F). These conserved surface features are very likely to be important for protein–protein interactions and ligand binding. Two regions of conservation stand out: first, five residues, Glu-89, Leu-231, Asp-232, Glu-264, and Tyr-268, form a shallow pocket on the negatively charged side of the doughnut (Fig. 5E); second, four residues, Arg-122, Lys-123, Arg-246, and Gly-247, are distributed closely together around the outer perimeter (Fig. 5F). In contrast, only one invariant residue, Glu-117, is located inside the channel. With the knowledge of the p32 three-dimensional structure, rational mutations of these conserved residues should help to test the proposed functions of the protein.
DISCUSSION
We solved the three-dimensional structure of human p32 in the hope of revealing structural similarities with other proteins of known functions, which would thus shed light on the functions of p32. However, the structure of p32 as reported here turned out to have an entirely new fold. In retrospect, it is perhaps not surprising that a protein with unusual properties has an unusual structure. To date, nearly 8,000 protein structures have been solved (29), which fall into approximately 600 different types of fold (26). As additional structures are solved, the total number of topological folds is expected to increase somewhat, but already most new structures tend to fall into known topological families. Therefore, each new fold provides a valuable prototype for understanding the architecture, folding, and evolution of proteins.
Although the structure of p32 did not reveal structural similarities to other proteins or motifs, this study provides valuable information for understanding and dissecting the functions of p32 and related proteins in the future. First, the structure revealed that oligomerization is an inherent feature of this class of proteins. Second, the conservation of specific amino acids clustered at discrete locations on the p32 trimer surface points to the likely functional importance of these regions. Third, the unusual asymmetric charge distribution suggests distinct functions for the two sides of the trimeric protein complex.
Many diverse functions have been proposed for p32, often involving very different subcellular locations, and it is highly unlikely that p32 can fulfill all these roles in a physiological setting. Many of the links to these functions were originally proposed on the basis of physical interactions of p32 with other molecules, often using two-hybrid or pull-down strategies, and these techniques have also been used to map smaller regions of p32 that mediate the observed interactions. However, in the absence of structural information, this type of deletion analysis could not take into account the structural integrity of the protein, including the folding of monomers and the stability of the trimer.
Concerning the normal cellular functions of p32, a recent study reported that disruption of the p32 homologous gene in Saccharomyces cerevisiae slowed down growth in glycerol medium but not in glucose medium, suggesting that p32 is involved in mitochondrial oxidative phosphorylation (16). In another report, the yeast p32 homologue was found to interact with the sorting signal of cytochrome b2 in vitro (18). According to this study, gene disruption did not appear to affect the sorting of cytochrome b2, nor did it affect cell growth in media with either fermentable or nonfermentable carbon sources. Further studies will be necessary to reconcile these discrepancies and to understand the mitochondrial functions of p32. In the meantime, the crystal structure already suggests some interesting clues for further functional studies of p32.
One interesting possibility is that p32 may serve as a high-capacity divalent cation storage protein to modulate mitochondrial matrix cation concentration, because 23% (48 amino acids) of its residues are aspartic and glutamic acids, and they are distributed on the surface of the trimer. This acidic surface characteristic is somewhat reminiscent of calsequestrin, a major Ca2+ storage protein found in the interior of sarcoplasmic reticulum vesicles (34). Calsequestrin is also rich in aspartic and glutamic acids (about 30% Asp and Glu, and a pI value around 4) and can bind and release 40–50 Ca2+ ions per molecule in each muscle contraction–relaxation cycle. Modulation of intracellular Ca2+ concentration by such proteins may directly affect the rate of pumping of Ca2+ into sarcoplasmic reticulum vesicles. Despite the structural differences between calsequestrin (35) and p32, the two proteins may act similarly in modulating Ca2+ ion levels, with the latter doing so in the mitochondrial matrix. Mitochondrial Ca2+ ion levels are strongly coupled with oxidative metabolism (36). For example, the activities of several mitochondrial dehydrogenases, such as pyruvate dehydrogenase, NAD-isocitrate dehydrogenase, and oxoglutarate dehydrogenase, are directly or indirectly regulated by intramitochondrial Ca2+ levels (37). We speculate that p32 may regulate mitochondrial oxidative phosphorylation by modulating the concentrations of divalent cations such as Ca2+.
An interesting feature of p32 is its high negative-charge density and asymmetric charge distribution, which suggest a possible mode of association with the inner mitochondrial membrane. The negatively charged side of p32 may potentially bind to the inner mitochondrial membrane in the presence of divalent metal ions, as negatively charged phospholipids can chelate divalent cations. Indeed, circumstantial evidence suggests that p32 is capable of associating with membranes (10, 13, 14, 18). If p32 is indeed localized proximally to the inner mitochondrial membrane, the central channel in the p32 trimer may have important functions. The channel is large enough to allow passage of a particle of moderate size. Depending on the degree of blockage by the loops connecting β6 and β7, which may undergo conformational changes, the opening would have a diameter ranging from 10 to 20 Å. This channel size would correspond to a range of molecular mass cutoffs for globular molecules of about 0.4–3 kDa, assuming an average density of about 1.35 g/cm3 (average protein density). The inner mitochondrial membrane has various types of pores of this size for protein import and exchange of solute molecules. Protein import into mitochondria has been extensively studied in recent years, and a number of trans-membrane proteins (TIMs) have been shown to be involved in protein import through the inner membrane (38). However, there is so far no evidence of the involvement of p32 in mitochondrial protein import or of its interaction with TIMs, except that p32 can interact with the sorting signal of cytochrome b2 in vitro (18).
Interestingly, in addition to the protein import channel, the mitochondrial inner membrane has a large conductance channel, known as the permeability transition pore (PTP) (39). The PTP allows free diffusion of solute molecules with a molecular mass of up to 1.5 kDa when the pore is fully open. Furthermore, high Ca2+ concentration appears to promote pore opening. This process plays an important role in triggering programmed cell death (40, 41). An intriguing speculation is that p32 may participate in apoptosis by interacting with the matrix side of the integral membrane component of PTP and regulating the pore opening in a divalent-metal concentration-dependent manner.
Curiously, most of the reported interactions of p32 should take place outside of mitochondria. For p32 to interact with the globular head domain of C1q (10), H-kininogen (13), vitronectin (14), and hyaluronic acid (15), it should be located on the extracellular side of the plasma membrane or in the extracellular matrix. Although p32 has been detected in the supernatants of several cultured cells and in normal human and rat sera (42), there is no direct evidence that it is specifically targeted to the cell surface or secreted. Small amounts of cell surface or plasma p32 may actually originate from lysed cells. An interesting question is whether the externalization of p32 is the result of an apoptotic process, as we have speculated above that p32 might be involved in apoptosis.
p32 is capable of interactions with numerous viral proteins, and many of these reported interactions take place in the nucleus. It is now clear that p32 is mainly localized in mitochondria at steady state and under normal circumstances. However, localization of a small fraction of p32 in the nucleus and/or dynamic shuttling of p32 between mitochondria and the nucleus remain possible. In fact, some nuclear localization has been reported by using different fixation procedures or different cell lines (9). More strikingly, it was observed that the localization of p32 in HeLa cells is altered during adenovirus infection (9). This finding led to the suggestion that p32 might be a part of a continuous nucleus–mitochondrion network that allows shuttling of macromolecules, and that adenovirus might take advantage of this host process to deliver its genome to the nucleus (9). The validity of this hypothesis remains to be tested.
In summary, we have reported the crystal structure of human p32 at 2.25 Å resolution. The crystal structure revealed that p32 has a novel fold. Three p32 molecules form a doughnut-shaped quaternary structure, with a sizable central channel and an unusual asymmetric charge distribution on the surface. Based on the structure, we have proposed several possible roles that p32 might play inside mitochondria. These and other proposals for the function of p32 and related proteins can now be further studied with the benefit of a detailed structure of the p32 trimer.
Acknowledgments
We thank M. Capel and R. M. Sweet for help with data collection at the National Synchrotron Light Source, Brookhaven National Laboratory, A. G. Murzin for help with the analysis of the protein fold, I. Watakabe for valuable technical assistance, and T. Messick for comments on the manuscript. This work was supported in part by the W. M. Keck foundation and by National Institutes of Health Grant GM55874 to R.-M.X.; A.R.K. was supported in part by Grant CA13106 from the National Cancer Institute.
ABBREVIATIONS
- MAD
multiwavelength anomalous diffraction
- SeMet
selenylmethionine
Footnotes
Data deposition: The atomic coordinates have been deposited with Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1p32).
References
- 1.Krainer A R, Mayeda A, Kozak D, Binns G. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 2.Desai K, Loewenstein P M, Green M. Proc Natl Acad Sci USA. 1991;88:8875–8879. doi: 10.1073/pnas.88.20.8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu L, Zhang Z, Loewenstein P M, Desai K, Tang Q, Mao D, Symington J S, Green M. J Virol. 1995;69:3007–3016. doi: 10.1128/jvi.69.5.3007-3016.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fridell R A, Harding L S, Bogerd H P, Cullen B R. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- 5.Luo Y, Yu H, Peterlin B M. J Virol. 1994;68:3850–3856. doi: 10.1128/jvi.68.6.3850-3856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tange T O, Jensen T H, Kjems J. J Biol Chem. 1996;271:10066–10072. doi: 10.1074/jbc.271.17.10066. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Finan J E, Middeldorp J M, Hayward SD. Virology. 1997;236:18–29. doi: 10.1006/viro.1997.8739. [DOI] [PubMed] [Google Scholar]
- 8.Bruni R, Roizman B. Proc Natl Acad Sci USA. 1996;93:10423–10427. doi: 10.1073/pnas.93.19.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews D A, Russell W C. J Gen Virol. 1998;79:1677–1685. doi: 10.1099/0022-1317-79-7-1677. [DOI] [PubMed] [Google Scholar]
- 10.Ghebrehiwet B, Lim B L, Peerschke E I B, Willis A C, Reids K B M. J Exp Med. 1994;179:1809–1821. doi: 10.1084/jem.179.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu L, Lowenstein P M, Zhang Z, Green M. J Virol. 1995;69:3017–3023. doi: 10.1128/jvi.69.5.3017-3023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simos G, Georgatos S D. FEBS Lett. 1994;346:225–228. doi: 10.1016/0014-5793(94)00479-x. [DOI] [PubMed] [Google Scholar]
- 13.Herwald H, Dedio J, Kellner R, Loos M, Müller-Esterl W. J Biol Chem. 1996;271:13040–13047. doi: 10.1074/jbc.271.22.13040. [DOI] [PubMed] [Google Scholar]
- 14.Lim B L, Reid K B M, Ghebrehiwet B, Peerschke E I B, Leigh L A E, Preissner K T. J Biol Chem. 1996;271:26739–26744. doi: 10.1074/jbc.271.43.26739. [DOI] [PubMed] [Google Scholar]
- 15.Deb T B, Datta K. J Biol Chem. 1996;271:2206–2212. doi: 10.1074/jbc.271.4.2206. [DOI] [PubMed] [Google Scholar]
- 16.Muta T, Kang D, Kitajima S, Fujiwara T, Hamasaki N. J Biol Chem. 1997;272:24363–24370. doi: 10.1074/jbc.272.39.24363. [DOI] [PubMed] [Google Scholar]
- 17.Dedio J, Jahnen-Dechent W, Bachmann M, Müller-Esterl W. J Immunol. 1998;160:3534–3542. [PubMed] [Google Scholar]
- 18.Seytter T, Lottspeich F, Neupert W, Schwarz E. Yeast. 1998;14:303–310. doi: 10.1002/(SICI)1097-0061(19980315)14:4<303::AID-YEA217>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson W A, Horton J R, LeMaster D M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z. In: Data Collection and Processing Proceedings of the CCP4 Study Weekend. Sawyer L, Issacs N, Bailey S, editors. Daresbury Laboratory, Warrington, U.K.: Science and Engineering Research Council; 1993. pp. 56–62. [Google Scholar]
- 21.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1997;53:571–579. doi: 10.1107/S0907444997005398. [DOI] [PubMed] [Google Scholar]
- 22.Furey W, Swaminathan S. Methods Enzymol. 1996;277:590–620. doi: 10.1016/s0076-6879(97)77033-2. [DOI] [PubMed] [Google Scholar]
- 23.Jones T A, Zou J Y, Cowan S W, Kjeldgard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 24.Brünger A T, Adams P D, Clore G M, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, Read R J, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr A. 1993;42:140–149. [Google Scholar]
- 26.Orengo C A, Michie A D, Jones S, Jones D T, Swindells M B, Thornton J M. Structure (London) 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 27.Holm L, Sander C. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 29.Abola E E, Sussman J L, Prilusky J, Manning N O. Methods Enzymol. 1997;277:556–571. doi: 10.1016/s0076-6879(97)77031-9. [DOI] [PubMed] [Google Scholar]
- 30.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 31.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 32.Nicholls A, Sharp K, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 33.Chothia C, Murzin A G. Structure (London) 1993;1:217–222. doi: 10.1016/0969-2126(93)90010-e. [DOI] [PubMed] [Google Scholar]
- 34.Yano K, Zarain-Herzberg A. Mol Cell Biochem. 1994;135:61–70. doi: 10.1007/BF00925961. [DOI] [PubMed] [Google Scholar]
- 35.Wang S, Trumble W R, Liao H, Wesson C R, Dunker A K, Kang C H. Nat Struct Biol. 1998;5:476–483. doi: 10.1038/nsb0698-476. [DOI] [PubMed] [Google Scholar]
- 36.McCormack, J. G. & Denton, R. M. (1993) Biochem. Soc. Trans.21 (Pt 3), 793–799. [DOI] [PubMed]
- 37.Nichols B J, Denton R M. Mol Cell Biochem. 1995;149:203–212. doi: 10.1007/BF01076578. [DOI] [PubMed] [Google Scholar]
- 38.Neupert W. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 39.Bernardi P, Broekemeier K M, Pfeiffer D R. J Bioenerg Biomembr. 1994;26:509–517. doi: 10.1007/BF00762735. [DOI] [PubMed] [Google Scholar]
- 40.Petit P X, Susin S A, Zamsami N, Mignotte B, Kroemer G. FEBS Lett. 1996;396:7–13. doi: 10.1016/0014-5793(96)00988-x. [DOI] [PubMed] [Google Scholar]
- 41.Green D R, Reed J C. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 42.van den Berg R H, Prins F, Faber-Krol M C, Lynch N J, Schwaeble W, van Es L A, Daha M R. J Immunol. 1997;158:3909–3916. [PubMed] [Google Scholar]