Abstract
Background
Most studies of the vaginal microflora have been based on culture or on qualitative molecular techniques. Here we applied existing real-time PCR formats for Lactobacillus crispatus, L. gasseri and Gardnerella vaginalis and developed new formats for Atopobium vaginae, L. iners and L. jensenii to obtain a quantitative non culture-based determination of these species in 71 vaginal samples from 32 pregnant and 28 non-pregnant women aged between 18 and 45 years.
Results
The 71 vaginal microflora samples of these women were categorized, using the Ison and Hay criteria, as refined by Verhelst et al. (2005), as follows: grade Ia: 8 samples, grade Iab: 10, grade Ib: 13, grade I-like: 10, grade II: 11, grade III: 12 and grade IV: 7.
L. crispatus was found in all but 5 samples and was the most frequent Lactobacillus species detected. A significantly lower concentration of L. crispatus was found in grades II (p < 0.0001) and III (p = 0.002) compared to grade I. L. jensenii was found in all grades but showed higher concentration in grade Iab than in grade Ia (p = 0.024). A. vaginae and G. vaginalis were present in high concentrations in grade III, with log10 median concentrations (log10 MC), respectively of 9.0 and 9.2 cells/ml. Twenty (38.5%) of the 52 G. vaginalis positive samples were also positive for A. vaginae. In grade II we found almost no L. iners (log10 MC: 0/ml) but a high concentration of L. gasseri (log10 MC: 8.7/ml). By contrast, in grade III we found a high concentration of L. iners (log10 MC: 8.3/ml) and a low concentration of L. gasseri (log10 MC: 0/ml). These results show a negative association between L. gasseri and L. iners (r = -0.397, p = 0.001) and between L. gasseri and A. vaginae (r = -0.408, p < 0.0001).
Conclusion
In our study we found a clear negative association between L. iners and L. gasseri and between A. vaginae and L. gasseri. Our results do not provide support for the generally held proposition that grade II is an intermediate stage between grades I and III, because L. gasseri, abundant in grade II is not predominant in grade III, whereas L. iners, abundant in grade III is present only in low numbers in grade II samples.
Background
Bacterial vaginosis (BV) is considered to be the most frequent vaginal infectious disorder in women of childbearing age. Prevalences of 4.9% to 36% have been reported from European and American studies [1]. The condition is symptomatic in half of the women and also represents a psychological burden. More importantly, this disturbed vaginal microflora can cause serious sequelae such as non-gonococcal, non-chlamydial PID (Pelvic Inflammatory Disease) [2,3], postpartum endometritis [4] and preterm birth [5-7]. Indeed, 40% of the cases of spontaneous preterm labor and preterm birth are thought to be associated with BV [8]. BV has also been associated with increased susceptibility to HIV and to genital tract infection with Chlamydia trachomatis and Neisseria gonorrhoeae [9-11].
BV is characterized by an overgrowth of many different, mostly anaerobic, bacteria. Gardnerella vaginalis has been considered to be the most characteristic microorganism in BV, but its pathogenic role was unclear, until recently, when the establishment of bacterial biofilm as a potential cause of recurrent BV [12], with a predominant presence of G. vaginalis, has clearly established a pathogenic role for this bacterium. Also recently, Atopobium vaginae has been strongly associated with BV, independently by different groups [13-15] and with biofilm in BV [12]. The suggestion that its association with BV is stronger than that of G. vaginalis, basically because G. vaginalis is more frequently detected in a normal vaginal microflora than A. vaginae [14] was confirmed by more recent studies [16,17]. The insight that biofilm is formed in BV [12] has strong explanatory power with regard to the high recurrence rate of BV, despite initial relief after antibiotic treatment [18] and is in perfect correlation with the presence of the characteristic cells or 'clue cells', which are vaginal epithelial cells covered with layers of bacteria.
Four species of lactobacilli are now considered to be predominantly linked to the vaginal microflora: Lactobacillus crispatus, L. jensenii, L. gasseri and L. iners, with the latter only recently being recognized as it was long overlooked because it does not grow on De Man Rogosa Sharpe agar, the medium typically used to culture lactobacilli [19,20].
In 1983 Spiegel et al. [21] devised a method for the diagnosis of bacterial vaginosis by direct Gram stain. Nugent et al. [22] proposed a modification of these criteria. By counting the different cell types (Lactobacillus spp., Gardnerella vaginalis/Bacteroides spp., Mobiluncus spp.) a score between zero and ten is obtained, whereby a score of 7 or higher corresponds to bacterial vaginosis and a score between 3 and 7 is considered intermediate between undisturbed vaginal microflora and bacterial vaginosis. Ison and Hay [23] suggested a simplification of this time-consuming approach, through an estimation of the ratios of the observed cellular types rather than the determination of the exact number of the bacteria and distinguished five grades (0 (no bacteria), I (normal), II (intermediate), III (bacterial vaginosis) and IV (streptococci)). Verhelst et al. [24] recently suggested, on the basis of Gram stain, terminal restriction fragment length polymorphism analysis of the 16S rRNA gene (T-RFLP) and molecular identification (tDNA-PCR) of cultured isolates, a new classification of the undisturbed vaginal microflora (grade I) whereby grade I microflora could be split up into four categories, designated grade Ia, Ib, Iab and I-like. Grade Ia was shown to contain predominantly L. crispatus, grade Ib predominantly L. gasseri and L. iners, grade Iab a mixture of these three species and grade I-like Bifidobacterium spp. rather than Lactobacillus spp. Grade I-like microflora, of which the Gram stain at first glance indicates the presence of lactobacilli, was considered as not representative for normal vaginal micoflora, but as probably an unrecognized type of disturbed vaginal microflora [25] that had previously not been distinguished from a healthy vaginal microflora.
In this study, we developed real-time PCR primers for L. iners, L. jensenii and A. vaginae and used these, together with described real-time PCR formats for L. crispatus [26], L. gasseri [26] and G. vaginalis [27], in an attempt to quantify some of the important bacterial species in the normal and disturbed vaginal microflora.
Results
Grading of vaginal microflora
In this study, Gram-stained smears of 71 vaginal swabs from 60 women, 32 of whom were pregnant, were examined microscopically. For 11 pregnant women the Gram stain based grading of their vaginal microflora differed between two trimesters and therefore swabs from both trimesters were included. Eight women had a vaginal microflora categorized by Gram stain as grade Ia, 10 as grade Iab, 13 as grade Ib, 10 as grade I-like, 11 as grade II, 12 as grade III and 7 as grade IV, according to criteria described previously [24].
Primer development and real-time PCR format
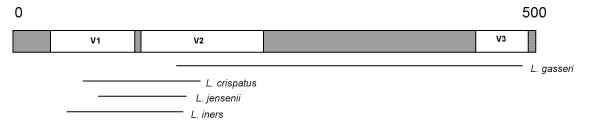
Primers for real-time PCR for Atopobium vaginae, Lactobacillus iners and L. jensenii were developed during this study (Tables 1, 2, 3). Primer positions relative to the 16S rRNA gene of Escherichia coli are presented in Figure 1. Table 4 lists all primers used and the real-time PCR formats. No cross-reactivity was detected with DNA from any of the tested species, including closely-related Lactobacillus and Atopobium species. In addition we found no cross reactivity for Bifidobacterium bifidum, B. breve and B. infantis with the G. vaginalis primers developed by Zariffard et al. [27].
Table 1.
Alignment of the 16S rRNA gene primers of L. iners with the sequences of other important vaginal lactobacilli
| Forward primer (position 70–85) | 5' | 3' | |||||||||||||||||||
| L. iners | G | T | C | T | G | C | C | T | T | G | A | A | G | A | T | C | G | G | |||
| L. crispatus | A | C | . | . | . | . | . | C | C | A | T | . | . | T | C | T | . | . | |||
| L. gasseri | A | C | . | . | . | . | . | C | A | A | G | . | . | C | T | . | . | ||||
| L. jensenii | A | C | . | . | . | . | . | C | . | T | . | . | . | T | C | T | . | . | |||
| L. vaginalis | A | C | . | . | . | . | . | C | . | . | . | . | . | C | G | G | . | . | |||
| Reverse primer (position 228–210) | 5' | 3' | |||||||||||||||||||
| L. iners | A | C | A | G | T | T | G | A | T | A | G | G | C | A | T | C | A | T | C | ||
| L. crispatus | . | . | . | C | . | . | A | G | C | . | C | T | . | . | . | . | G | . | T | ||
| L. gasseri | . | . | . | C | . | . | A | G | C | . | C | T | . | . | . | . | G | . | T | ||
| L. jensenii | . | . | . | C | . | . | A | G | C | . | C | T | . | . | . | . | G | . | T | ||
| L. vaginalis | . | . | . | C | C | . | A | G | C | . | C | T | . | . | . | . | G | . | T |
Table 2.
Alignment of the 16S rRNA gene primers of L. jensenii, with the sequences of other important vaginal lactobacilli
| Forward primer (position 117–137) | 5' | 3' | |||||||||||||||||||||
| L. jensenii | C | C | T | T | A | A | G | T | C | T | G | G | G | A | T | A | C | C | A | T | T | ||
| L. crispatus | . | . | C | A | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | ||
| L. gasseri | . | . | A | A | G | . | . | A | . | . | . | . | . | . | . | . | A | . | . | C | C | ||
| L. iners | . | . | A | A | G | . | . | A | T | C | . | . | . | . | . | . | A | . | . | C | C | ||
| L. vaginalis | . | . | . | G | . | . | . | C | G | G | . | . | . | . | . | . | A | . | . | . | C | ||
| Reverse primer (position 207–187) | 5' | 3' | |||||||||||||||||||||
| L. jensenii | A | C | G | C | C | G | C | C | T | T | T | T | A | A | A | C | T | T | C | T | T | ||
| L. crispatus | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | G | C | T | G | A | ||
| L. gasseri | . | - | A | . | . | A | T | . | . | . | . | . | . | . | . | . | . | C | T | A | G | ||
| L. iners | . | - | A | . | . | A | T | . | . | . | . | . | . | . | . | . | A | G | T | . | G | ||
| L. vaginalis | . | A | A | . | . | A | T | . | . | . | . | G | . | . | . | . | G | A | A | A | A |
Table 3.
Alignment of the 16S rRNA gene primers of A. vaginae with the sequences of the described Atopobium species
| Forward primer (position 1006–1025) | 5' | 3' | ||||||||||||||||||||
| A. vaginae | G | G | T | G | A | A | G | C | A | G | T | G | G | A | A | A | C | A | C | T | ||
| A. fossor | . | . | . | . | . | . | . | . | G | . | C | . | . | . | . | . | . | G | T | C | ||
| A. minutum | . | . | . | . | . | . | . | . | G | . | C | . | . | . | . | . | . | G | T | C | ||
| A. parvulum | . | . | . | . | . | . | . | . | G | . | C | . | . | . | . | . | . | G | T | C | ||
| A. rimae | . | . | . | . | . | . | . | . | G | . | C | . | . | . | . | . | . | G | T | C | ||
| Reverse primer (position 1282–1265) | 5' | 3' | ||||||||||||||||||||
| A. vaginae | A | T | T | C | G | C | T | T | C | T | G | C | T | C | G | C | G | C | A | |||
| A. fossor | . | . | . | G | . | . | . | . | A | C | T | T | . | . | . | . | A | A | G | |||
| A. minutum | . | . | . | G | . | . | . | . | A | C | T | T | . | . | . | . | A | A | G | |||
| A. parvulum | . | . | . | . | . | . | . | C | A | C | C | . | . | . | . | . | . | G | G | |||
| A. rimae | . | . | . | . | . | . | . | C | G | C | C | . | . | T | . | . | . | G | G |
Figure 1.
Localisation of the primers used in this study on the 16S rRNA gene.
Table 4.
Overview of the primer sequences and PCR conditions as used in this study
| Specificity | Name | Primer sequence (5'-3') | 16S rDNA positiona (5'-3') | Cycling conditions | Reference |
| A. vaginae | ATOVAGRT3Fw | GGTGAAGCAGTGGAAACACT | 1006–1025 | 10' 95°C, (15" 95°C, 20" 62°C, 40" 72°C) × 40 | This study |
| ATOVAGRT3Rev | ATTCGCTTCTGCTCGCGCA | 1282–1265 | |||
| G. vaginalis | F-GV1 | TTACTGGTGTATCACTGTAAGG | 16S-23S spacer | 10' 95°C, (45" 94°C, 45" 55°C, 45" 72°C) × 50 | [27] |
| R-GV3 | CCGTCACAGGCTGAACAGT | 16S-23S spacer | |||
| L. crispatus | LcrisF | AGCGAGCGGAACTAACAGATTTAC | 65–89 | 10' 95°C, (15" 95°C, 1' 60°C) × 40 | [26] |
| LcrisR | AGCTGATCATGCGATCTGCTT | 205–185 | |||
| L. gasseri | LactoF | TGGAAACAGRTGCTAATACCG | 157–177 | 10' 95°C, (15" 95°C, 1' 60°C) × 40 | [26] |
| LgassR | CAGTTACTACCTCTATCTTTCTTCACTAC | 470–442 | |||
| L. iners | InersFw | GTCTGCCTTGAAGATCGG | 70–85 | 10' 95°C, (1' 95°C, 1' 55°C, 1' 65°C) × 35 | This study |
| InersRev | ACAGTTGATAGGCATCATC | 228–210 | |||
| L. jensenii | LABJENTR2Fw | CCTTAAGTCTGGGATACCATT | 117–137 | 10' 95°C, (15" 95°C, 10" 54°C, 30" 72°C) × 40 | This study |
| LABJENRT2Rev | ACGCCGCCTTTTAAACTTCTT | 207–187 |
aRelative to the position in the Escherichia coli 16S rDNA sequence.
Using Qiagen extracted DNA from pure cultures; the detection limit for A. vaginae is 1.3 pg/ml, for G. vaginalis 43.2 pg/ml, for L. crispatus 4.7 pg/ml, for L. gasseri 10.7 pg/ml, for L. iners 6.4 pg/ml and for L. jensenii 15.0 pg/ml.
Qualitative results
Table 5 presents the overall presence of the six species studied in the different grades of vaginal microflora. L. crispatus was found in all but 5 samples (93%) and was the most frequent Lactobacillus species detected. L. jensenii was found in 33 samples (46%) but especially in grade Iab where it was present in 8 of 10 samples (80%).
Table 5.
Percentage of samples of each grade for which the tested species were detected
| Grade Ia | Grade Iab | Grade Ib | Grade I | Grade I-like | Grade II | Grade III | Grade IV | Total (%) | |
| Number of samples | 8 | 10 | 13 | 31 | 10 | 11 | 12 | 7 | 100 |
| A. vaginae | 38 | 20 | 15 | 23 | 30 | 18 | 83 | 14 | 32 |
| G. vaginalis | 88 | 50 | 61.5 | 65 | 70 | 63 | 100 | 29 | 68 |
| L. crispatus | 100 | 100 | 100 | 100 | 80 | 82 | 92 | 100 | 93 |
| L. gasseri | 88 | 90 | 85 | 87 | 60 | 100 | 33 | 43 | 72 |
| L. iners | 50 | 90 | 92 | 81 | 70 | 27 | 100 | 86 | 75 |
| L. jensenii | 13 | 80 | 46 | 48 | 60 | 55 | 50 | 0 | 47 |
Forty-eight samples (68%) were positive for G. vaginalis and 20/48 (42%) of these were also positive for A. vaginae. Only three samples were positive for A. vaginae but negative for G. vaginalis. All grade III samples were positive for G. vaginalis and 10/13 (83%) were also positive for A. vaginae.
L. gasseri was detected in 52/71 samples (73%) and 27/31 (87%) were grade I, 7/10 (70%) grade I-like, 11/11 (100%) grade II, 4/12 (33%) grade III and 3/7 (43%) grade IV.
L. iners was detected in 53/71 samples (75%) from which 25/31 (81%) were grade I, 7/10 (70%) grade I-like, 3/11 (27%) grade II, 12/12 (100%) grade III and 6/7 (86%) grade IV.
Quantitative results
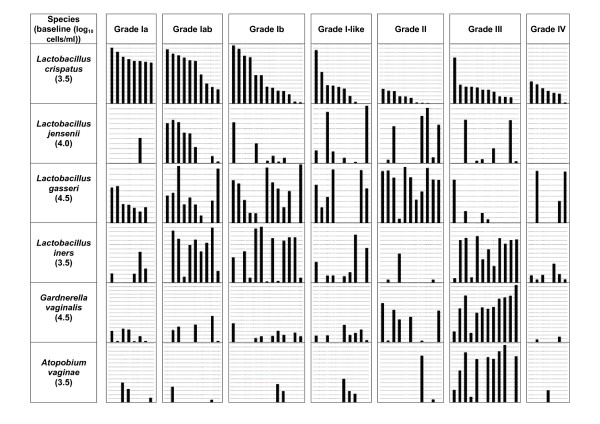
Figure 2 and Table 6 give an overview of the results obtained with real-time PCR for four vaginal Lactobacillus species and for Gardnerella vaginalis and Atopobium vaginae.
Figure 2.
Overview of the results obtained with real-time PCR for L. crispatus, L. jensenii, L. gasseri, L. iners, G. vaginalis and A. vaginae. x-axis: each bar represents a different sample. y-axis: logarithmic scale of cells/ml. Spaces in between horizontal lines represent log10 0.5 differences. Detection limits are 3.5 for L. crispatus, 4.0 for L. jensenii, 4.5 for L. gasseri, 3.5 for L. iners, 4.5 for G. vaginalis and 3.5 for A. vaginae. Data sorted per grade according to decreasing concentration of L. crispatus.
Table 6.
Quantitative determination of the six species in the different grades expressed as Median log10 cells/ml (Interquartile Range)
| Grade I | Grade I-like | Grade II | Grade III | Grade IV | |
| A. vaginae | 0 (0 – 0) | 0 (0 – 4.7) | 0 (0 – 0) | 9.0 (4.4 – 9.7) | 0 (0 – 0) |
| G. vaginalis | 5.1 (0 – 6.0) | 5.4 (0 – 5.9) | 4.7 (0 – 8.8) | 9.2 (8.8 – 10.7) | 0 (0 – 4.9) |
| L. crispatus | 8.8 (5.6 – 9.5) | 5.5 (2.8 – 6.2) | 4.2 (3.6 – 5.1) | 5.2 (4.3 – 5.6) | 5.1 (4.7 – 5.9) |
| L. gasseri | 6.6 (5.6 – 7.9) | 6.4 (0 – 8.3) | 8.7 (7.4 – 9.3) | 0 (0 – 5.3) | 0 (0 – 9.2) |
| L. iners | 6.5 (3.7 – 8.6) | 4.3 (3.9 – 4.5) | 0 (0 – 3.8) | 8.3 (5.6 – 8.7) | 4.3 (3.9 – 4.5) |
| L. jensenii | 4.0 (0 – 6.0) | 4.3 (0 – 6.3) | 4.3 (0 – 7.9) | 2.1 (0 – 5.2) | 0 (0 – 0) |
The median log10 cells/ml were expressed as per 1 ml elution buffer.
We found a significantly higher level of L. crispatus in grade I (log10 Median Concentration (MC) = 8.8 cells/ml) compared to the other grades, i.e. II: 4.2, p < 0.0001, III: 5.2, p = 0.002, IV: 5.1, p = 0.004 and I-like: 5.5, p = 0.014. L. crispatus was also present in significantly higher amounts in grade III compared to grade II (p = 0.023).
L. crispatus was present in high levels (i.e. at least log10 8.5 cells/ml) in all 8 grade Ia samples (log10 MC = 9.0 cells/ml), in most (i.e. 6/10) grade Iab samples (log10 MC = 8.9 cells/ml), but only in 3/12 grade Ib samples (log10 MC = 5.6 cells/ml).
L. jensenii was found in all grades but showed higher concentration in grade Iab (log10 MC = 7.0 cells/ml) than in grade Ia (log10 MC = 0 cells/ml, p = 0.024)
The level of L. gasseri in grade I (log10 MC = 6.6 cells/ml) was significantly lower than in grade II (log10 MC = 8.7 cells/ml, p < 0.01) and significantly higher than in grade III (log10 MC = 0 cells/ml, p < 0.0001). The quantity of L. gasseri in grade II was significantly higher than in grade III (p < 0.0001) and than in grade IV (log10 MC = 0 cells/ml, p < 0.05).
The level for L. iners did not differ either between grades I and III or between grades I and I-like. L. iners was present in grade II (log10 MC = 0 cells/ml) in significantly lower amounts than in grade III (log10 MC = 8.3 cells/ml, p < 0.0001), in grade I (log10 MC = 6.5 cells/ml, p < 0.0001), in grade IV (log10 MC = 4.3 cells/ml, p < 0.05) and in grade I-like (log10 MC = 4.3 cells/ml, p < 0.05).
The concentrations of A. vaginae and G. vaginalis were significantly higher in grade III (respectively log10 MC = 9.0 cells/ml and 9.2 cells/ml) than in grade I (respectively log10 MC = 0 cells/ml and 5.1 cells/ml, p < 0.0001), grade II (respectively log10 MC = 0 cells/ml and 4.7 cells/ml p < 0.005), grade IV (respectively log10 MC = 0 cells/ml and 0 cells/ml, p < 0.005 and p < 0.0001) and grade I-like (respectively log10 MC = 0 cells/ml and 5.4 cells/ml, p < 0.005 and p < 0.0001). Concentrations of G. vaginalis were also higher in grade I than in grade IV (p < 0.05).
Significant positive correlations were found in our data set between L. jensenii and L. gasseri (r = 0.244, p < 0.05), between G. vaginalis and L. iners (r = 0.351, p < 0.05) and between G. vaginalis and A. vaginae (r = 0.469, p < 0.0001). Negative correlations were found between L. gasseri and L. iners (r = -0.397, p = 0.001) and between A. vaginae and L. gasseri (r = -0.408, p < 0.0001).
Gram staining of L. iners and L. gasseri
Figure 3 represents the Gram stains of 6 strains of L. gasseri and 6 strains of L. iners. Gram stains of L. iners and L. gasseri showed that many of these strains displayed pleiomorphic cell morphology, i.e. different cell types could be observed within one microscopic field. Also, the cell morphologies of strains differed within the same species. Most strikingly, all L. iners strains had a relatively Gram-negative stain appearance and were mostly very short rods. By contrast, the L. gasseri were clearly Gram-positive in all cases. Some strains of L. gasseri formed streptobacillar chains, consisting of long to extra-long rods.
Figure 3.
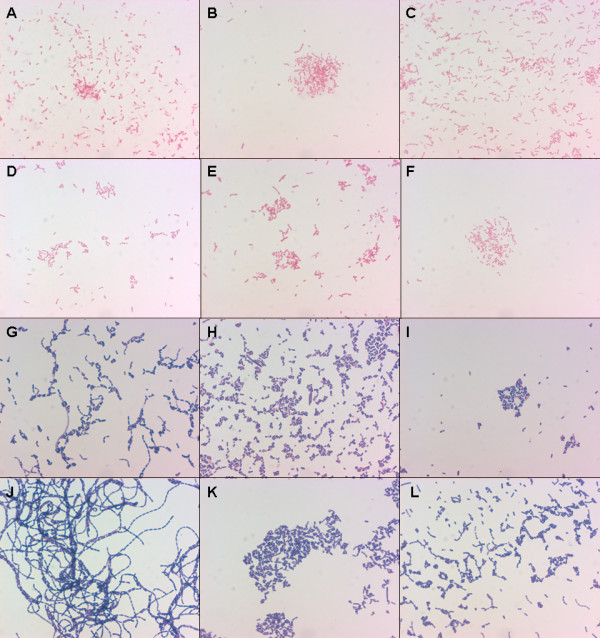
Microscopic images (100 ×) of Gram stains of L. iners and L. gasseri. A-F: L. iners; A: BVS11, B: FB04-04, C: FB06-01, D: FB17-07, E: FB94-05b and F: FB30-01AN; G-L: L. gasseri: FB02-02, H: FB33-01, I: FB34-01AN, J: FB89-3, K: FB103-2, L: PB88-T2-1.
MIC of L. gasseri and L. iners for clindamycin
The clindamycin MIC value for ten strains of L. iners and L. gasseri was determined using the agar dilution method. Eight of the ten L. iners had a MIC value of 0.125 μg/ml, one strain had a MIC value of 2 μg/ml and one strain of >8 μg/ml. For L. gasseri one strain had a MIC value of 0.5 μg/ml, two strains had MIC values of 2 μg/ml, six strains had MIC values of 4 μg/ml and for one strain the MIC was more than 8 μg/ml. In summary, 80% of L. iners isolates were inhibited by 0.125 μg of clindamycin/ml, with MIC50 = 0.125 μg/ml, whereas 90% of the L. gasseri isolates was resistant to 1 μg of clindamycin/ml, with MIC50 = 4 μg/ml.
Mutual inhibition of L. gasseri and L. iners
No inhibition zones were observed with the techniques we used.
Discussion
In this study we used real-time PCR to obtain a quantitative estimation of the presence of the four predominant Lactobacillus species known to occupy the vaginal econiche and of Atopobium vaginae and Gardnerella vaginalis.
Real-time PCR has been applied previously for the description of changes in the vaginal microflora. Zariffard et al. [27] and Sha et al. [28,29] used real-time PCR on frozen cervicovaginal lavage samples to quantify the presence of Mycoplasma hominis, G. vaginalis and the combined presence of L. crispatus and L. jensenii, in comparison with the microscopic interpretation of the Gram stain according to the criteria of Nugent [22]. Bradshaw et al. [16] applied real-time PCR for A. vaginae for a follow-up study of recurrent BV before and after treatment with oral metronidazole. Ferris et al. [17] applied real-time PCR for A. vaginae to samples of six BV patients before and after treatment with a topical metronidazole gel. Thus far, only one group (Byun et al. [26]), studying advanced dental caries, used real-time PCR to quantify the presence of specific Lactobacillus species.
Compared to microscopy and culture, molecular methods such as PCR may provide less observer-dependent and culture medium dependent information and a more direct and detailed view of the quantity of certain species in a sample. E.g., A. vaginae was only recently recognized as strongly linked to disturbed vaginal microflora because of its fastidious growth requirements in vitro. A possible additional advantage of PCR, compared to culture, is that it permits detection of dead or metabolically inactive bacteria, and as such it can be applied for the detection of bacteria in a dormant state, as is the case in biofilms, which occur also in BV as has been shown by means of FISH [12].
Besides culture-associated bias, difficulties with regard to the standardization of vaginal sampling make quantification of the vaginal microflora cumbersome and make bacterial counts reported in different publications difficult to compare. Some groups have attempted to standardize sampling by cervicovaginal lavage (CVL), whereby the volume of saline that is recovered from the vagina can be measured and used for calibration of the bacterial load/ml. Drawbacks are that CVL samples also from the cervix and does not represent a pure vaginal sample. Also it cannot be excluded that biofilm, that may be present in cases of BV, is recalcitrant with regard to dissolving in the saline that is used and is not sampled in the same manner as loosely associated microflora. In the condition of BV there may be more vaginal cell debris so it is feasible that there is more non-bacterial material present in the sample. In conclusion, vaginal sampling remains difficult to standardize. In this study we used unweighted vaginal swabs taken by the same gynecologist so there is no standardization for the volume of the sample, but standardization of the way samples are taken.
Besides sampling bias and culture bias also real-time PCR may be biased, because the conversion of the PCR positivity threshold value to a concentration of bacteria, depends on the manner the standard curve is calibrated. The standard curve, which is prepared by extracting DNA from a dense bacterial suspension and by serial tenfold dilution of this DNA extract, can be compared to i) the bacterial colony count, as determined by plating out tenfold serial dilutions of the initial suspension, or to ii) the number of bacterial genomes in the initial suspension, determined on the basis of measurement of the DNA-concentration present in the extract from the initial suspension. We found differences between calculation of number of genomes by measurement of DNA-concentration and determination of number of cells by dilution culture from less to one log10 unit (L. jensenii) to more than 3 log10 units (L. crispatus) (data not presented). A possible explanation may be that species like L. jensenii are more easily cultured on blood plates than for example L. crispatus and therefore show a lower discrepancy between DNA-concentration and culture. In conclusion, when preparation of the standard curve in this study had been based on cultured cells, L. jensenii would have been overestimated relative to the other species.
L. crispatus and L. jensenii
L. crispatus can be found in samples of all grades but the log10 median concentration/ml (MC) of this species is much higher in grade I (8.8) compared to grades II (4.2) and III (5.2). Although the difference between the log10 quantity of L. crispatus in grade Ia and grade Ib is not significantly different (p = 0.096), possibly because of the low sample size, a different profile is observed for these grades (Figure 2 and 3).
L. crispatus is present in high inoculum (i.e. at least) in all 8 grade Ia samples (log10 MC 9.0), in most (i.e. 6/10) grade Iab samples (log10 MC 8.9), but only in 3/12 grade Ib samples (log10 MC = 5.6). Our study confirms that a grade Ia microflora is characterized by high concentrations of L. crispatus, whereas grades Iab and Ib consist of a more diverse microflora, with different Lactobacillus species present in comparable amounts.
Also, in grade Ib, high concentrations of L. crispatus can be found but on Gram stain especially long and small Lactobacillus cell types, which were suggested to be more typical for L. gasseri [24], are observed.
Based on cloning of the 16S rDNA library from the indigenous microbiota of three women with normal vaginal microflora, Verhelst et al. [14] found an almost pure population of L. jensenii in one of them and concluded that this species was associated with a normal vaginal microflora, but in this study (71 swabs) we can not correlate L. jensenii with either normal or disturbed vaginal microflora since it is present sporadically in samples of all grades, and in both high and low numbers. Its presence in 8/10 grade Iab samples, compared to 1/8 grade Ia samples is striking (p = 0.024).
G. vaginalis and A. vaginae
A positive correlation between G. vaginalis and A. vaginae could be established (p < 0.0001). G. vaginalis can be found in all grades but in much higher concentration in a disturbed vaginal microflora. Delaney et al. [30] performed a comparative study in which they quantified G. vaginalis, Lactobacillus spp., Prevotella spp. and Peptostreptococcus spp. in vaginal swabs by means of culture. The mean log10 number of G. vaginalis in BV was determined to be 9.64 cfu/g, which may compare well to the value we obtained, i.e. 9.2 cells/ml in grade III.
A. vaginae is found less frequently than G. vaginalis in non-grade III samples, but in high concentrations in 10 of the 12 grade III samples, confirming that the presence of A. vaginae seems to be a diagnostically more valuable marker for BV than the presence of G. vaginalis, as suggested by Verhelst et al. [14] and confirmed by several other studies [31-33]. A recent study using T-RFLP for the characterization of normal (n = 20) and BV (n = 50) microflora found that the terminal restriction fragment (TRF) with the length to that corresponding of A. vaginae was present in the vagina of 48 of the 50 women with BV (grade II and III lumped together) and absent from all 20 women with normal microflora [32]. Bradshaw et al. [16] and Ferris et al. [17] monitored the changes in the concentration of A. vaginae before and after treatment with metronidazole and both found that very high vaginal concentrations of A. vaginae were predictive for patients for whom treatment failed partially or completely, although Bradshaw et al. [16] found a significant reduction of A. vaginae after metronidazole treatment. This might be due to a lower susceptibility of some strains of A. vaginae for metronidazole [34] but a more plausible explanation might be the existence of biofilm as shown by Swidsinski et al. [12], and in which G. vaginalis was shown to be associated with A. vaginae.
L. gasseri and L. iners
Overall L. gasseri and L. iners are abundantly present in most grades. Strikingly, L. gasseri is virtually absent in grade III whereas L. iners is virtually absent in grades Ia and II.
In our study we found a high concentration of L. iners to be clearly associated with low concentrations of L. gasseri, which is more prevalent in normal (grade Ia, grade Ib, grade Iab) and mildly disturbed flora (grade II). This was confirmed by Spearman rank testing, which showed that L. gasseri and L. iners are significantly negatively correlated to each other in grades II and III (r = -0.793, p < 0.0001), but also when all samples are considered together (r = -0.397, p = 0.001).
It can be noted that even the single grade III sample with a high concentration of L. gasseri was also one of the two grade III samples with low concentration of L. iners. In addition, we found a positive correlation between G. vaginalis and L. iners. The distribution frequencies over the different grades of L. iners and G. vaginalis are quite similar, i.e. present in most grades but predominantly in grade III, but different from A. vaginae, i.e. present almost exclusively in grade III, which may explain why L. iners does not correlate with A. vaginae. This distribution of L. iners was also observed in the T-RFLP study of Thies et al. [32], with 32 out of 50 BV samples and 11 out of 20 normal microflora samples containing the L. iners TRF. The L. gasseri group TRF was observed only twice, in women with normal microflora. Unfortunately, these authors did not distinguish between grade II and III.
According to some research groups [35], L. iners is the most abundant vaginal Lactobacillus species, not only in black African women (64% of 241 healthy premenopausal Nigerian women) but also in white women from Sweden, the US and Canada and is considered as even more typical for normal vaginal microflora than L. crispatus. Although L. iners was present in high numbers in grades Iab and Ib, we found L. crispatus and L. gasseri to be more abundant in grade Ia. Ferris et al. [17] concluded that L. iners is a more transitional species, present in recently cured patients. They reported the predominance of L. iners in 5 patients with BV after metronidazole treatment, whereas for the sixth patient, the only one with a complete treatment failure, L. iners was present but not predominant. These results were confirmed by Jakobsson and Forsum [36].
Interestingly, it has been observed by the group of Taylor-Robinson that grade II microflora responds poorly to clindamycin treatment whereas most subjects with grade III microflora readily reverted to grade I [37,38] and also that women with clindamycin treated grade III microflora had a better pregnancy outcome than women with clindamycin treated grade II microflora [37]. In agreement, the predominance of clindamycin susceptible L. iners in grade III might explain the observations of Taylor-Robinson et al. [38].
Finally, in case grade II would be an intermediate stage in between transition from grade I to grade III, one would expect to see an increase in L. iners and a decrease in L. gasseri, since the former is present in high numbers in grade III and the latter almost absent, but we observe exactly the opposite, i.e. nearly complete absence of L. iners and high numbers of L. gasseri in grade II.
In order to establish whether this could be related to the differential presence of L. gasseri and L. iners in these grades, with L. gasseri predominant in grade II, we determined the MIC for clindamycin for 10 strains of each. We found that 8 out of 10 L. iners strains were inhibited by less than 0.25 μg/ml, whereas 9 out of 10 L. gasseri strains had a MIC value of 2 or more μg clindamycin/ml. Although L. gasseri cannot be considered definitely as the causative micro-organism responsible for the adverse pregnancy outcome that has been associated with grade II, our data indicate that it can be considered as contributing to the poor response to clindamycin treatment.
To test the hypothesis that L. gasseri and L. iners are mutually exclusive, e.g. by the production of bacteriocins, we carried out inhibition tests between both species. However, with the method we used we could not observe mutual inhibition between both species.
With the use of molecular techniques it becomes clear that the definition for bacterial vaginosis in which it is claimed that the lactobacilli are replaced by anaerobic bacteria is not quite correct. Our data indicate that rather L. crispatus, L. gasseri and L. jensenii present in a healthy vaginal microflora and grade II microflora are replaced largely by L. iners in grade III vaginal microflora. Accordingly, the qualitative culture study by Delaney & Onderdonk [30] found a weak negative correlation between Nugent score and numbers of all lactobacilli, meaning that the number of lactobacillar cells did not decrease with high Nugent score, i.e. disturbed microflora. On the other hand, Zariffard et al. [27] and Sha et al. [28,29] who quantified lactobacilli in cervicovaginal lavage samples, using a real-time PCR format which picked up only L. crispatus and L. jensenii, found a decline in the number of lactobacilli in BV. The discrepancy may be explained because we take also L. iners and L. gasseri into account.
To further resolve these discrepancies we carried out Gram staining of several strains of each L. gasseri and L. iners. It is of importance to note that L. iners has not only long escaped from our attention due to its inability to grow on MRS agar, but also hides as a Lactobacillus from Gram stains, because it stains Gram-negative and its cell morphology is rather coccobacillar than bacillar. Since it is the predominant Lactobacillus species in BV microflora – even present in high numbers – this has led to the false assumption that the BV microflora is devoid of lactobacilli.
The very long lactobacillar cells observed for some of the L. gasseri isolates remind of earlier observations. Pahlson & Larsson [39] reported the occurrence of very long fusiform lactobacilli in 7.4% of the vagina of 981 women studied. Horowitz et al. [40] reported the occurrence of 'vaginal lactobacillosis', a pathological condition characterized by the presence of extremely long lactobacilli (up to 60 μm).
Grade I-like was described only recently by Verhelst et al. [24] as a grade that, on Gram stain, resembles grade I because of the Gram positive rods, but with the use of molecular methods it was shown that these rods were Bifidobacterium spp. instead of Lactobacillus spp.
Again this study confirms the separate nature of grade I-like vaginal microflora. A significantly lower concentration of L. crispatus (log10 MC = 5.5 cells/ml) is present compared to grade I (log10 MC = 8.8 cells/ml, p = 0.014) and much lower concentrations of G. vaginalis (log10 MC = 4.0 cells/ml, p < 0.0001) and A. vaginae (log10 MC = 1.6 cells/ml, p = 0.003) are present in grade I-like microflora compared to grade III (respectively log10 MC = 9.2 cells/ml and log10 MC = 9.0 cells/ml).
Conclusion
We report a strong negative association between L. iners and L. gasseri, whereas L. gasseri is predominant in grade II and L. iners in grade III. It has been mentioned by others [5] that a grade II microflora is not necessarily intermediate between grade I and III.
L. iners and L. gasseri do not inhibit each other by production of bacteriocins, according to the technique we used. In addition we found L. iners to be very sensitive for clindamycin (MIC50 = 0.125 μg/ml) compared to the higher MIC value for L. gasseri (MIC50 = 4 μg/ml). Comparison of these results with findings of other research groups is very difficult because L. iners has long been overlooked in culture and Gram stain. The unexpected Gram negative pleiomorphic cell shape of L. iners, as established in this study, and its inability to grow on MRS agar made the estimation of the occurrence of this species almost impossible until the recent development of molecular techniques.
Methods
Strains and culture conditions
The specificity of the primer set for Lactobacillus iners was tested using the following strains: L. iners (CCUG 28746T), L. brevis (LB4), L. delbrueckii (LB31), L. crispatus (ATCC 33199T), L. casei (ATCC 393T), L. fermentum (ATCC 23271, ATCC 11739T, SAV1, SAV 3, SAV5), L. gasseri (ATCC 33323T), L. jensenii (ATCC 25258T), L. johnsonii (DSM 20553), L. plantarum (ATCC 8014, ATCC 10012, ATCC 14917T), L. reuteri (ATCC 23272T, RC-14), L. rhamnosus (ATCC 7469T, GR-1, GG) and L. salivarius (SAV6).
The specificity of the primer set for Lactobacillus jensenii was tested using the following strains:L. acidophilus (LMG 11430), L. animalis (LMG 9843T), L. bifermentans (LMG 11431), L. brevis (LMG 7761), L. casei (FB86-16), L. coleohominis (PB2003/024-T1-1a), L. crispatus (LMG 9479T), L. curvatus (0606 93811c), L. delbrueckii (LMG 6412T), L. fermentum (LMG 6902T), L. gasseri (LMG 9203T), L. helveticus (LBP152), L. iners (BVS11, PB2003/044-T1-1, PB2003/53-T1-1), L. intestinalis (LMG 14196T), L. jensenii (PB2003/204-T1-1, FB65-6, FB122-BP-1, FB146-BA-5, FB154-CAN-2), L. johnsonii (LBP151), L. plantarum (LMG 9212), L. reuteri (LMG 9213T), L. rhamnosus (LMG 8153, LMG18243), L. salivarius (LMG 9477T) and L. vaginalis (LMG 12891T).
The specificity of the primer set for Gardnerella vaginalis was tested using the following strains: Bifidobacterium infantis (PB2003/126-T1-3), B. breve (PB2003/108-T1-1) and B. bifidum (PB2003/99-T1-2).
The specificity of the primer set for Atopobium vaginae was tested using the following strains: A. minutum (CCUG 31167), A. parvulum (CCUG 32760), A. rimae (CCUG 31168) and A. vaginae (CCUG 38953T, CCUG 42099, CCUG 44116, CCUG 44125, PB2003/189-T1-4). All strains were cultured on Trypticase soy agar with 5% sheep blood (Becton Dickinson, Erembodegem, Belgium) in anaerobic conditions (Gaspak, Becton Dickinson) for 72 hours at 37°C.
Samples
A total of 142 swabs (Copan, Brescia, Italy) were collected by sampling 60 women of childbearing age (32 pregnant women, 28 non-pregnant women). Sampling was carried out by insertion of two sterile cotton swabs into the vaginal vault, after placement of a non-lubricated speculum. The swabs were rotated against the vaginal wall at the mid-portion of the vault and were carefully removed to prevent contamination with microflora of the vulva and introitus. The first swab was used to prepare a smear on a glass slide for the purpose of grading as described by Verhelst et al. [14]. The second swab was returned to a sterile tube for the purpose of DNA extraction (dry swab). Both swabs were sent to the microbiology laboratory and were processed within 4 hours.
The study protocol was approved by the ethical committee of the Ghent University Hospital and individual written informed consent was obtained.
DNA extraction of bacteria and samples
DNA was extracted from cultured bacteria by simple alkaline lysis. One colony was suspended in 20 μl of lysis buffer (0.25% SDS, 0.05 N NaCl and 95 ml sterile distilled water) and heated for 15 minutes at 95°C. After briefly centrifugating, 180 μl of HPCL-water was added and this was centrifuged again for 5 minutes at 16300 g. Concentrations of DNA-extracts ranged between 2.1 and 87.5 ng/μl.
For dry vaginal swabs, the QIAamp DNA mini kit (Qiagen, Hilden, Germany) was used according to the manufacturer's recommendations, with minor modifications. The dry swab specimen from each patient was swirled for 15 s in 400 μl of lysis buffer (20 mM Tris-HCl, pH 8.0; 2 mM EDTA; 1.2% Triton). Fifty units of mutanolysin (25 U/μl) (Sigma, Bornem, Belgium) were added and the samples were incubated for 30 min at 37°C. After the addition of 20 μl Proteinase K (20 mg/ml) and 200 μl AL buffer (Qiagen), samples were incubated for 30 min at 56°C. Next, 200 μl of ethanol was added and DNA was purified by adding the lysate to the Qiagen columns as described by the manufacturer. Finally, the total bacterial DNA was eluted with 100 μl of AE buffer (Qiagen). DNA extracts were aliquoted and stored at -20°C.
Primers
PCR primer sets targeting L. iners, L. jensenii and A. vaginae were designed by aligning the sequences of the 16S rRNA gene of different strains of both species by CLUSTALW [41]. Specificity of the primer sets (Eurogentec, Seraing, Belgium) was analyzed by using the BLAST algorithm [42]. Primer sequences for L. jensenii and L. iners alignment to the most common vaginal lactobacilli are shown in Table 1 and Table 2. Primer sequences for A. vaginae and alignment to other members of the Atopobium group are shown in Table 3. Position of the primers on the 16S rRNA-gene is shown on Figure 1. Optimal annealing temperature of each primer set was established by gradient PCR and analysis on an agarose gel. In addition, already described primers for L. crispatus and L. gasseri [26] and G. vaginalis [27] were used. Primer sequences and cycling conditions are summarized in Table 4. Primer concentrations were 100 nM each except for the assays for G. vaginalis and L. iners for which the primer concentrations were 200 nM.
Construction of the standard curves for real-time PCR
To construct standard curves for the real-time PCR's, A. vaginae (CCUG 38953T), L. crispatus (PB2003/125-T1-1), L. gasseri (FB102-1) and L. jensenii (PB2003/204-T1-1) were cultured on TSA + 5% sheep blood (Becton Dickinson) and G. vaginalis (LMG 7832T) and L. iners (BVS11) were cultured on CNA agar (Becton Dickinson) + 5% human blood. A suspension was made in MRS Broth (Oxoid, Drongen, Belgium) and DNA was extracted. The DNA concentration of this stock was determined ten times by using the Nanodrop ND-1000 (Nanodrop Technologies, Wilmington, USA) and the mean value was used for further calculations. For each strain a tenfold dilution series was prepared by dilution of the DNA stock in HPLC grade water. Dilutions were aliquoted and stored frozen at -20°C.
Real-time PCR
The qPCR Core Kit for SYBR Green I (Eurogentec) was applied and analysis was performed on the ABI 7300 real-time PCR system (Applied Biosystems, Foster City, CA).
Reactions were done in PCR mixtures containing 2.5 μl of DNA extract, 2.5 μl of 10 × Reaction Buffer, 3.5 mM MgCl2, 0.2 mM dNTP mixture, 0.625 U HotGoldStar Taq polymerase, 0.75 μl SYBR® Green I, diluted 10-fold in DMSO and the appropriate primer concentration, adjusted with HPLC grade water to 25 μl. Primer concentrations are summarized in Table 4. Each run included a standard curve and each sample was run in triplicate. In case the result was not in the range of the standard curve, the samples were diluted tenfold and analyzed in triplicate again. The median log10 cells/ml were expressed as per 1 ml elution buffer.
Agar dilution method
Clindamycin (Sigma, Bornem, Belgium) minimal inhibitory concentrations (MIC) were determined for 10 strains of L. gasseri (FB02-02, FB13-01, FB29-6, FB34-01AN, FB86-6, FB89-3, FB102-1b, FB103-2, PB88-T2-1 and PB327-1) and 10 strains of L. iners (BVS11, FB04-04, FB05-01, FB06-01, FB07-01, FB17-07, FB30-01AN, FB77-03, FB94-05b and FB123-CNA-4) using the agar dilution method on Trypticase Soy Agar (Becton Dickinson) + 5% human blood and incubation anaerobically during 72 hours at 37°C in the Anaerobic Workstation BugBoxPlus (LED Techno, Heusden-Zolder, Belgium). Concentrations of 8, 4, 2, 1, 0.5, 0.250, 0.125 and 0.0625 μg/ml, were tested for each strain in triplicate.
Mutual inhibition test
The same strains used for MIC-determination were grown in Trypticase Soy Broth (Becton Dickinson) + 5% human blood for 72 hours at 37°C in the anaerobic workstation BugBoxPlus (LED Techno, Heusden-Zolder, Belgium). Supernatant of the liquid cultures and of the negative control were sterilized trough a 0.22 μm filter (Spin-X Centrifuge Tube Filter, Costar, Corning, NY14831). Plates were inoculated with a swab to obtain confluent growth. Volumes of 20 μl of the filter sterilized supernatants were dropped onto the inoculated plates, followed by anaerobic incubation at 37°C for 72 hours, whereafter inhibition zones were recorded.
Data analysis
For the calculation of number of cells in each dilution, genome size and % G+C of the bacterial species was taken into account. The genome size of G. vaginalis (ATCC 14019, ATCC 14018, GVP 007 and GVP 004) has been approximated to 1.7 Mb [43] and % G+C was 43 [44]. For L. gasseri data described for strain ATCC 33323 (genome size: 1.9 Mb, 35.3% G+C) [45] were used. Also for A. vaginae strain CCUG 38953T (44% G+C) [46], L. crispatus strain ATCC 33820T (35% G+C) [47], L. iners strain CCUG 28746T (34.4% G+C) [20] and L. jensenii strain ATCC 25258T (36.1% G+C) [48] a genome size of 1.9 Mb was assumed.
Statistical analysis
Data were analyzed under the non-parametric assumption, taking into consideration the log10 [count] distributions of species under study did not approximate the normal distribution. For any given category (grades I-IV) the distribution of concentrations (log10 cells/ml) of each species was expressed as the median count and the accompanying interquartile range (IQR). Between group comparisons of distributions were performed with the Mann-Whitney U-test for two groups and with the Kruskal Wallis test for multiple groups. Correlations between the different species were determined by the Spearman (rank) test and reported as Spearman's rho value (r). All analyses were performed using SPSS v15 software (Chicago, Illinois).
Authors' contributions
EDB, RV and MV participated in the development of the study design, the analysis of the study samples, the collection, analysis and interpretation of the data, and in the writing of the report. HV and MT participated in the development of the study design, the collection of the study samples, the collection, analysis and interpretation of the data, and in the writing of the report. MAQ, JB and JRT developed the L. iners primers, participated in analysis and interpretation of the data, and in the writing of the report. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
Ellen De Backer is indebted for a PhD Research funded by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen).
Contributor Information
Ellen De Backer, Email: ellekendb@yahoo.com.
Rita Verhelst, Email: Rita.Verhelst@UGent.be.
Hans Verstraelen, Email: Hans.Verstraelen@UGent.be.
Mohammed A Alqumber, Email: alqmo407@student.otago.ac.nz.
Jeremy P Burton, Email: jeremy.burton@blis.co.nz.
John R Tagg, Email: john.tagg@stonebow.otago.ac.nz.
Marleen Temmerman, Email: Marleen.Temmerman@UGent.be.
Mario Vaneechoutte, Email: Mario.Vaneechoutte@UGent.be.
References
- Morris M, Nicoll A, Simms I, Wilson J, Catchpole M. Bacterial vaginosis: a public health review. BJOG. 2001;108:439–450. doi: 10.1016/S0306-5456(00)00124-8. [DOI] [PubMed] [Google Scholar]
- Haggerty CL, Hillier SL, Bass DC, Ness RB. Bacterial vaginosis and anaerobic bacteria are associated with endometritis. Clin Infect Dis. 2004;39:990–995. doi: 10.1086/423963. [DOI] [PubMed] [Google Scholar]
- Ness RB, Kip KE, Hillier SL, Soper DE, Stamm CA, Sweet RL, Rice P, Richter HE. A cluster analysis of bacterial vaginosis-associated microflora and pelvic inflammatory disease. Am J Epidemiol. 2005;162:585–590. doi: 10.1093/aje/kwi243. [DOI] [PubMed] [Google Scholar]
- Watts DH, Krohn MA, Hillier SL, Eschenbach DA. Bacterial vaginosis as a risk factor for post-cesarean endometritis. Obstet Gynecol. 1990;75:52–58. [PubMed] [Google Scholar]
- Hay PE, Lamont RF, Taylor-Robinson D, Morgan DJ, Ison C, Pearson J. Abnormal bacterial colonisation of the genital tract and subsequent preterm delivery and late miscarriage. BMJ. 1994;308:295–298. doi: 10.1136/bmj.308.6924.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier SL, Nugent RP, Eschenbach DA, Krohn MA, Gibbs RS, Martin DH, Cotch MF, Edelman R, Pastorek JG, 2nd, Rao AV, et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. N Engl J Med. 1995;333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- Leitich H, Bodner-Adler B, Brunbauer M, Kaider A, Egarter C, Husslein P. Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am J Obstet Gynecol. 2003;189:139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- Pretorius C, Jagatt A, Lamont RF. The relationship between periodontal disease, bacterial vaginosis, and preterm birth. J Perinat Med. 2007;35:93–99. doi: 10.1515/JPM.2007.039. [DOI] [PubMed] [Google Scholar]
- Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B, Mandaliya K, Ndinya-Achola JO, Bwayo J, Kreiss J. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis. 1999;180:1863–1868. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- Sewankambo N, Gray RH, Wawer MJ, Paxton L, McNaim D, Wabwire-Mangen F, Serwadda D, Li C, Kiwanuka N, Hillier SL, Rabe L, Gaydos CA, Quinn TC, Konde-Lule J. HIV-1 infection associated with abnormal vaginal flora morphology and bacterial vaginosis. Lancet. 1997;350:546–550. doi: 10.1016/S0140-6736(97)01063-5. [DOI] [PubMed] [Google Scholar]
- Hawes SE, Hillier SL, Benedetti J, Stevens CE, Koutsky LA, Wolner-Hanssen P, Holmes KK. Hydrogen peroxide-producing lactobacilli and acquisition of vaginal infections. J Infect Dis. 1996;174:1058–1063. doi: 10.1093/infdis/174.5.1058. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Mendling W, Loening-Baucke V, Ladhoff A, Swidsinski S, Hale LP, Lochs H. Adherent biofilms in bacterial vaginosis. Obstet Gynecol. 2005;106:1013–1023. doi: 10.1097/01.AOG.0000183594.45524.d2. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Masztal A, Aldridge KE, Fortenberry JD, Fidel PL, Jr., Martin DH. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect Dis. 2004;4:5. doi: 10.1186/1471-2334-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Delanghe J, Van Simaey L, De Ganck C, Temmerman M, Vaneechoutte M. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 2004;4:16. doi: 10.1186/1471-2180-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JP, Devillard E, Cadieux PA, Hammond JA, Reid G. Detection of Atopobium vaginae in postmenopausal women by cultivation-independent methods warrants further investigation. J Clin Microbiol. 2004;42:1829–1831. doi: 10.1128/JCM.42.4.1829-1831.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw CS, Tabrizi SN, Fairley CK, Morton AN, Rudland E, Garland SM. The association of Atopobium vaginae and Gardnerella vaginalis with bacterial vaginosis and recurrence after oral metronidazole therapy. J Infect Dis. 2006;194:828–836. doi: 10.1086/506621. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Norori J, Zozaya-Hinchliffe M, Martin DH. Cultivation-independent analysis of changes in bacterial vaginosis flora following metronidazole treatment. J Clin Microbiol. 2007;45:1016–1018. doi: 10.1128/JCM.02085-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay PE. Recurrent bacterial vaginosis. Dermatol Clin. 1998;16:769–73, xii-xiii. doi: 10.1016/S0733-8635(05)70044-9. [DOI] [PubMed] [Google Scholar]
- Antonio MA, Hawes SE, Hillier SL. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J Infect Dis. 1999;180:1950–1956. doi: 10.1086/315109. [DOI] [PubMed] [Google Scholar]
- Falsen E, Pascual C, Sjoden B, Ohlen M, Collins MD. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int J Syst Bacteriol. 1999;49 Pt 1:217–221. doi: 10.1099/00207713-49-1-217. [DOI] [PubMed] [Google Scholar]
- Spiegel CA, Amsel R, Holmes KK. Diagnosis of bacterial vaginosis by direct gram stain of vaginal fluid. J Clin Microbiol. 1983;18:170–177. doi: 10.1128/jcm.18.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol. 1991;29:297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ison CA, Hay PE. Validation of a simplified grading of Gram stained vaginal smears for use in genitourinary medicine clinics. Sex Transm Infect. 2002;78:413–415. doi: 10.1136/sti.78.6.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Van Simaey L, De Ganck C, De Backer E, Temmerman M, Vaneechoutte M. Comparison between Gram stain and culture for the characterization of vaginal microflora: definition of a distinct grade that resembles grade I microflora and revised categorization of grade I microflora. BMC Microbiol. 2005;5:61. doi: 10.1186/1471-2180-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraelen H, Verhelst R, Roelens K, Claeys G, Weyers S, De Backer E, Vaneechoutte M, Temmerman M. Modified classification of Gram-stained vaginal smears to predict spontaneous preterm birth: a prospective cohort study. Am J Obstet Gynecol. 2007;196:528 e1–6. doi: 10.1016/j.ajog.2006.12.026. [DOI] [PubMed] [Google Scholar]
- Byun R, Nadkarni MA, Chhour KL, Martin FE, Jacques NA, Hunter N. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J Clin Microbiol. 2004;42:3128–3136. doi: 10.1128/JCM.42.7.3128-3136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariffard MR, Saifuddin M, Sha BE, Spear GT. Detection of bacterial vaginosis-related organisms by real-time PCR for Lactobacilli, Gardnerella vaginalis and Mycoplasma hominis. FEMS Immunol Med Microbiol. 2002;34:277–281. doi: 10.1111/j.1574-695X.2002.tb00634.x. [DOI] [PubMed] [Google Scholar]
- Sha BE, Chen HY, Wang QJ, Zariffard MR, Cohen MH, Spear GT. Utility of Amsel criteria, Nugent score, and quantitative PCR for Gardnerella vaginalis, Mycoplasma hominis, and Lactobacillus spp. for diagnosis of bacterial vaginosis in human immunodeficiency virus-infected women. J Clin Microbiol. 2005;43:4607–4612. doi: 10.1128/JCM.43.9.4607-4612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha BE, Zariffard MR, Wang QJ, Chen HY, Bremer J, Cohen MH, Spear GT. Female genital-tract HIV load correlates inversely with Lactobacillus species but positively with bacterial vaginosis and Mycoplasma hominis. J Infect Dis. 2005;191:25–32. doi: 10.1086/426394. [DOI] [PubMed] [Google Scholar]
- Delaney ML, Onderdonk AB. Nugent score related to vaginal culture in pregnant women. Obstet Gynecol. 2001;98:79–84. doi: 10.1016/S0029-7844(01)01402-8. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Masztal A, Martin DH. Use of species-directed 16S rRNA gene PCR primers for detection of Atopobium vaginae in patients with bacterial vaginosis. J Clin Microbiol. 2004;42:5892–5894. doi: 10.1128/JCM.42.12.5892-5894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies FL, Konig W, Konig B. Rapid characterization of the normal and disturbed vaginal microbiota by application of 16S rRNA gene terminal RFLP fingerprinting. J Med Microbiol. 2007;56:755–761. doi: 10.1099/jmm.0.46562-0. [DOI] [PubMed] [Google Scholar]
- Burton JP, Chilcott CN, Al-Qumber M, Brooks HJ, Wilson D, Tagg JR, Devenish C. A preliminary survey of Atopobium vaginae in women attending the Dunedin gynaecology out-patients clinic: is the contribution of the hard-to-culture microbiota overlooked in gynaecological disorders? Aust N Z J Obstet Gynaecol. 2005;45:450–452. doi: 10.1111/j.1479-828X.2005.00456.x. [DOI] [PubMed] [Google Scholar]
- De Backer E, Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Temmerman M, Vaneechoutte M. Antibiotic susceptibility of Atopobium vaginae. BMC Infect Dis. 2006;6:51. doi: 10.1186/1471-2334-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anukam KC, Osazuwa EO, Ahonkhai I, Reid G. Lactobacillus vaginal microbiota of women attending a reproductive health care service in Benin city, Nigeria. Sex Transm Dis. 2006;33:59–62. doi: 10.1097/01.olq.0000175367.15559.c4. [DOI] [PubMed] [Google Scholar]
- Jakobsson T, Forsum U. Lactobacillus iners; a marker of changes in the vaginal flora? J Clin Microbiol. 2007 doi: 10.1128/JCM.00558-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein IJ, Morgan DJ, Lamont RF, Sheehan M, Dore CJ, Hay PE, Taylor-Robinson D. Effect of intravaginal clindamycin cream on pregnancy outcome and on abnormal vaginal microbial flora of pregnant women. Infect Dis Obstet Gynecol. 2000;8:158–165. doi: 10.1002/1098-0997(2000)8:3/4<158::AID-IDOG11>3.0.CO;2-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Robinson D, Morgan DJ, Sheehan M, Rosenstein IJ, Lamont RF. Relation between Gram-stain and clinical criteria for diagnosing bacterial vaginosis with special reference to Gram grade II evaluation. Int J STD AIDS. 2003;14:6–10. doi: 10.1258/095646203321043183. [DOI] [PubMed] [Google Scholar]
- Pahlson C, Larsson PG. The ecologically wrong vaginal lactobacilli. Med Hypotheses. 1991;36:126–130. doi: 10.1016/0306-9877(91)90253-U. [DOI] [PubMed] [Google Scholar]
- Horowitz BJ, Mardh PA, Nagy E, Rank EL. Vaginal lactobacillosis. Am J Obstet Gynecol. 1994;170:857–861. doi: 10.1016/s0002-9378(94)70298-5. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Lim D, Trivedi H, Nath K. Determination of Gardnerella vaginalis genome size by pulsed-field gel electrophoresis. DNA Res. 1994;1:115–122. doi: 10.1093/dnares/1.3.115. [DOI] [PubMed] [Google Scholar]
- Eschenbach DA. Bacterial vaginosis and anaerobes in obstetric-gynecologic infection. Clin Infect Dis. 1993;16 Suppl 4:S282–7. doi: 10.1093/clinids/16.supplement_4.s282. [DOI] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Jovita M, Collins MD, Sjoden B, Falsen E. Characterization of a novel Atopobium isolate from the human vagina: description of Atopobium vaginae sp. nov. Int J Syst Bacteriol. 1999;49 Pt 4:1573–1576. doi: 10.1099/00207713-49-4-1573. [DOI] [PubMed] [Google Scholar]
- Cato EP, C MWE, Johnson JL. Synonymy of strains of "Lactobacillus acidophilus" group A2 (Johnson et al. 1980) with the type strain of Lactobacillus crispatus (Brygoo and Aladame 1953) Moore and Holdeman 1970. Int J Syst Bacteriol. 1983;33:426–428. [Google Scholar]
- Gasser F, Mandel M, Rogosa M. Lactobacillus jensenii sp.nov., a new representative of the subgenus Thermobacterium. J Gen Microbiol. 1970;62:219–222. doi: 10.1099/00221287-62-2-219. [DOI] [PubMed] [Google Scholar]