Abstract
While hard ticks (Ixodidae) take several days to feed on their hosts, soft ticks (Argasidae) feed faster, usually taking less than one hour per meal. Saliva assists in the feeding process by providing a cocktail of anti-hemostatic, anti-inflammatory and immunomodullatory compounds. Saliva of hard ticks has been shown to contain several families of genes each having multiple members, while those of soft ticks are relatively unexplored.
Analysis of the salivary transcriptome of the soft tick Ornithodorus parkeri, the vector of the relapsing fever agent Borrelia parkeri, indicates that gene duplication events have led to a large expansion of the lipocalin family, as well as of several genes containing Kunitz domains indicative of serine protease inhibitors, and several other gene families also found in hard ticks. Novel protein families with sequence homology to insulin growth factor-binding protein (prostacyclin-stimulating factor), adrenomedulin, serum amyloid A protein precursor and similar to HIV envelope protein were also characterized for the first time in the salivary gland of a blood-sucking arthropod.
The sialotranscriptome of O. parkeri confirms that gene duplication events are an important driving force in the creation of salivary cocktails of blood-feeding arthropods, as was observed with hard ticks and mosquitoes. Most of the genes coding for expanded families are homologous to those found in hard ticks, indicating a strong common evolutionary path between the two families. As happens to all genera of blood-sucking arthropods, several new proteins were also found, indicating the process of adaptation to blood feeding still continues to recent times.
Keywords: Ornithodorus parkeri, Ixodidae, Argasidae, Sialotranscriptomes, salivary gland transcriptome, sialome, Tick salivary glands
1. Introduction
Ticks are specialized mites in the suborder Ixodida of the order Parasitiformes. They are unique among mites by being larger, having specialized mouthparts and being obligate ectoparasites of terrestrial vertebrates including amphibians, reptiles, birds and mammals.
Two major families exist among ticks, the Argasidae (soft ticks) and the Ixodidae (hard ticks) (Black and Piesman, 1994). Phylogeny based on 16S rDNA sequences indicated that the two families are monophyletic, and indicated a divergence time no earlier than the late Jurassic (140 million years ago) (Black and Piesman, 1994), although other authors suggest earlier divergence (late Permian, 245 million years ago) based on the radiation of reptiles (Hoogstraal and Aechlimann, 1982)
The adaptation to blood feeding involves evolution of a complex cocktail of salivary components that help the parasite to overcome their host’s defenses against blood loss (hemostasis) and the development of inflammatory reactions at the feeding site that may disrupt blood flow or trigger host-defensive behavior by the sensation of pain or itching. Accordingly, saliva of blood-sucking arthropods contain anti-clotting, anti-platelet, vasodilatory, anti-inflammatory and immunomodullatory components, usually in redundant amounts (Ribeiro and Francischetti, 2003).
Salivary transcriptomes (sialotranscriptomes) of several hard tick species have been characterized to date (Alarcon-Chaidez et al., 2007; Francischetti et al., 2005; Lambson et al., 2005; Nene et al., 2004b; Ribeiro et al., 2006; Santos et al., 2004). These databases indicate a large expansion of genes coding for several protein families, including metalloproteases, cysteine-rich proteins similar to metalloprotease domains (the ixostatins and ixodegrins), lipocalins, Kunitz-domain containing proteins, RGD containing peptides, defensins, and many novel protein families that may include antimicrobial proteins. In Ixodes scapularis and I. pacificus alone, several hundred putative secreted salivary peptides were identified, most of which belong to nearly one dozen expanded gene families (Francischetti et al., 2005; Ribeiro et al., 2006). However, comparative data on Argasidae sialomes are nonexistent. In this paper, we report the transcriptome and proteome analysis of Ornithodorus parkeri, a vector of Borrelia parkeri, an agent of relapsing fever in the Western United States.
2. Materials and methods
2.1. Ticks
Tick salivary gland extracts were prepared by collecting glands from O. parkeri late instar nymphs and adult ticks. Glands were dissected by first bisecting the tick and then teasing the salivary glands away from the other internal organs and the tick exoskeleton. Glands were rinsed by immersion in PBS and added to 10 ml of PBS, DW or RNA later and stored frozen at −75 °C until further analysis.
2.2. Chemicals
Standard laboratory chemicals were purchased from Sigma Chemicals (St. Louis, MO) if not specified otherwise. Formic acid and trifluoroacetic acid (TFA) were obtained from Fluka (Milwaukee, WI). Trypsin was purchased from Promega (Madison, WI). HPLC-grade acetonitrile was from EM Science (Darmstadt, Germany), and water was purified by a Barnstead Nanopure system (Dubuque, IA).
2.3. Salivary gland isolation and library construction
Ornithodorus parkeri salivary gland mRNA from 50 pairs of glands was isolated using the Micro-FastTrack mRNA isolation kit (Invitrogen, San Diego, CA). The PCR-based cDNA library was made following the instructions for the SMART cDNA library construction kit (Clontech, Palo Alto, CA). This system utilizes oligoribonucleotide (SMART IV) to attach an identical sequence at the 5’ end of each reverse-transcribed cDNA strand. This sequence is then utilized in subsequent PCR reactions and restriction digests.
First-strand synthesis was carried out using PowerScript reverse transcriptase at 42 °C for 1 hr in the presence of the SMART IV and CDS III (3’) primers. Second-strand synthesis was performed by a long distance (LD) PCR-based protocol, using Advantage ™ Taq Polymerase (Clontech) mix in the presence of the 5’ PCR primer and the CDS III (3’) primer. The cDNA synthesis procedure resulted in the creation of SfiI A & B restriction enzyme sites at the ends of the PCR products that are used for cloning into the phage vector. The PCR conditions were: 95 °C for 20 sec; 24 cycles of 95 °C for 5 sec., 68 °C for 6 min. A small portion of the cDNA obtained by PCR was analysed on a 1.1% agarose gel to check for the quality and range of cDNA synthesised. Double-stranded cDNA was immediately treated with proteinase K (0.8 μg/ml) at 45 °C for 20 min and the enzyme was removed by ultrafiltration though a Microcon (Amicon) YM-100 centrifugal filter device. The cleaned, double-stranded cDNA was then digested with SfiI at 50 °C for 2 hrs, followed by size fractionation on a ChromaSpin– 400 column (Clontech, Palo Alto, CA). The profile of the fractions was checked on a 1.1% agarose gel and fractions containing cDNAs of more than 400 bp were pooled and concentrated using a Microcon YM-100.
The cDNA mixture was ligated into the λ TriplEx2 vector (Clontech, Palo Alto, CA) and the resulting ligation mixture was packaged using the GigaPack® III Plus packaging extract (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. The packaged library was plated by infecting log phase XL1- Blue E. coli cells (Clontech, Palo Alto, CA). The percentage of recombinant clones was determined by performing a blue-white selection screening on LB/MgSO4 plates containing X-gal / IPTG. Recombinants were also determined by PCR, using vector primers (5’ λ TriplEx2 Sequencing Primer and 3’ λ TriplEx2 Sequencing) flanking the inserted cDNA and visualising the products on a 1.1% agarose / EtBr gel.
2.4. Sequencing of the O. parkeri cDNA library
The O. parkeri salivary gland cDNA library was plated on LB/MgSO4 plates containing X-gal / IPTG, to an average of 250 plaques per 150 mm Petri plate. Recombinant (white) plaques were randomly selected and transferred to 96-well MICROTEST ™ U Bottom plates (BD BioSciences, Franklin Lakes, NJ), containing 100 μl of SM buffer [0.1 M NaCl; 0.01 M MgSO4; 7 H2 O; 0.035 M Tris-HCl (pH 7.5); 0.01% gelatin] per well. The plates were covered and placed on a gyrating shaker for 30 min at room temperature. The phage suspension was either immediately used for PCR or stored at 4 °C for future use.
To amplify the cDNA using a PCR reaction, four microliters of the phage sample was used as a template. The primers were sequences from the λ TriplEx2 vector and named pTEx2 5seq (5’-TCC GAG ATC TGG ACG AGC - 3’) and pTEx2 3LD (5’- ATA CGA CTC ACT ATA GGG CGA ATT GGC - 3’), positioned at the 5’ end and the 3’ end of the cDNA insert, respectively. The reaction was carried out in 96-well flexible PCR plates (Fisher Scientific, Pittsburgh, PA) using the TaKaRa EX Taq polymerase (TAKARA Mirus Bio, Madison, WI), on a Perkin Elmer GeneAmp® PCR system 9700 (Perkin Elmer Corp., Foster City, CA). The PCR conditions were: one hold of 95 °C for 3 min; 25 cycles of 95 °C for 1 min, 61 °C for 30 sec; 72 °C for 2 min. The amplified products were analysed on a 1.5% Agarose / EtBr gel. 1100 cDNA library clones were PCR amplified and the ones showing single band were selected for sequencing. Approximately 200–250 ng of each PCR product was transferred to Thermo-Fast 96-well PCR plates (ABgene Corp., Epsom, Surray, UK) and frozen at - 20 °C, before cycle sequencing using an ABI3730XL machine.
2.5. Bioinformatic tools and procedures used
Expressed sequence tags (EST) were trimmed of primer and vector sequences, clusterised, and compared with other databases as described (Valenzuela et al., 2003). The BLAST tool (Altschul et al., 1990), CAP3 assembler (Huang and Madan, 1999), ClustalW (Thompson et al., 1997), and Treeview software (Page, 1996) were used to compare, assemble, and align sequences and to visualise alignments. For functional annotation of the transcripts we used the tool BlastX (Altschul et al., 1997) to compare the nucleotide sequences to the NR protein database of the National Center for Biotechnology Information (NCBI) and to the Gene Ontology (GO) database (Ashburner et al., 2000). The tool, Reverse Position Specific Blast (RPSBLAST), (Altschul et al., 1997) was used to search for conserved protein domains in the Pfam (Bateman et al., 2000), SMART (Schultz et al., 2000), Kog (Tatusov et al., 2003), and Conserved Domains Databases (CDD) (Marchler-Bauer et al., 2002). We have also compared the transcripts with other subsets of mitochondrial and rRNA nucleotide sequences downloaded from NCBI, and to several organism proteomes downloaded from NCBI (yeast), Flybase (Drosophila melanogaster), or ENSEMBL (An. gambiae). Segments of the three-frame translations of the EST (because the libraries were unidirectional we did not use six-frame translations), starting with a methionine found in the first 300 predicted amino acids (AA), or to the predicted protein translation in the case of complete coding sequences, were submitted to the SignalP server (Nielsen et al., 1997) to help identify translation products that could be secreted. O-glycosylation sites on the proteins were predicted with the program NetOGlyc (Hansen et al., 1998). Functional annotation of the transcripts was based on all the comparisons above. Following inspection of all these results, transcripts were classified as either Secretory (S), Housekeeping (H) or of Unknown (U) function, with further subdivisions based on function and/or protein families. Phylogenetic analysis and statistical Neighbor Joining (NJ) bootstrap tests of the phylogenies were done with the Mega package (Kumar et al., 2004).
2.6. Assays for proteolytic activities of fXa and thrombin
Proteolytic activities were measured spectrophotometrically using protease specific chromogenic substrates as described previously (Francischetti et al. 2002a). Briefly, inhibition of thrombin activity by SGE was measured by incubating 10 μl SGE and 90 μl alpha-thrombin (9 mU total, Enzyme Research Laboratories, IN) in incubation buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.4, 0.1% BSA) at 37°C for 15 min. Chromogenic substrate (100 μl S2238 – diluted in incubation buffer to give a final concentration of 250 μM, Chromogenix) was added and the reaction measured at 405 nm for 15 minutes with constant mixing every 12 seconds at 37°C. Inhibition of fXa activity was measured by incubating 10 μl SGE and 90 μl fXa (0.7nM final concentration diluted in incubation buffer) for 10 minutes at 37°C before addition of S2222 (250 μM final concentration, Chromogenix) and monitoring the reaction as described above. In both cases the slope of the reaction was used to calculate the enzyme activity.
2.7. Gel-shift assay to detect interaction with GPIIbIIIa
Gel shift assays were performed as previously described (Mans and Neitz, 2004b). Briefly, 2 μl of GPIIbIIIa (20 pmoles, Enzyme Research Laboratories) were incubated with SGE or buffer for 30 minutes at room temperature. This mixture was then loaded onto a 10% NuPAGE SDS-PAGE (Novagen) gel and run in the presence of running buffer (NuPAGE-MES SDS running buffer, Novagen) without prior boiling.
2.8. Proteome characterization using Edman sequencing
Five pairs of O. parkeri salivary glands were dissected, placed in 50 μl water and immediately frozen at -80°C. The glands were sonicated and centrifuged at 16,000 g for 10 min. The supernatant (soluble fraction, 20 μgs), and the pellet (insoluble fraction, resuspended in 20 μl water, ~ 20 μg) of O. parkeri salivary gland homogenate was mixed with LDS loading buffer in the presence of β-mercaptoethanol (2%, v/v) and electrophoresed in a 4–12% NuPAGE gel (MES buffer). Then, the proteins in the gel were electroblotted to PVDF membranes in 10 mM CAPS-methanol (10%, v/v) buffer, pH 11. The membrane was stained with 0.025% Commassie blue, destained in 50% methanol without acetic acid, and the bands sequenced by Edman degradation. Identification of proteins was performed using in-house software. The matches can be verified in the supplemental spreadsheets S1 and S2 where a lower case letter represents a mismatch and the upper case single letter representing a amino acid represents a match. Notice that the insoluble fraction can contain otherwise soluble proteins that precipitated in the absence of ions (solubilized in water), or that had affinity for membrane fractions.
2.9. Proteome characterization using two-dimensional gel electrophoresis and tandem mass spectrometry
Two dimensional gel electrophoresis was performed using ZOOM IPGRunner System (Invitrogen) under manufacturer’s recommended running conditions. Briefly, approximately 130 μg of sample proteins (6 pairs of SG) were solubilized with 155 μl rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 20 mM DTT, 0.5% carrier ampholytes, pH 3–10). The samples were absorbed by rehydration ZOOM strips (7 cm; pH 3–10 NL) overnight at room temperature and then focused under manufacturer’s recommended conditions. The focused IPG strips were reduced/alkylated/equilibrated with reducing and then alkylation reagents dissolved in the sample buffer. The strips were then applied onto NuPAGE 4–12% Bis Tris ZOOM gels (Invitrogen). The gels were run under MOPS buffer and stained with SeeBlue staining solution (Bio Rad). A total of 60 spots were selected for tryptic digestion, based on their staining intensity. Protein identification of 2D gel-separated proteins was performed on reduced and alkylated trypsin-digested samples prepared by standard mass spectrometry protocols. Tryptic digests were analyzed by coupling the Nanomate (Advion BioSciences)—an automated chip based nano electrospray interface source—to a quadrupole time of flight mass spectrometer, QStarXL MS/MS System (Applied Biosystems/Sciex). Computer controlled data dependent automated switching to MS/MS provided peptide sequence information. AnalystQS software (Applied Biosystems/Sciex) was used for data acquisition. Data processing and databank searching were performed with Mascot software (Matrix Science). The NR protein database from the NCBI, National Library of Medicine, NIH, was used for the search analysis, as was a protein database generated during the course of this work.
2.10. Proteomic characterization using one-dimensional gel electrophoresis and tandem mass spectrometry
The soluble and the insoluble protein fractions (see section 2.8 above) from salivary gland homogenates from O. parkeri corresponding to 100 micrograms of protein were both brought up in reducing Laemmli gel- loading buffer. The samples were boiled for 10 min and 50 μg of protein was loaded and resolved on a NuPAGE 4–12% Bis-Tris precast gel with antioxidants and visualized by staining with SimplyBlue (Invitrogen). Each gel lane was sliced into about 25 gel slices, destained and digested overnight with trypsin at 37 °C. Peptides were subsequently extracted and desalted using ZipTips (Millipore, Bedford, MA) and resuspended in 0.1% TFA prior to MS analysis.
Nanoflow reversed-phase liquid chromatography (RPLC)-MS/MS was performed using an Agilent 1100 nanoflow LC system (Agilent Technologies, Palo Alto, CA) coupled online with a linear ion-trap (LIT) MS (LTQ, ThermoElectron, San Jose, CA). NanoRPLC columns were slurry-packed in-house with 5 μm, 300 Å pore size C-18 phase (Jupiter, Phenomenex, CA) in a 75 μm i.d. × 10 cm fused silica capillary (Polymicro Technologies, Phoenix, AZ) with a flame pulled tip. After sample injection, the column was washed for 20 min with 98% mobile phase A (0.1% formic acid in water) at 0.5 μl/min and peptides were eluted using a linear gradient of 2% mobile phase B (0.1% formic acid in acetonitrile) to 42% mobile phase B in 40 min at 0.25 μl/min, then to 98% B in an additional 10 min. The LIT-mass spectrometer was operated in a data-dependent MS/MS mode in which each full MS scan was followed by seven MS/MS scans where the seven most abundant molecular ions were dynamically selected for collision-induced dissociation (CID) using a normalized collision energy of 35%. Dynamic exclusion was applied to minimize repeated selection of peptides previously selected for CID.
Tandem mass spectra were searched using SEQUEST on a 20 node Beowulf cluster against an O. parkeri proteome database with methionine oxidation included as dynamic modification. Only tryptic peptides with up to two missed cleavage sites meeting a specific SEQUEST scoring criteria [delta correlation (ΔCn) ≥ 0.08 and charge state dependent cross correlation (Xcorr) ≥ 1.9 for [M+H]1+, ≥ 2.2 for [M+2H]2+ and ≥ 3.1 for [M+3H]3+] were considered as legitimate identifications.
3. Results and discussion
3.1. cDNA library characteristics
A total of 1,529 clones were sequenced and used to assemble a database [see additional File1] that yielded 649 clusters of related sequences, 380 of which contained only one expressed sequence tag (EST). The consensus sequence of each cluster is named either a contig (deriving from two or more sequences) or a singleton (deriving from a single sequence). For simplicity sake, this paper uses ‘cluster’ to denote sequences deriving both from consensus sequences and from singletons. The 649 clusters were compared using the program BlastX, BlastN, or RPSBLAST (Altschul et al., 1997) to the nonredundant protein database of the National Center of Biological Information (NCBI), to a gene ontology database (Ashburner et al., 2000), to the conserved domains database of the NCBI (Marchler-Bauer et al., 2002), and to a custom-prepared subset of the NCBI nucleotide database containing either mitochondrial or rRNA sequences.
Because the libraries used are unidirectional, the three-frame translations of the dataset were also derived, and open reading frames (ORF) starting with a methionine and longer than 40 AA residues were submitted to SignalP server (Nielsen et al., 1997) to help identify putative-secreted proteins. The EST assembly, BLAST, and signal peptide results were loaded into an Excel spreadsheet for manual annotation, and can be browsed as the Supplemental table S1.
Four categories of expressed genes derived from the manual annotation of the contigs (Table 1). The putatively secreted (S) category contained 20% of the clusters and 40% of the sequences, with an average number of 4.7 sequences per cluster. The housekeeping (H) category had 31% and 33% of the clusters and sequences, respectively, and an average of 2.5 sequences per cluster. Forty-nine percent of the clusters, containing 27 % of all sequences, were classified as unknown (U), because no functional assignment could be made. This category had an average of 1.3 sequences per cluster. A good proportion of these transcripts could derive from 3/ or 5/ untranslated regions of genes of the above two categories, as was recently indicated for a sialotranscriptome of An. gambiae (Arca et al., 2005).
Table 1.
Transcript abundance according to functional class
| Class | Clusters* | Sequences | Sequences / Cluster |
|---|---|---|---|
| Housekeeping | 204 (31.4) | 511 (33.4) | 2.5 |
| Secreted | 130 (20.0) | 707 (40.0) | 4.71 |
| Unknown | 315 (48.5) | 406 (26.7) | 1.29 |
| Total | 649 | 1529 |
Number (Percent of total)
3.2. Housekeeping (H) genes
The 204 clusters (comprising 511 EST) attributed to H genes expressed in the salivary glands of O. parkeri were further characterized into 16 subgroups according to function (Table 2). Not surprisingly, for an organ specialized in secreting polypeptides, the two larger sets were associated with protein synthesis machinery (298 EST in 89 clusters) and energy metabolism (21 clusters containing 38 EST), a pattern also observed in other sialotranscriptomes (Francischetti et al., 2002b; Ribeiro et al., 2004a; Ribeiro et al., 2004b). We have arbitrarily included in the H category a group of 18 EST that grouped into ten clusters, representing highly conserved proteins of unknown function, presumably associated with cellular function, but still uncharacterized. They are named conserved proteins of unknown function in Supplemental table S1, immediately preceding the clusters of the Unknown class. These sets may help functional identification of the “Conserved hypothetical” proteins as previously reviewed in (Galperin and Koonin, 2004). The complete list of all 204 gene clusters, along with further information about each, is given in Supplemental file S1.
Table 2.
Functional classification of housekeeping transcripts
| Function | Clusters | Sequences | Sequences / Cluster |
|---|---|---|---|
| Protein synthesis | 89 | 298 | 3.35 |
| Energy metabolism | 21 | 38 | 1.81 |
| Transcription machinery | 16 | 29 | 1.81 |
| Signal transduction | 13 | 32 | 2.46 |
| Protein export | 11 | 15 | 1.36 |
| Protein modification | 10 | 19 | 1.90 |
| Unknown conserved | 10 | 18 | 1.80 |
| Nuclear regulation | 6 | 8 | 1.33 |
| Proteasome machinery | 6 | 8 | 1.33 |
| Cytoskeletal | 6 | 11 | 1.83 |
| Lipid metabolism | 4 | 6 | 1.50 |
| Oxidant metabolism | 4 | 5 | 1.25 |
| Transporters/Storage | 3 | 17 | 5.67 |
| Transcription factors | 2 | 4 | 2.00 |
| Amino acid metabolism | 2 | 2 | 1.00 |
| Carbohydrate metabolism | 1 | 1 | 1.00 |
|
| |||
| Total | 204 | 511 | |
3.3. Possibly secreted (S) class of expressed genes
Inspection of Supplemental file S1 indicates the expression of several expanded gene families, including those coding for lipocalins, Kunitz-domain containing polypeptides and mucins, among others (Table 3). Lipocalin members alone represent 39 clusters and 246 EST, or 16% of the total number of sequenced clones, a pattern observed previously in hard ticks (Ribeiro et al., 2006) and in Argas monolakensis (Mans et al., submitted). Similarly, Kunitz-domain containing peptides account for 26 clusters and 194 EST. Several other peptide families, described in greater detail below, were also observed.
Table 3.
Functional classification of transcripts coding for secreted proteins
| Family | Clusters | Sequences | Sequences / Cluster |
|---|---|---|---|
| Lipocalins | 39 | 246 | 6.31 |
| Basic tail/Kunitz | 26 | 194 | 7.46 |
| Other | 25 | 72 | 2.88 |
| Similar to Argas proteins | 9 | 40 | 4.44 |
| Mucins | 7 | 19 | 2.71 |
| Lipases | 6 | 11 | 1.83 |
| Fibrinogen-related | 4 | 8 | 2.00 |
| 5’ nucleotidase/apyrase | 4 | 7 | 1.75 |
| Antimicrobials | 4 | 6 | 1.50 |
| Serine proteases | 3 | 4 | 1.33 |
| Metalloproteases | 3 | 5 | 1.67 |
| Total | 130 | 612 |
3.4. Analysis of the Ornithodorus parkeri sialotranscriptome
Several clusters of sequences coding for housekeeping and putative secreted polypeptides indicated in Supplemental file S1 are abundant and complete enough to extract consensus sequences of novel sequences. Additionally, we have performed primer extension studies in several clones to obtain full- or near-full length sequences of products of interest. A total of 159 novel sequences, 78 of which code for putative secreted proteins, are grouped together in Supplemental file S2. Table 4 has a summary of the secreted subset, with links to GenBank.
Table 4.
Description of salivary secreted polypeptides deposited in GenBank, with summary of proteomic experiments
| Seq name | Gi accession number and Link to NCBI | Status | Description | Match to 1D gel Edman (Figure 5) | Match to 2D gel MS/MS result (Figure 6) | Match to 1D PAGE HPLC MS/MS results (Figure 7) |
|---|---|---|---|---|---|---|
|
Secreted polypeptides
| ||||||
|
Lipocalins
| ||||||
| OP-468 | gi∣149287076 | Complete | salivary lipocalin | Y | ||
| OP-168 | gi∣149286978 | Complete | salivary lipocalin | Y | ||
| OP-509 | gi∣149287102 | Complete | salivary lipocalin | Y | ||
| OP-113 | gi∣149286916 | Complete | salivary lipocalin | Y | ||
| OP-316 | gi∣149287038 | Complete | salivary lipocalin | Y | Y | |
| OP-49 | gi∣149287088 | Complete | salivary secreted lipocalin | Y | Y | |
| OP-49a | gi∣149287092 | Complete | Salivary lipocalin | Y | Y | |
| OP-24 | gi∣149287008 | Complete | salivary lipocalin | Y | Y | |
| OP-485 | gi∣149287084 | Complete | salivary lipocalin | Y | ||
| OP-1 | gi∣149286896 | Complete | Moubatin homologue 1 | Y | Y | Y |
| OP-30 | gi∣149287032 | Complete | short salivary moubatin | Y | Y | |
| OP-15 | gi∣149286962 | Complete | moubatin homologue 6 | Y | Y | |
| OP-16 | gi∣149286972 | Complete | salivary lipocalin | Y | Y | |
| OP-55a | gi∣149287118 | Complete | moubatin homologue 5 variant | Y | Y | |
| OP-55 | gi∣149287116 | Complete | moubatin homologue 5 | Y | Y | |
| OP-3 | gi∣149287030 | Complete | Moubatin homolog 3 | Y | Y | Y |
| OP-2 | gi∣149287000 | Complete | Moubatin 1 homolog 2 | Y | Y | Y |
| OP-6 | gi∣149287126 | Complete | Moubatin homolog 4 | Y | Y | |
| OP-540 | gi∣149287112 | Complete | salivary secreted lipocalin | Y | Y | |
| OP-88 | gi∣149287170 | Complete | Moubatin homolog 7 | Y | Y | |
| OP-177 | gi∣149286990 | 5’ truncated | truncated salivary lipocalin | |||
|
Mucins
| ||||||
| OP-57 | gi∣149287122 | Complete | salivary mucin | |||
| OP-90 | gi∣149287174 | Complete | Salivary mucin with basic tail | |||
| OP-56 | gi∣149287120 | Complete | salivary mucin | |||
| OP-58 | gi∣149287120 | Complete | salivary mucin | |||
|
Kunitz-domain containing polipeptides
| ||||||
| OP-9 | gi∣149287172 | Complete | savignygrin homologue 1 | Y | Y | |
| OP-10 | gi∣149286898 | Complete | savignygrin homologue 2 | |||
| OP-463 | gi∣149287074 | Complete | putative salivary secreted peptide | |||
| OP-175a | gi∣149286988 | Complete | Kunitz salivary protein | |||
| OP-26 | gi∣149287018 | Complete | putative salivary secreted peptide | Y | ||
| OP-26a | gi∣149287024 | Complete | putative salivary secreted peptide variant | Y | Y | |
| OP-27 | gi∣149287026 | 5’ truncated | acid tail salivary peptide | Y | ||
| OP-175 | gi∣149286986 | Complete | Dual Kunitz salivary protein | |||
| OP-23 | gi∣149287026 | Complete | Basic tail salivary secreted protein | Y | Y | |
| OP-74 | gi∣149287150 | Complete | Dual Kunitz salivary protein | Y | ||
| OP-441 | gi∣149287064 | Complete | Kunitz domain-containing salivary protein | Y | ||
|
Cystatin
| ||||||
| op-new-431 | gi∣149287198 | 5’ truncated | truncated salivary cystatin | Y | ||
|
Metalloprotease
| ||||||
| op-new-266 | gi∣149287192 | 5’ truncated | truncated metalloprotease | |||
|
Phospholipase A2
| ||||||
| OP-525 | gi∣149287108 | Complete | phospholipase A2 | Y | ||
|
Possible immune-related peptides
| ||||||
| OP-104 | gi∣149286902 | Complete | salivary lipid interacting protein | Y | Y | |
| OP-129 | gi∣149286938 | Complete | Ficolin salivary secreted protein | Y | ||
| op-new-613 | gi∣149287206 | Complete | GYY secreted salivary peptide | |||
| OP-336 | gi∣149287050 | Complete | hebreain homolog | |||
|
Prostacyclin stimulating factor
| ||||||
| opn-339 | gi∣149287184 | Complete | Prostacyclin-stimulating factor | |||
|
Adrenomedulin-like peptide
| ||||||
| OP-501 | gi∣149287098 | Complete | adrenomodulin-like peptide | |||
|
Similar to serum amyloid protein
| ||||||
| OP-255 | gi∣149287014 | Complete | similar to serum amyloid A protein | |||
|
Basic/Acid tail proteins
| ||||||
| OP-11 | gi∣149286912 | Complete | acid tail salivary protein | Y | Y | |
| OP-11a | gi∣149286922 | Complete | acid tail salivary protein variant | Y | Y | |
| OP-12 | gi∣149286924 | Complete | acid tail salivary protein | Y | Y | |
| OP-4 | gi∣149287060 | Complete | Basic tail salivary tick peptide | Y | Y | |
| OP-8 | gi∣149287158 | Complete | Basic tail salivary tick peptide | Y | Y | |
| OP-173 | gi∣149286982 | Complete | salivary secreted basic tail protein | |||
|
7 Cys domain protein
| ||||||
| OP-148 | gi∣149286960 | Complete | salivary basic tailless peptide | Y | ||
|
7 DB soft tick family
| ||||||
| OP-482 | gi∣149287080 | Complete | hypothetical salivary secreted protein | |||
|
7-8 kDa family
| ||||||
| OP-67 | gi∣149287136 | Complete | putative salivary secreted peptide - member 1 | |||
| OP-68 | gi∣149287138 | Complete | putative salivary secreted peptide - member 2 | |||
| OP-124 | gi∣149286932 | Complete | putative salivary secreted peptide - member 3 | |||
| OP-32 | gi∣149287040 | Complete | putative salivary secreted peptide - member 4 | |||
|
8.4 kDa soft tick family
| ||||||
| OP-166 | gi∣149286976 | Complete | putative salivary secreted peptide | |||
| OP-390 | gi∣149287058 | Complete | putative salivary secreted peptide | |||
| OP-25 | gi∣149287012 | Complete | putative salivary secreted peptide | |||
|
17.7 soft tick family
| ||||||
| OP-60 | gi∣149287128 | Complete | putative salivary secreted protein | |||
| OP-96 | gi∣149287180 | Complete | putative salivary secreted protein | Y | ||
|
Similar to HIV envelope protein
| ||||||
| op-368 | gi∣149287054 | Complete | similar to HIV envelope glycoprotein - fragment | |||
|
Other putative salivary secreted proteins
| ||||||
| OP-140 | gi∣149286950 | Complete | Similar to tissue factor inhibitor | Y | Y | Y |
| OP-143 | gi∣149286956 | Complete | Putative salivary protein | Y | ||
| OP-155 | gi∣149286968 | Complete | hypothetical salivary secreted protein | |||
| OP-225 | gi∣149287002 | Complete | putative salivary peptide | |||
| OP-330 | gi∣149287048 | Complete | putative salivary secreted peptide | |||
| OP-447 | gi∣149287070 | Complete | putative salivary secreted peptide | Y | ||
| OP-83 | gi∣149287162 | Complete | putative salivary secreted peptide | |||
| op-new-154 | gi∣149287188 | Complete | putative salivary secreted peptide | |||
| OP-445 | gi∣149287068 | Complete | putative salivary secreted protein | |||
| OP-19 | gi∣149286998 | Complete | putative salivary secreted peptide | |||
| OP-72 | gi∣149287146 | Complete | basic salivary secreted protein | Y | ||
| OP-160a | gi∣149286974 | Fragment | similar to Argas phospholipase A2 | Y | Y | |
Following is a detailed description of the full-length, or near full-length, transcripts found in the salivary glands of O. parkeri:
3.5 Putative salivary secreted proteins having ubiquitous domains or with known function: Lipocalins
Both hard (Ribeiro et al., 2006) and soft (Mans et al., 2003) ticks, as well as triatomine bugs (Ribeiro et al., 2004a) have multiple members of the lipocalin family expressed in their salivary glands, only a few of which have a known function. The lipocalin family is widespread in animals, where it serves mostly as binder or carrier of small, usually hydrophobic, ligands. It is characterized by having an antiparallel β-barrel with a repeated +1 topology (Flower, 1995); however, primary sequence conservation is weak, where a single conserved tryptophane residue is identifiable (Flower, 1996). They serve multiple functions in blood-sucking arthropods: In Rhodnius, they function as heme-binding proteins that carry nitric oxide (nitrophorin lipocalins), and as binders of hemostasis mediators such as adenosine nucleotides, histamine and serotonin (Andersen et al., 2005). In ticks, histamine and serotonin binding lipocalins are also known (Paesen et al., 2000; Sangamnatdej et al., 2002). Remarkably, while lipocalins in general are binders of hydrophobic ligands, in all the cases mentioned above they bind hydrophilic molecules. In addition to these small ligand binding activity, some lipocalins also inhibit blood coagulation (Gudderra et al., 2005; Ribeiro et al., 1995; Zhang et al., 1998), platelet aggregation (Keller et al., 1993; Waxman and Connolly, 1993), alternative complement activation (Nunn et al., 2005), and have toxic properties (Mans et al., 2003; Mans et al., 2002c). Despite this accumulating functional knowledge on hematophagous arthropods’ salivary lipocalins, the vast majority of salivary lipocalins have unknown function.
Nineteen full length and two partial sequences of salivary lipocalins found in O. parkeri sialotranscriptome are shown in Supplemental file S2. Some of these may be alleles from the same gene, such as the pairs OP-55/ OP-55a and OP-2/OP-3, as they are more than 90% identical, or they may reflect recent or conserved gene duplication events. These 21 sequences were aligned by the program ClustalW (Thompson et al., 1997) with all other tick lipocalins existing in the nonredundant protein database of the NCBI, which were found by BlastP with a cutoff e value of 0.1. The alignment was submitted to the MEGA package (Kumar et al., 2004) to produce the circular dendrogram shown in Fig. 1. It is clear that five distinct branches contain 19 of the 21 proteins (numbered 1–5 in Fig. 1 - some of the branches contain lipocalins from other soft tick species), indicating their diversity, while the proximity of some members in the same branch indicate relatively recent gene duplication events, or gene conversion events that maintain sequence similarity in tandem repeated genes, or positive selection to maintain their primary structure (Fig.1).
Fig. 1.
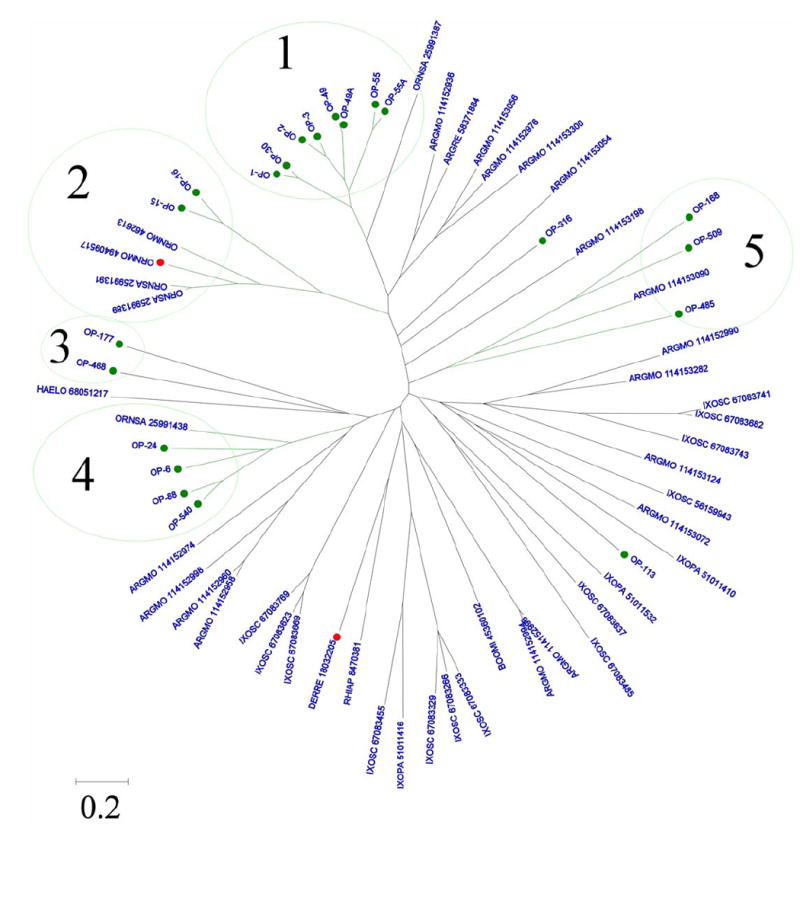
Dendrogram of the Ornithodoros parkeri salivary lipocalins with other tick salivary lipocalins. The O. parkeri sequences are indicated by OP-X where X is the number shown in Supplemental file S2. The remaining sequences derived from the nonredundant (NR) protein database of the National Center for Biotechnology Information (NCBI), and are represented by five letters followed by the NCBI gi∣ accession number. The five letters derive from the first three letters of the genus and the first two letters from the species name. The protein sequences were aligned by the Clustal program (Thompson et al., 1997), and the dendrogram was done with the Mega package (Kumar et al., 2004) after 10,000 bootstraps with the neighbor joining (NJ) algorithm. The bar at the bottom represents 20% amino acid substitution. The large numbers in the figure indicate the branches with O. parkeri proteins. O. parkeri sequences are marked with a filled green circle. Red circles indicate proteins for which a function has been determined as explained in the text.
Branch 1 (Fig. 1) contains the most abundantly expressed lipocalins in O. parkeri salivary glands and groups with TSGP1 a protein previously described for O. savignyi (Mans et al., 2003; Mans et al., 2001). These are good candidates for containing histamine/serotonin binding proteins because physiologically active concentrations of histamine and serotonin are near 1 μM concentrations, needing relatively large amounts of protein to sequester these biologically active amines. Indeed, several of these clade 1 proteins were recognized in the proteome experiments described below, and include OP-1, OP-2, OP-55a and OP-3 which are proteins with large amounts of coding transcripts (64, 25, 22 and 13 ESTs, respectively). OP-24, OP-6 and OP-540, belonging to clade 4 are also abundantly transcribed (17, 7 and 6 and ESTs, respectively), and were all identified in the proteomic experiments. Although lipocalins and other protein families, such as the odorant binding protein family, are known or “expected” to bind small ligands in the barrels they form, they are also known, as described above, to have additional functions such as anti-clotting (Gudderra et al., 2005; Isawa et al., 2002) and anti-complement functions (Nunn et al., 2005) that do not reside in their conventional binding pocket, but rather on the protein’s outer surface. Accordingly, it is possible that the remaining lipocalins not only may bind small compounds that are physiologically active at lower concentrations, such as prostaglandins and leukotrienes (Calvo et al., 2006), but they may also bind larger cytokines and chemokines. Indeed, uncharacterized cytokine-binding proteins are known to exist in tick saliva (Gillespie et al., 2001; Hajnicka et al., 2001; Hajnicka et al., 2005; Konik et al., 2006). However, a recently described chemokine binding protein from Rhipicephalus sanguineus belongs to a novel protein family (Frauenschuh et al., 2007).
Branch 2 contains 2 lipocalins from O. parkeri (OP-15 and OP-16) as well as 2 lipocalins from O. savigny named TSGP3 (gi∣25991391) and TSGP2 (gi∣25991389), this last one shown to be toxic to mice, as well as the salivary anti-complement activity of O. moubata and the platelet aggregation inhibitor, moubatin from the same species. Branch 4 contains several O. parkeri lipocalins as well as the protein named TSGP4 (gi∣25991438∣), which was also shown to be toxic to mice (Mans et al., 2002c). However, toxic reactions were not observed with closely related O. moubata salivary proteins, indicating that sequence similarity may be a poor predictor of function in tick lipocalins (Mans et al., 2003; Mans and Neitz, 2004a). Also indicated (with a red circle marker) are two histamine and serotonin binders that are closely related, DERRE_18032205 (gi∣18032205), from the hard tick Dermacentus reticulatus, and RHIAP_8470381 (gi∣8470381) from Rhipicephalus appendiculatus; these last two proteins have no close relatives among the soft ticks. Except for biogenic amine binding functions proposed for group 1 other lipocalins can work at low concentrations as ligands in host receptor-mediated interactions, as is the possible mode of action of the toxic lipocalin of the African soft tick, O. savigny (Mans et al., 2003; Mans et al., 2002c).
3.6 Mucins
Supplemental file S2 reports 4 mRNA sequences coding for basic proteins (pI of the mature protein 8.7–10.7) ranging in MW of the mature protein from 12.5–17.8 kDa. All are rich in threonine, and some in serine and have 13–34 putative galactosylation sites. Two of these proteins (OP-57 and OP-90) are related at a level of 50% similarity, and may be results of gene duplication events. OP-57 has also a weak Kunitz-domain signature indicated by comparison to the SMART database. None of these proteins produce a clear homolog from the NR database when compared by BlastP with the filter of low complexity turned off, except for other proteins having repeated Thr or Ser residues. These proteins may help to lubricate the tick mouthparts in addition to other possible interactions with the host cellular matrix proteins.
3.7 Kunitz-domain containing polypeptides
The Kunitz domain characterizes many protease inhibitors. It was first identified in the pancreatic Kunitz inhibitor, also known as aprotinin or bovine pancreatic trypsin inhibitor (BPTI) (Ascenzi et al., 2003). Multiple Kunitz domains may occur in a single protein, thus providing for inhibition in multiple protease amplification cascades such as occur with the vertebrate tissue-factor pathway inhibitor (TFPI) of the extrinsic blood coagulation cascade (Bajaj et al., 2001). Tick salivary proteins containing this domain are known in both hard and soft ticks, and were found abundantly expressed in hard tick sialotranscriptomes (Alarcon-Chaidez et al., 2007; Francischetti et al., 2005; Nene et al., 2004a; Ribeiro et al., 2006). Some of the hard proteins have been characterized functionally and have both expected and unexpected functions. Expected functions include inhibition of blood clotting at different target proteases, such as the proteins named ixolaris (Francischetti et al., 2004) and penthalaris (Francischetti et al., 2004). Unexpected functions are related to inhibition of platelet aggregation, as will be described further below.
In soft ticks, inhibitors of the blood coagulation proteases thrombin and Factor Xa also belong to the Kunitz-family. For fXa this includes fXaI and TAP from O. savignyi and O. moubata, respectively (Gaspar et al., 1996; Joubert et al., 1998; Waxman et al., 1990). For thrombin this includes monobin, ornithodorin and savignin, from Argas monolakensis, O. moubata and O. savignyi, respectively (Mans et al., submitted) (Nienaber et al., 1999) (van de Locht et al., 1996). Monobin, savignin and ornithodorin are orthologs, which indicates that the thrombin inhibitor is a conserved protein in the soft tick family. It is thus of interest that no close homologs of either the fXa or thrombin inhibitors were found in the cDNA library of O. parkeri. We thus tested salivary gland extracts (SGE) for direct inhibition of either thrombin or fXa (Fig. 2). Results indicated that no thrombin inhibitory activity is present in the glands of O. parkeri and even at 1 salivary gland equivalent only 50% inhibition was observed for fXa. These results suggest that both direct fXa and thrombin inhibitory activity were lost in O. parkeri. The partial inhibition of fXa could imply that an inhibitor exists that possibly targets the fXase complex, perhaps in a manner analogous to that of ixolaris or penthalaris (Francischetti et al., 2004; Francischetti et al., 2002a). Proteins with this possible function would show sequence similarity to TFPI-like molecules in the manner that ixolaris does. In the library of O. parkeri two potential candidates can be identified, which are OP-74 (GI∣149287150) and OP-441 (GI∣149287064) that both retrieve TFPI-like proteins as top hits and that both possess 2 Kunitz domains.
Fig. 2.
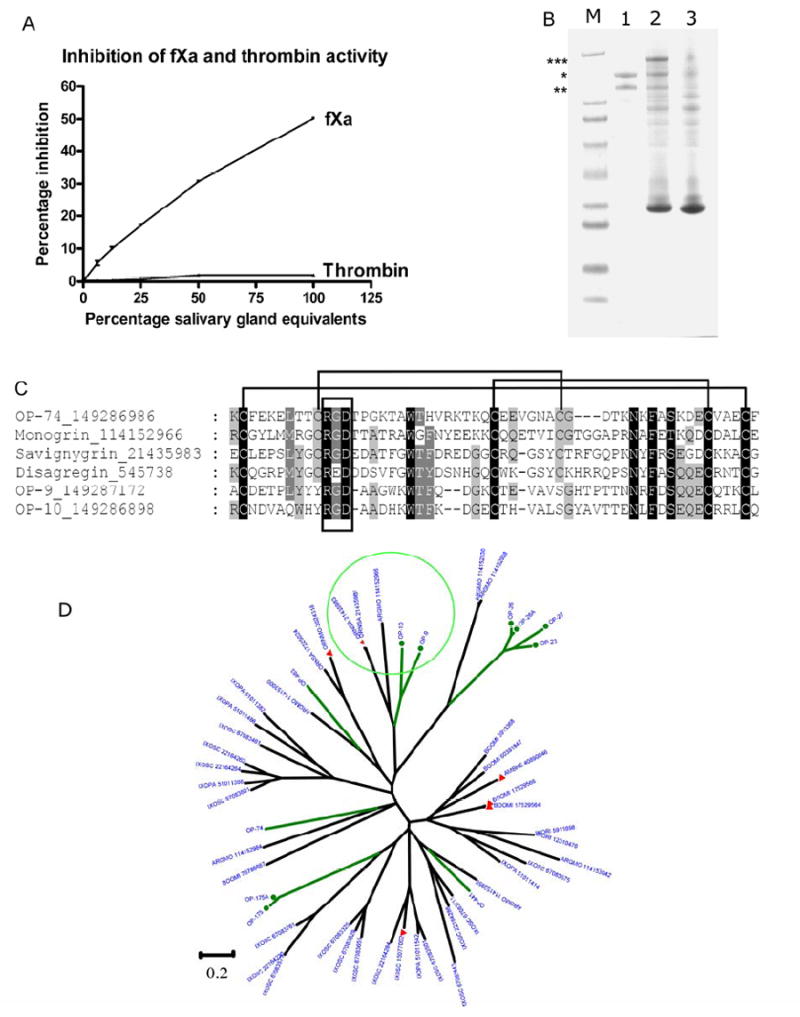
The Kunitz-family of Ornithodorus parkeri. A) Inhibition of fXa and thrombin amidolytic activity by crude salivary gland extract (SGE). B) Protection of SDS-induced dissociation of GPIIbIIIa by crude SGE. Lane 1 shows GPIIbIIIa in the absence of SGE (Notice dissociation into 2 bands, * and **), lane 2 GPIIbIIIa and SGE (notice appearance of larger band marked with ***) and lane 3 control SGE in the absence of GPIIbIIIa. Lane M are molecular weight markers. The two top markers are for 188 and 62 kDa. C) Alignment of O. parkeri Kunitz proteins that possess RGD-motifs with other known platelet aggregation inhibitors and GPIIbIIIa antagonists from soft ticks. Indicated are the canonical disulphide bond patterns of the Kunitz family as well as the RGD-motif. The proteins were truncated on either end to include only the Kunitz-domain that contains the RGD-motif. D) Dendrogram of the Ornithodoros parkeri salivary Kunitz domain containing polypeptides with other similar tick proteins. The O. parkeri sequences are indicated by OP-X, where X is the number shown in Supplemental file S2, and are marked with green branches. Proteins with a function previously determined are marked with a filled red triangle, as explained in the text. The circle in the top of the figure indicates the branch containing the savignygrins, which have a disintegrin RGD motif. See legend of Fig. 1 for additional information.
An unexpected function for Kunitz inhibitors is displayed by the protein named savignygrin, from the soft tick O. savigny, a platelet aggregation inhibitor that possesses the RGD integrin-recognition motif, which is located on the substrate-binding loop of the canonical BPTI inhibitors, but has no inhibitory activity against plasma serine proteases (Mans et al., 2002b). This activity indicates the unusual paths taken in the adaptation of salivary proteins to blood feeding in arthropods. Orthologs of savignygrin are found in O. parkeri (OP-9 and OP-10) and possess the RGD-motif. Of interest is their lack of the cysteine bond that restricts the substrate-binding presenting loop in other Kunitz proteins. Of interest is the presence of the RGD-motif in the second domain of OP-74, one of the potential factor Xase inhibitor domains. In the case of OP-74, the RGD-motif is at the conserved position found in savignygrin, but this protein still retains the conserved disulphide bond found in other Kunitz proteins. Of the potential fibrinogen receptor antagonists and platelet aggregation inhibitors found in the salivary glands of O. parkeri, OP-9 is one of the most abundant proteins found in the salivary glands (proteome section) and thus the most likely candidate. In order to find evidence for biological activity, we performed an assay that detects interaction with GPIIbIIIa (Mans and Neitz, 2004b). This assay relies on the formation of an SDS-resistant GPIIbIIIa-inhibitor complex that can be detected by SDS-PAGE electrophoresis (Mans and Neitz, 2004b). GPIIbIIIa was partially protected against SDS-induced dissociation in the presence of SGE from O. parkeri, while in the homogenate absence it dissociated (Fig. 2). This is indicative of a fibrinogen-receptor antagonist that possesses an RGD-motif.
Supplemental file 2 presents 11 proteins from O. parkeri having clear Kunitz domains, or being similar to proteins with such domains. They range in mature molecular weight (MW) from 6.5 to 16.1 kDa, the lower range containing a single Kunitz domain. The O. parkeri Kunitz-domain containing polypeptides were aligned with other similar tick proteins found in the NR database to produce the dendrogram found in Fig. 2. Similarly to the gene expansion found in Ixodes (Francischetti et al., 2005; Ribeiro et al., 2006), O. parkeri Kunitz-domain containing sequences are spread in several poorly connected families, shown as green branches in Fig. 2. Only a few of the Kunitz-containing tick sequences have been functionally evaluated, and they are marked with solid red triangles in Fig. 2. This includes the I. scapularis protein named Ixolaris, which is a tissue factor pathway inhibitor (Francischetti et al., 2002a), and the thrombin inhibitors of Rhipicephalus (Boophilus) microplus (boophilin - gi∣17529566) , Amblyomma hebraeum, named Amblin (Lai et al., 2004a), and from the soft tick O. moubata, named ornithodorin and savignin (van de Locht et al., 1996) (Mans et al., 2002a). The O. savigny Kunitz disintegrins (containing the RGD motif) appear in a branch marked with a circle in Fig. 2 where it co-segregates with OP-9, OP-10 and one protein from A. monolakensis, all of which have the consensual RGD motif flanked by cysteines. This RGD/Kunitz combination appears to be unique to soft ticks.
3.8 Cystatin
Cystatins are inhibitors of cysteine proteinases such as cathepsins B and L, for example. They have been previously described in the sialotranscriptome of ticks (Ribeiro et al., 2006; Valenzuela et al., 2002), from where a protein named sialostatin L has been characterized biochemically and shown to specifically inhibit cathepsin L (Kotsyfakis et al., 2006). Cystatins have been also characterized from other organs of both soft and hard ticks (Grunclova et al., 2006; Lima et al., 2006; Zhou et al., 2006). Supplemental file 2 provides for a partial O. parkeri sequence coding for the majority of the mature peptide of a member of the cystatin family. When compared with the NR database by the BLAST program, all top matches are from tick cystatins. The target of this cystatin remains to be identified.
3.9 Enzymes
A fibrinogen-specific metalloprotease has been characterized previously in the saliva of I. scapularis (Francischetti et al., 2003), consistent with the sialotranscriptome finding of an expanded gene family coding for this enzyme (Francischetti et al., 2005; Ribeiro et al., 2006). These metalloproteases are also a common finding in snake venoms (Takeya et al., 1993). Recently, 6 such enzymes were characterized from the tick Haemaphysalis longicornis (Harnnoi et al., 2007). Supplemental file S2 presents evidence for the presence of such salivary enzyme in O. parkeri in the form of OP-new-266, a 249 AA fragment of such enzyme containing the HEXXH zinc-binding motif of the enzyme active site, in addition to high similarity to proteins in the NR database annotated as metalloproteases from both soft and hard ticks. This enzyme may participate in fibrinolytic reactions in addition to other possible physiologic interventions.
Transcripts coding for secreted phospholipases of the A2 family have been found in sialotranscriptomes of mosquito, sand flies and ticks. However, only in the hard tick Amblyomma americanum was the activity demonstrated to occur in dopamine-stimulated saliva (Bowman et al., 1997; Zhu et al., 1998). It is proposed that the enzyme promotes hemolysis of the ingested red cells and thus facilitates digestion (Zhu et al., 1997). Evidence for a salivary phospholipase in O. parkeri is indicated by the proteins OP-525 (Supplemental file S2); however, no signal peptide indicative of secretion is found indicating this enzyme may be intracellular, or that the transcript may be truncated.
3.10 Possible immune-related polypeptides
Sialotranscriptomes of blood-sucking arthropods consistently detect the presence of antimicrobial peptides and other proteins that are potentially implicated with the innate immune response (Ribeiro and Francischetti, 2003). These proteins may prevent microbial contamination of the blood meal. Supplemental file S2 presents four proteins that might exercise such function. OP-104 has the SMART ML domain, implicated in lipid recognition, particularly in the recognition of pathogen-related products. These single-domain proteins were predicted to form a beta-rich fold containing multiple strands, and to mediate diverse biologic functions through interacting with specific lipids (Visintin et al., 2006). Other members of this protein family function in lysosomal lipid metabolism and if this is the case with OP-104, then it has a housekeeping role. However, OP-104 also has an RGD motif, indicating it may bind to integrins. Proteins of this family were reported previously in salivary and midgut transcriptome of the hematophagous fly, Culicoides sonorensis (Campbell et al., 2005).
OP-129 codes for a protein containing a fibrinogen domain found in ficolins and angiopoietins, which are known to be implicated in pathogen recognition in invertebrates (Grubhoffer et al., 2004; Wang et al., 2005; Zhang and Loker, 2003). This protein is related to previously described lectins of soft and hard ticks, including Dorin M which was shown to be expressed in hemocytes and salivary glands of Ornithodoros moubata (Kovar et al., 2000; Rego et al., 2005; Rego et al., 2006). It may be possible also that this protein may have been co-opted to provide an antihemostatic or anti-inflammatory role during tick feeding.
OP-new-613 codes for a short peptide (54 AA, 34 AA on the mature peptide) containing GYY repeats, which have been shown in the worm C. elegans to be a novel antimicrobial (Couillault et al., 2004). Short peptides of this type have been repeatedly found in mosquito and tick sialotranscriptomes and appear ubiquitous in invertebrates.
OP-336 is homologous and of similar size to the histidine-rich tick antimicrobial peptides named here in (Lai et al., 2004b) and microplusin (Fogaca et al., 2004) found in soft and hard ticks.
3.11 Peptides similar to vertebrate proteins not found previously in invertebrate sialomes
Three putative polypeptides with high similarity to vertebrate proteins might exert pharmacologic activities on their hosts. OPN-339 represents a truncated protein that is >60 % similar to vertebrate proteins containing the cdd IB domain of insulin growth factor-binding protein homologues, and has the IGcam domain of cell adhesion molecule subfamily, which includes close similarity to the mammalian protein named prostacyclin-stimulating factor (gi∣861521) that induces the endothelium cells to produce this vasodilatory prostaglandin (Yamauchi et al., 1994; Yamauchi et al., 1993).
OP-501 produces a best match to the NR database (over 60% similarity) to vertebrate adrenomedulin, a vasodilatory and immunomodullatory peptide (Zudaire et al., 2006). Vertebrate pre-pro adrenomedullin is processed by a peptidase to cleave the propeptide at the amino terminus, and further processed by a serine protease and carboxypeptidase B to produce the mature peptide (Zudaire et al., 2006). The predicted tick mature peptide, after signal secretion peptide removal, starts at the same position as mature vertebrate adrenomedullin, and the basic residues match the carboxiterminus of the processing region of the vertebrate peptides, suggesting a conserved mechanism of processing. OP-501 and OPN-339 are vasodilatory candidates in the salivary repertoire of O. parkeri.
OP-255 produces BlastP matches to similar-sized vertebrate proteins annotated as serum amyloid A protein precursor, which are blood proteins that increase their concentration over 1,000 fold in inflammatory conditions and have various physiologic effects including anti-platelet activity (Urieli-Shoval et al., 2000; Zimlichman et al., 1990).
3.12 Putative salivary secreted proteins belonging to tick-specific polypeptide families: Basic or acidic tail proteins found in both Argasidae and Ixodidae
This class of proteins, thus far unique to hard ticks (Francischetti et al., 2005; Ribeiro et al., 2006; Valenzuela et al., 2002), but also found in A. monolakensis (Mans, submitted), range from 10 to 20 kDa, and have in common the presence of basic (usually lysine) or acidic (usually glutamate) residues on their carboxyterminus, although a subfamily lacking the charged tail also exists. The sialotranscriptome of O. parkeri is rich in this class of proteins, six full-length versions of which are shown in Supplemental file S2. BLAST comparisons with the NR protein database show similarities to proteins from Argas monolakensis, I. pacificus and I. scapularis, indicating that this unique family of proteins exists in both soft and hard ticks. The radial phylogram of this family (Fig. 3) shows the expansion of the proteins in Ixodes due to polymorphism and/or gene duplication, a situation that might be occurring also with O. parkeri and A. monolakensis. Only one member of this family, from I. scapularis and named Salp14 (gi∣15428308), has been shown to exert anticlotting activity (Narasimhan et al., 2002). Alignment of the selected basic tail proteins of soft and hard ticks (Fig. 4) allows identification of the framework motif of six cysteines with a few other interspersed conserved amino acids to produce the block C-X(11-14)-C-X-[FY]-C-X(5,7)-W-X(2,7)-G-X(2,7)-C-X-Y-X(5,12)-G-X-C-X(2)-G-X-C. Search of the NR database for this motif produces only matches to tick proteins, indicating it can function as a pattern match to identify proteins of this type eventually found in the future. Interestingly, no members of this family have been found thus far in the Rhipicephalinae, suggesting a closer relationship between the Argasidae and the prostriate ticks. Here, we call attention that OP-12 exceptionally has an RGD motif flanked by cysteines, indicating this protein may additionally have anti-platelet activity.
Fig. 3.
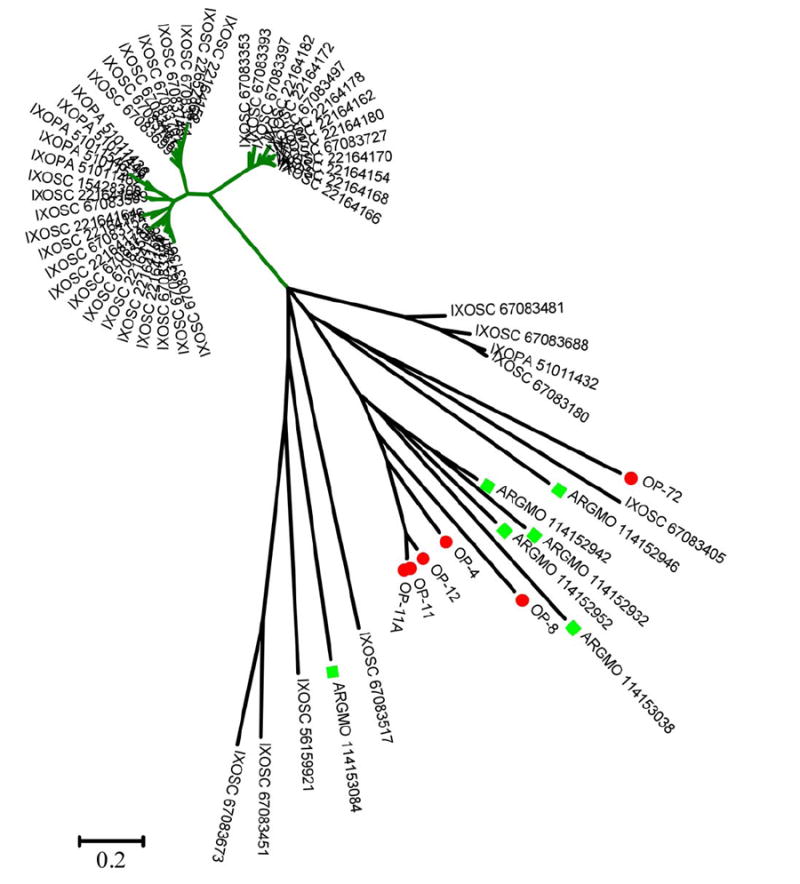
Dendrogram of the Ornithodoros parkeri salivary basic tail polypeptides with other similar tick proteins. The O. parkeri sequences are indicated by OP-X where X is the number shown in Supplemental file S2, and are marked with a filled red circle. Argas monolakensis proteins are shown with a square marker. See legend of Fig. 2 for additional information.
Fig. 4.

Alignment of soft and hard tick basic tail polypeptides. The O. parkeri sequences are indicated by OP-X where X is the number shown in Supplemental file S2. Additional sequences are from Argas monolakensis and Ixodes scapularis, indicated by their name and GenBank numbers. The alignment leads to the unique pattern C-X(11-14)-C-X-[FY]-C-X(5,7)-W-X(2,7)-G-X(2,7)-C-X-Y-X(5,12)-G-X-C-X(2)-G-X-C.
3.13 7 Cys domain family found in Argasids
OP-148 is similar to two previously described polypeptides found in the salivary glands of A. monolakensis, and to no other proteins found in the NR database. When OP-148 is submitted to PSI-BLAST against the nonredundant protein database, it collects members of the basic and acidic tail proteins to which the 7 Cys domain proteins may be related.
3.14 7 DB family found in Argasids
OP-482 is similar to Argas salivary proteins of the DB family, and to no ther proteins found in the NR database. Several rounds of PSI-BLAST additionally retrieves Ixodes proteins of the previously described 18 kDa family (Ribeiro et al., 2006), as well as, with less confidence, basic and acid tail proteins. It may be possible that the 18 kDa, 7 Cys and 7 DB families may all be ancestrally related to the large tick basic-tail proteins.
3.15 7.9 kDa family found in both Argasids and Ixodids
The O. parkeri sialotranscriptome contain four distantly related polypeptides of similar size that are weakly similar to similar-sized peptides found in the sialotranscritpomes of Argas, Ixodes, Rhipicephalus and Haemaphysalis ticks. Their function is unknown.
3.16 8.4 kDa family found in Argasids
Supplemental file 2 lists three polypeptides of 7.3–8.2 kDa and mature MW of 4.8–6.3 kDa that are similar to a 8.4 kDa peptide from the soft tick A. monolakensis. The PSI-BLAST tool retrieves only Argas proteins from the NR protein database (not shown). This protein family contains the unique motif E-x(6)-[RK]-x(2,3)-C-x(3,4)-C-x(4,5)-C-x(3)-C-x-C-x(10,13)-C, as revealed by no matches to the nonredundant protein database when the program Seedtop was used as a pattern search tool. The function of this protein family is unknown.
3.17 17 kDa family found in Argasids
Two related full-length polypeptides of 16 kDa shown in Supplemental file S2 are similar to three other polypeptides found in the sialotranscriptome of A. monolakensis. No similarities are found among these proteins and any other known proteins. This tick protein family has six cysteines with the pattern C-x(12,13)-C-x(4,5)-C-x(41,44)-C-x(8)-x(34,35)-C that is ubiquitously found in vertebrates and invertebrates. OP-96 has a weak match to the Der-p2-like domain, which are Group 2 allergen proteins belonging to the ML domain family; these may play a role in natural immunity. In concordance with this finding, the PSI-BLAST tool retrieved from the NR database a family of mite allergens of ~150 amino acids in length, and also similar-sized proteins annotated as Nieman-Pick proteins, from mosquitoes and Culicoides. This protein family is related to the ML family that has lipid recognizing motifs and can function in innate immunity or lipid transport, as described above in the sub-group of immunity-related proteins.
3.18 Similar to HIV envelope protein and Ixodes protein
OP-368 is a truncated protein similar to a previously described I. scapularis secreted protein, and, curiously, to a segment of the envelope protein of human and simian immunodeficiency virus. Submission of OP-368 to PSI-Blast against the NR protein database retrieves many variants of the GP-120 viral envelope glycoprotein that is associated with viral cell entry and apoptotic-inducing activities (Castedo et al., 2003; Perfettini et al., 2005). The role of these proteins remains to be identified.
3.19 Putative salivary secreted proteins belonging to novel polypeptide families
Supplemental file S2 presents 14 polypeptides containing a signal peptide indicative of secretion. They have no significant similarities to other known proteins. Accumulating data on tick sialotranscriptomes may eventually find similarities among these proteins that are now otherwise orphans.
3.20 Housekeeping proteins
Supplemental file S2 describes 81 full-length sequences coding for housekeeping proteins, including five conserved hypothetical proteins without a known function, and one protein that is similar to tick lipocalins but has a KDEL signature of ER retention (OP-141). This salivary lipocalin may have been recruited to some granule biogenesis function specific to salivary gland function (Mans and Neitz, 2004c; Mans et al., 2001).
3.21 Preliminary characterisation of the salivary proteome of O. parkeri
To obtain information on protein expression in the salivary glands of O. parkeri, we performed a redundant and complementary proteomic approach using (1) Single dimension polyacrylamide gel electrophoresis (PAGE) protein separation followed by blotting the proteins into a PVDF membrane and performing Edman degradation of the bands (Fig. 5); (2) Two dimensional (2D) gel electrophoresis separation of the homogenate followed by tryptic digest and tamdem mass spectrometry (MS/MS) of the Coomassie blue-stained bands (Fig. 6); and (3) High density one dimension gel electrophoresis followed by tryptic digestion and reverse phase HPLC/MS/MS of the indicated bands (Fig. 7), to attempt increased coverage of lower molecular mass peptides. The data of these three experiments are summarized below, and in Table 4.
Fig. 5.
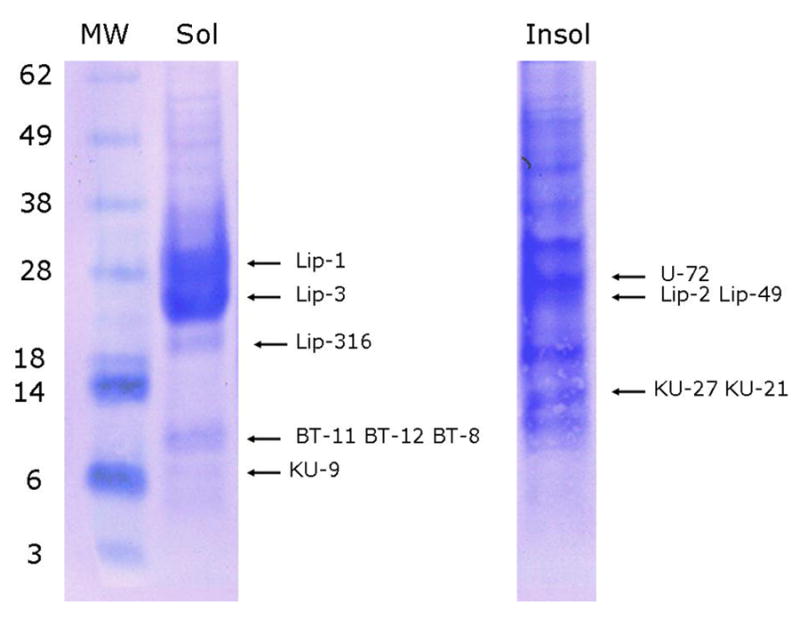
– 1D gel electrophoresis of Ornithodorus parkeri salivary gland homogenates. Soluble (Sol) and insoluble (Insol) salivary gland homogenate of O. parkeri were dissolved in sodium dodecyl sulfate buffer and applied to a polyacrylamide gel. After electrophoresis, the proteins were transferred to a PVDF membrane, stained with Coomassie blue and bands submitted to Edman degradation. Numbers on the left indicate molecular weight marker positions in the gel. The bands labeled with Lip-X correspond to lipocalins where X are the number of the OP-X protein found in Supplemental file S2. Similrly, BT are for basic tail proteins, KU for Kunitz domain proteins and U for unknown function protein. For experimental details, see Materials and Methods.
Fig. 6.
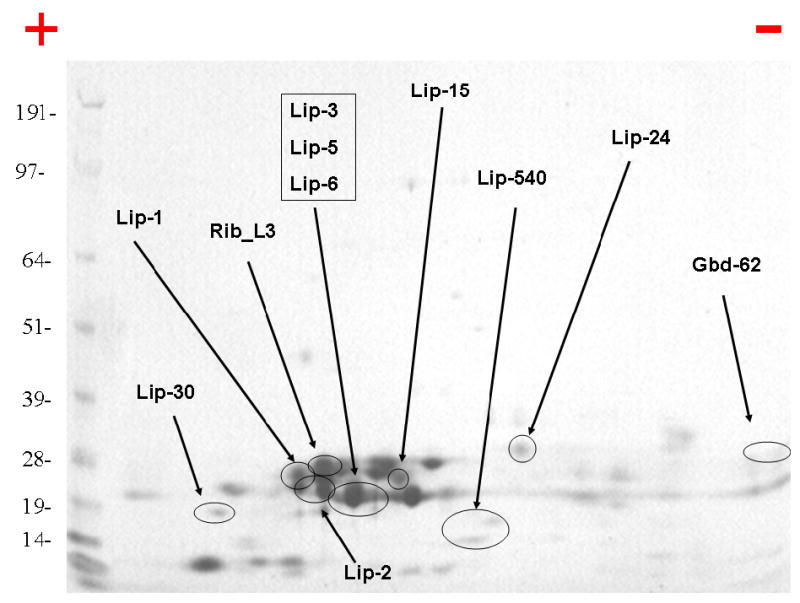
– 2D gel electrophoresis of Ornithodorus parkeri salivary gland homogenates. Numbers on the left indicate molecular weight marker positions in the gel. The + and − signs indicate the anode or cathode side of the isoelectrophocusing dimension, which ranged from pH 3–10. Gel bands that were identified to a protein (following tryptic digestion and mass spectrometry) are shown in the gel. In some cases, more than one band accounted for the same protein, possibly due to trailing or multiple isoforms. The bands labeled with Lip-X correspond to lipocalins where X is the number of the OP-X protein found in Supplemental file S2. Rib_L3 is for ribosomal protein L3, and Gbd-62 is for the guanosine binding protein OP-62. For experimental details, see Materials and Methods.
Fig. 7.
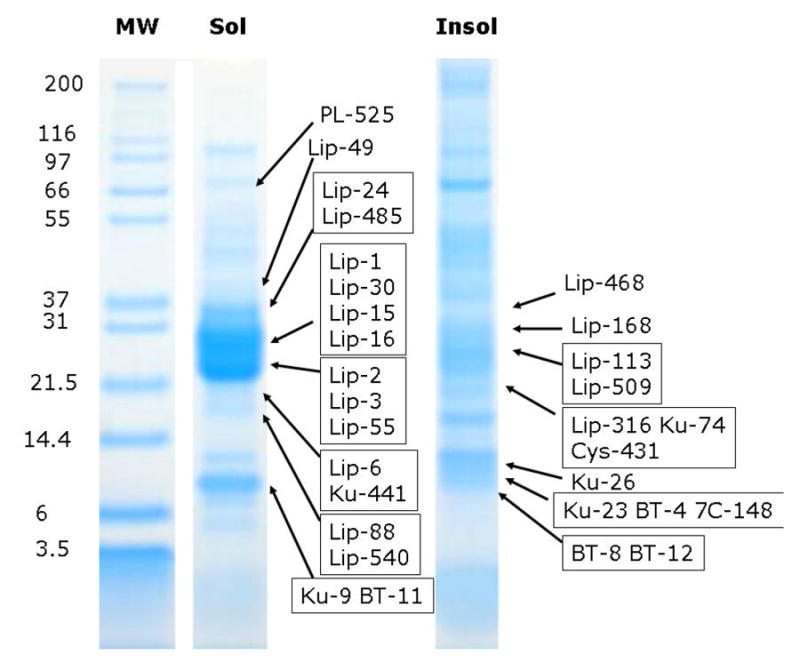
– 1D gel electrophoresis of Ornithodorus parkeri salivary gland homogenates followed by tryptic digestion and HPLC MS/MS of results. Soluble (Sol) and insoluble (Insol) salivary gland homogenate of O. parkeri were dissolved in reducing sodium dodecyl sulfate buffer and applied to a polyacrylamide gel. After electrophoresis, each gel was sliced into 26 (Sol) or 25 (Insol) fractions, submitted to tryptic digest and reverse phase HPLC tandem mass spectrometry. Numbers on the left indicate molecular weight marker positions in the gel. The bands labeled with Lip-X correspond to lipocalins where X is the number of the OP-X protein found in Supplemental file S2. Similarly, BT is for basic tail proteins, KU for Kunitz domain proteins PL for Phospholipase, Cys for Cystatin, 7C for 7Cys domain protein. Other protein matches can be found in Supplemental file S2.
3.22 1D-PAGE followed by blotting to PVDF membrane and Edman degradation
Two gel experiments were run, consisting of soluble and insoluble proteins following homogenization. These two fractions were treated with sodium dodecyl sulfate buffer before applying to the gel beds. Two bands deriving from the soluble fraction were strongly stained by Coomassie blue near the 28 kDa marker, yielding Edman products for the lipocalins OP-1 and OP-3 (Fig. 5). In the same region, the insoluble fraction yielded signals for OP-2 and OP-49. Note that the gel location corresponds to a larger MW than that predicted to the mature proteins, which range from 17.4–17.7 kDa, suggesting these lipocalins may have further translation modifications. The basic tail peptides OP-11, OP-12, OP-8 and the Kunitz-containing peptides OP-9, OP-27 and OP-21 were identified in gel locations corresponding with their expected MW ranges. The Edman products for all these proteins were exactly as predicted by the SignalP server. We additionally found Edman products for the lipocalin OP-316 and for the protein of unknown function OP-72, but these sequences did not correspond to their predicted mature N termini: OP-316 amino terminus was near 60 residues internal to the predicted site, indicating processing of this protein. Consistent with this possible modification, OP-316 appears detached from the remaining identified lipocalins at a gel location of lower MW. OP-72 Edman product was only two residues internally to the predicted cleavage site, but was found at a gel location consistent with a MW twice its predicted size of 13.3 kDa of the mature protein.
3.23 2D-PAGE followed by tryptic digestion and MS-MS
Salivary gland homogenates of five pairs of salivary glands (90 μg protein) were separated by 2D-SDS-PAGE. The stained protein bands were excised from the gel and submitted to tryptic digestion followed by MS/MS identification. The resulting electropherogram, similar to the 1D gel, is dominated by a cluster of bands indicating a MW ranging from 19–28 kD and slightly acidic pI. Several of these bands were identified as lipocalins by tryptic digestion and tandem mass spectrometry of the products. Additionally, a guanylate-binding protein and a ribosomal protein were also identified. These results are summarized in Fig. 6, and in column AL of Supplemental file S2.
3.24 1D-PAGE followed by tryptic digestion, RP-HPLC and MS-MS
To further explore the expression of polypeptides in O. parkeri salivary glands, we submitted both the soluble (S) and insoluble (I) fractions of salivary gland homogenates to PAGE followed by in-gel tryptic digestion of 25 (precipitate) or 26 (soluble) gel fragments, and reverse phase HPLC/MS/MS of the digested products. Results of this experiment lead to a much greater identification of polypeptides, as shown in Fig. 7 and Supplemental file S2. Twenty of the 21 lipocalins indicated in Supplemental file S2 were identified by at least two unique peptides derived from the same gel fraction. Peptide coverage greater than 40% of the mature protein was found for five lipocalins (OP-1, OP-2, OP-3, OP-30 and OP-55a). As indicated in the previous gel experiments of Figs. 5 and 6, these lipocalins are also located in gel positions consistent with a larger molecular mass than that predicted by their sequences.
Six Kunitz-domain proteins were identified, five of which were identified by peptides covering greater than 38% of their predicted sequences. The O. parkeri homolog of savignygrin (Mans et al., 2002b), coded by OP-9, had almost 100% peptide coverage and was found in a gel fraction having a strong Coomassie-stained band with gel location between the 6 and 14.4 kDa markers, while the predicted mass of the mature peptide is 6.5 kDa. The high peptide coverage and positive identification in the two other experiments indicate that the savignygrin homolog is a strongly expressed protein. OP-26 is another Kunitz-domain containing protein that was identified with > 90% peptide coverage at a gel location consistent with a molecular mass of 17 kDa, almost twice its expected mass of 9.6 kDa.
Five of the six basic tail proteins indicated in Supplemental file S2 were identified in this experiment, the same identified by Edman degradation results (Fig. 5). Three of these are identified by more than 50% peptide coverage, two of which were found in the same fraction as the savignygrin homolog. The locations of these proteins in the gel are consistent with their predicted mature MW. The 7Cys domain protein coded by OP-148 was identified by three peptides from a gel fragment located at the 14.4 kDa marker, a MW larger than that predicted of 10.6 kDa. The protein coded by OP-96, a member of the 17.7 kDa family, was identified by three peptides in the S gel and two in the I gel, at similar locations between the 14.4 and 21.5 markers, slightly larger than its predicted mass of 14.8 kDa.
The cystatin determined by the truncated OP-new-431 was also identified with over 60% peptide coverage, in a gel location consistent with a mass of ~18 kDa. The fibrinogen- domain containing protein OP-129 was identified (40% coverage) at a gel location compatible with a dimer (expected 18.8 kDa, appearing near the 37 kDa marker in both S and I gel experiments). The phospholipase A2 coded by the truncated OP-525 was identified by the same two fragments in adjacent gel bands between for 55.4 and 66.3 kDa MW markers, which is in accordance with the molecular weight obtained by gel filtration of a previously described salivary phospholipase A2 from the hard tick, Amblyomma americanum (Zhu et al., 1997).
Four of the 13 proteins indicated in Supplemental file S2 as not belonging to any known protein family were identified by two or more peptide fingerprints originating in the same gel fraction, including 44 % coverage (from 7 peptides) for OP-143, a predicted secreted protein with 19.8 kDa mature MW and located in the gel just at and above the 21.5 kDa marker. This protein has a weak BLAST result to an Argas lipocalin, spanning a similar range of the protein, including the conserved W found in this family of proteins, and a similar size as lipocalins, suggesting OP-143 may be a divergent lipocalin member.
Supplemental file S2 identifies in columns AM-AP additional proteins of putative housekeeping function that were identified by this proteomic experiment.
Conclusion
Analysis of the salivary transcriptome of the soft tick Ornithodorus parkeri indicates conservation of the main protein families found in Ixodes scapularis ticks, in particular the large expansion of the lipocalin, Kunitz, and basic tail superfamilies. This last family may have originated divergent protein lineages in soft ticks, known as the 7Cys and 7DB protein families previously described from Argas monolakensis. Protein expression for many members of these families was confirmed. Metalloproteases and phospholipase A2 were also found, in common with Ixodidae. Less well defined is the highly divergent 7–8 kDa family, found both in hard and soft ticks. Additionally, protein families found thus far only in soft ticks were identified, namely the 8.4 and 17.7 kDa families. A number of possible secreted proteins do not belong to previously described salivary gland families, and include homologs of the vertebrate proteins adrenomedulin, serum amyloid protein and prostacyclin- stimulating factor. The triple proteomic approach to identify expressed proteins was complementary, although the 1D-PAGE/tryptic digestion/RPLC/MS/MS approach was clearly more sensitive and provided greater proteomic coverage. Together, these experiments confirmed expression of all major protein families and indicate OP-1 and OP-3 as the major secreted lipocalins, and the savignygrin homolog the most abundant Kunitz-domain peptide. Despite their different strategies of blood feeding, hard and soft ticks overall share most of their salivary repertoire, in concordance with the proposed close relationship between these two Acari families (Hoogstraal and Aechlimann, 1982). As in all sialotranscriptome studies done thus far, the vast majority of the identified proteins have no confirmed or known function.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the government of the United States of America. We are grateful to the NIAID Core facility lead by Dr. Robert Hohman for their support, Nancy Shulman and Brenda Marshall for editorial assistance and Chuong Huynh from NCBI for help with posting the data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ivo M.B. Francischetti, Email: ifrancischetti@niaid.nih.gov.
Ben J. Mans, Email: bmans@niaid.nih.gov.
Zhaojing Meng, Email: mengz@ncifcrf.gov.
Nanda Guderra, Email: ngudderr@gmu.edu.
Timothy D. Veenstra, Email: tv52i@nih.gov.
Van M. Pham, Email: VPHAM@niaid.nih.gov.
José M.C. Ribeiro, Email: jribeiro@nih.gov.
References
- Alarcon-Chaidez FJ, Sun J, Wikel SK. Transcriptome analysis of the salivary glands of Dermacentor andersoni Stiles (Acari: Ixodidae) Insect Biochem Mol Biol. 2007;37:48–71. doi: 10.1016/j.ibmb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JF, Gudderra NP, Francischetti IM, Ribeiro JM. The role of salivary lipocalins in blood feeding by Rhodnius prolixus. Arch Insect Biochem Physiol. 2005;58:97–105. doi: 10.1002/arch.20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JM. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–86. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Ascenzi P, Bocedi A, Bolognesi M, Spallarossa A, Coletta M, De Cristofaro R, Menegatti E. The bovine basic pancreatic trypsin inhibitor (Kunitz inhibitor): a milestone protein. Curr Protein Pept Sci. 2003;4:231–51. doi: 10.2174/1389203033487180. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj MS, Birktoft JJ, Steer SA, Bajaj SP. Structure and biology of tissue factor pathway inhibitor. Thromb Haemost. 2001;86:959–72. [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–6. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WC, IV, Piesman J. Phylogeny of hard- and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc Natl Acad Sci U S A. 1994;91:10034–8. doi: 10.1073/pnas.91.21.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AS, Gengler CL, Surdick MR, Zhu K, Essenberg RC, Sauer JR, Dillwith JW. A novel phospholipase A2 activity in saliva of the lone star tick, Amblyomma americanum (L.) Exp Parasitol. 1997;87:121–32. doi: 10.1006/expr.1997.4201. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281:1935–42. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Vandyke KA, Letchworth GJ, Drolet BS, Hanekamp T, Wilson WC. Midgut and salivary gland transcriptomes of the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae) Insect Mol Biol. 2005;14:121–36. doi: 10.1111/j.1365-2583.2004.00537.x. [DOI] [PubMed] [Google Scholar]
- Castedo M, Perfettini JL, Andreau K, Roumier T, Piacentini M, Kroemer G. Mitochondrial apoptosis induced by the HIV-1 envelope. Ann N Y Acad Sci. 2003;1010:19–28. doi: 10.1196/annals.1299.004. [DOI] [PubMed] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–94. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Flower DR. Multiple molecular recognition properties of the lipocalin protein family. J Mol Recognit. 1995;8:185–95. doi: 10.1002/jmr.300080304. [DOI] [PubMed] [Google Scholar]
- Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogaca AC, Lorenzini DM, Kaku LM, Esteves E, Bulet P, Daffre S. Cysteine-rich antimicrobial peptides of the cattle tick Boophilus microplus: isolation, structural characterization and tissue expression profile. Dev Comp Immunol. 2004;28:191–200. doi: 10.1016/j.dci.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Mather TN, Ribeiro JM. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem Biophys Res Commun. 2003;305:869–75. doi: 10.1016/s0006-291x(03)00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Mather TN, Ribeiro JM. Penthalaris, a novel recombinant five-Kunitz tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick vector of Lyme disease, Ixodes scapularis. Thromb Haemost. 2004;91:886–98. doi: 10.1160/TH03-11-0715. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, My Pham V, Mans BJ, Andersen JF, Mather TN, Lane RS, Ribeiro JM. The transcriptome of the salivary glands of the female western black-legged tick Ixodes pacificus (Acari: Ixodidae) Insect Biochem Mol Biol. 2005;35:1142–61. doi: 10.1016/j.ibmb.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Andersen JF, Mather TN, Ribeiro JM. Ixolaris, a novel recombinant tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick, Ixodes scapularis: identification of factor X and factor Xa as scaffolds for the inhibition of factor VIIa/tissue factor complex. Blood. 2002a;99:3602–12. doi: 10.1182/blood-2001-12-0237. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J Exp Biol. 2002b;205:2429–51. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Frauenschuh A, Power CA, Deruaz M, Ferreira BR, da Silva JM, Teixeira MM, Dias J, Martin T, Wells TN, Proudfoot AE. Molecular cloning and characterization of a highly selective chemokine binding protein from the tick Rhipicephalus sanguineus. J Biol Chem. 2007 doi: 10.1074/jbc.M704706200. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004;32:5452–63. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar AR, Joubert AM, Crause JC, Neitz AW. Isolation and characterization of an anticoagulant from the salivary glands of the tick, Ornithodoros savignyi (Acari: Argasidae) Exp Appl Acarol. 1996;20:583–98. doi: 10.1007/BF00052809. [DOI] [PubMed] [Google Scholar]
- Gillespie RD, Dolan MC, Piesman J, Titus RG. Identification of an IL-2 binding protein in the saliva of the Lyme disease vector tick, Ixodes scapularis. J Immunol. 2001;166:4319–26. doi: 10.4049/jimmunol.166.7.4319. [DOI] [PubMed] [Google Scholar]
- Grubhoffer L, Kovar V, Rudenko N. Tick lectins: structural and functional properties. Parasitology. 2004;129(Suppl):S113–25. doi: 10.1017/s0031182004004858. [DOI] [PubMed] [Google Scholar]
- Grunclova L, Horn M, Vancova M, Sojka D, Franta Z, Mares M, Kopacek P. Two secreted cystatins of the soft tick Ornithodoros moubata: differential expression pattern and inhibitory specificity. Biol Chem. 2006;387:1635–44. doi: 10.1515/BC.2006.204. [DOI] [PubMed] [Google Scholar]
- Gudderra NP, Ribeiro JM, Andersen JF. Structural determinants of factor IX(a) binding in nitrophorin 2, a lipocalin inhibitor of the intrinsic coagulation pathway. J Biol Chem. 2005;280:25022–8. doi: 10.1074/jbc.M504386200. [DOI] [PubMed] [Google Scholar]
- Hajnicka V, Kocakova P, Slavikova M, Slovak M, Gasperik J, Fuchsberger N, Nuttall PA. Anti-interleukin-8 activity of tick salivary gland extracts. Parasite Immunol. 2001;23:483–9. doi: 10.1046/j.1365-3024.2001.00403.x. [DOI] [PubMed] [Google Scholar]
- Hajnicka V, Vancova I, Kocakova P, Slovak M, Gasperik J, Slavikova M, Hails RS, Labuda M, Nuttall PA. Manipulation of host cytokine network by ticks: a potential gateway for pathogen transmission. Parasitology. 2005;130:333–42. doi: 10.1017/s0031182004006535. [DOI] [PubMed] [Google Scholar]
- Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998;15:115–30. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- Harnnoi T, Sakaguchi T, Nishikawa Y, Xuan X, Fujisaki K. Molecular characterization and comparative study of 6 salivary gland metalloproteases from the hard tick, Haemaphysalis longicornis. Comp Biochem Physiol B Biochem Mol Biol. 2007;147:93–101. doi: 10.1016/j.cbpb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Hoogstraal H, Aechlimann A. Tick host specificity. Bull Societe Entomol Suisse. 1982;55:5–32. [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isawa H, Yuda M, Orito Y, Chinzei Y. A mosquito salivary protein inhibits activation of the plasma contact system by binding to factor XII and high molecular weight kininogen. J Biol Chem. 2002;13:13. doi: 10.1074/jbc.M203505200. [DOI] [PubMed] [Google Scholar]
- Joubert AM, Louw AI, Joubert F, Neitz AW. Cloning, nucleotide sequence and expression of the gene encoding factor Xa inhibitor from the salivary glands of the tick, Ornithodoros savignyi. Exp Appl Acarol. 1998;22:603–19. doi: 10.1023/a:1006198713791. [DOI] [PubMed] [Google Scholar]
- Keller PM, Waxman L, Arnold BA, S LD, Condra C, Connolly TM. Cloning of the cDNA and expression of moubatin, an inhibitor of platelet-aggregation. J Biol Chem. 1993;268:5445–5449. [PubMed] [Google Scholar]
- Konik P, Slavikova V, Salat J, Reznickova J, Dvoroznakova E, Kopecky J. Anti-tumour necrosis factor-alpha activity in Ixodes ricinus saliva. Parasite Immunol. 2006;28:649–56. doi: 10.1111/j.1365-3024.2006.00899.x. [DOI] [PubMed] [Google Scholar]
- Kotsyfakis M, Sa-Nunes A, Francischetti IM, Mather TN, Andersen JF, Ribeiro JM. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick Ixodes scapularis. J Biol Chem. 2006;281:26298–307. doi: 10.1074/jbc.M513010200. [DOI] [PubMed] [Google Scholar]
- Kovar V, Kopacek P, Grubhoffer L. Isolation and characterization of Dorin M, a lectin from plasma of the soft tick Ornithodoros moubata. Insect Biochem Mol Biol. 2000;30:195–205. doi: 10.1016/s0965-1748(99)00107-1. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lai R, Takeuchi H, Jonczy J, Rees HH, Turner PC. A thrombin inhibitor from the ixodid tick, Amblyomma hebraeum. Gene. 2004a;342:243–9. doi: 10.1016/j.gene.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Lai R, Takeuchi H, Lomas LO, Jonczy J, Rigden DJ, Rees HH, Turner PC. A new type of antimicrobial protein with multiple histidines from the hard tick, Amblyomma hebraeum. Faseb J. 2004b;18:1447–9. doi: 10.1096/fj.03-1154fje. [DOI] [PubMed] [Google Scholar]
- Lambson B, Nene V, Obura M, Shah T, Pandit P, Ole-Moiyoi O, Delroux K, Welburn S, Skilton R, de Villiers E, et al. Identification of candidate sialome components expressed in ixodid tick salivary glands using secretion signal complementation in mammalian cells. Insect Mol Biol. 2005;14:403–14. doi: 10.1111/j.1365-2583.2005.00571.x. [DOI] [PubMed] [Google Scholar]
- Lima CA, Sasaki SD, Tanaka AS. Bmcystatin, a cysteine proteinase inhibitor characterized from the tick Boophilus microplus. Biochem Biophys Res Commun. 2006;347:44–50. doi: 10.1016/j.bbrc.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Louw AI, Neitz AW. Amino acid sequence and structure modeling of savignin, a thrombin inhibitor from the tick, Ornithodoros savignyi. Insect Biochem Mol Biol. 2002a;32:821–8. doi: 10.1016/s0965-1748(01)00169-2. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Louw AI, Neitz AW. Savignygrin, a platelet aggregation inhibitor from the soft tick Ornithodoros savignyi, presents the RGD integrin recognition motif on the Kunitz-BPTI fold. J Biol Chem. 2002b;277:21371–8. doi: 10.1074/jbc.M112060200. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Louw AI, Neitz AW. The major tick salivary gland proteins and toxins from the soft tick, Ornithodoros savignyi, are part of the tick Lipocalin family: implications for the origins of tick toxicoses. Mol Biol Evol. 2003;20:1158–67. doi: 10.1093/molbev/msg126. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Neitz AW. Adaptation of ticks to a blood-feeding environment: evolution from a functional perspective. Insect Biochem Mol Biol. 2004a;34:1–17. doi: 10.1016/j.ibmb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Neitz AW. The mechanism of alphaIIbbeta3 antagonism by savignygrin and its implications for the evolution of anti-hemostatic strategies in soft ticks. Insect Biochem Mol Biol. 2004b;34:573–84. doi: 10.1016/j.ibmb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Neitz AW. Molecular crowding as a mechanism for tick secretory granule biogenesis. Insect Biochem Mol Biol. 2004c;34:1187–93. doi: 10.1016/j.ibmb.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Steinmann CM, Venter JD, Louw AI, Neitz AW. Pathogenic mechanisms of sand tampan toxicoses induced by the tick, Ornithodoros savignyi. Toxicon. 2002c;40:1007–16. doi: 10.1016/s0041-0101(02)00098-3. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Venter JD, Vrey PJ, Louw AI, Neitz AW. Identification of putative proteins involved in granule biogenesis of tick salivary glands. Electrophoresis. 2001;22:1739–46. doi: 10.1002/1522-2683(200105)22:9<1739::AID-ELPS1739>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–3. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S, Koski RA, Beaulieu B, Anderson JF, Ramamoorthi N, Kantor F, Cappello M, Fikrig E. A novel family of anticoagulants from the saliva of Ixodes scapularis. Insect Mol Biol. 2002;11:641–50. doi: 10.1046/j.1365-2583.2002.00375.x. [DOI] [PubMed] [Google Scholar]
- Nene V, Lee D, Kang’a S, Skilton R, Shah T, de Villiers E, Mwaura S, Taylor D, Quackenbush J, Bishop R. Genes transcribed in the salivary glands of female Rhipicephalus appendiculatus ticks infected with Theileria parva. Insect Biochem Mol Biol. 2004a;34:1117–28. doi: 10.1016/j.ibmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Nene V, Lee D, Kang’a S, Skilton R, Shah T, de Villiers E, Mwaura S, Taylor D, Quackenbush J, Bishop R. Genes transcribed in the salivary glands of female Rhipicephalus appendiculatus ticks infected with Theileria parva. Insect Biochem Mol Biol. 2004b;34:1117–28. doi: 10.1016/j.ibmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nienaber J, Gaspar AR, Neitz AW. Savignin, a potent thrombin inhibitor isolated from the salivary glands of the tick Ornithodoros savignyi (Acari: Argasidae) Exp Parasitol. 1999;93:82–91. doi: 10.1006/expr.1999.4448. [DOI] [PubMed] [Google Scholar]
- Nunn MA, Sharma A, Paesen GC, Adamson S, Lissina O, Willis AC, Nuttall PA. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J Immunol. 2005;174:2084–91. doi: 10.4049/jimmunol.174.4.2084. [DOI] [PubMed] [Google Scholar]
- Paesen GC, Adams PL, Nuttall PA, Stuart DL. Tick histamine-binding proteins: lipocalins with a second binding cavity. Biochim Biophys Acta. 2000;1482:92–101. doi: 10.1016/s0167-4838(00)00168-0. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Perfettini JL, Castedo M, Roumier T, Andreau K, Nardacci R, Piacentini M, Kroemer G. Mechanisms of apoptosis induction by the HIV-1 envelope. Cell Death Differ. 2005;12(Suppl 1):916–23. doi: 10.1038/sj.cdd.4401584. [DOI] [PubMed] [Google Scholar]
- Rego RO, Hajdusek O, Kovar V, Kopacek P, Grubhoffer L, Hypsa V. Molecular cloning and comparative analysis of fibrinogen-related proteins from the soft tick Ornithodoros moubata and the hard tick Ixodes ricinus. Insect Biochem Mol Biol. 2005;35:991–1004. doi: 10.1016/j.ibmb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Rego RO, Kovar V, Kopacek P, Weise C, Man P, Sauman I, Grubhoffer L. The tick plasma lectin, Dorin M, is a fibrinogen-related molecule. Insect Biochem Mol Biol. 2006;36:291–9. doi: 10.1016/j.ibmb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Alarcon-Chaidez F, Francischetti IM, Mans BJ, Mather TN, Valenzuela JG, Wikel SK. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem Mol Biol. 2006;36:111–29. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Andersen J, Silva-Neto MA, Pham VM, Garfield MK, Valenzuela JG. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem Mol Biol. 2004a;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004b;34:543–63. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Schneider M, Guimaraes JA. Purification and characterization of Prolixin S (Nitrophorin 2), the salivary anticoagulant of the blood sucking bug, Rhodnius prolixus. Biochem J. 1995;308:243–249. doi: 10.1042/bj3080243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangamnatdej S, Paesen GC, Slovak M, Nuttall PA. A high affinity serotonin- and histamine-binding lipocalin from tick saliva. Insect Mol Biol. 2002;11:79–86. doi: 10.1046/j.0962-1075.2001.00311.x. [DOI] [PubMed] [Google Scholar]
- Santos IK, Valenzuela JG, Ribeiro JM, de Castro M, Costa JN, Costa AM, da Silva ER, Neto OB, Rocha C, Daffre S, et al. Gene discovery in Boophilus microplus, the cattle tick: the transcriptomes of ovaries, salivary glands, and hemocytes. Ann N Y Acad Sci. 2004;1026:242–6. doi: 10.1196/annals.1307.037. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–4. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeya H, Miyata T, Nishino N, Omori-Satoh T, Iwanaga S. Snake venom hemorrhagic and nonhemorrhagic metalloendopeptidases. Meth Enzymol. 1993;223:365–378. doi: 10.1016/0076-6879(93)23058-u. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urieli-Shoval S, Linke RP, Matzner Y. Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr Opin Hematol. 2000;7:64–9. doi: 10.1097/00062752-200001000-00012. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–32. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IMB, Pham VM, Garfield MK, Mather TN, Ribeiro JMC. Exploring the sialome of the tick, Ixodes scapularis. J Exp Biol. 2002;205:2843–64. doi: 10.1242/jeb.205.18.2843. [DOI] [PubMed] [Google Scholar]
- van de Locht A, Stubbs MT, B W, Friedrich T, B C, Hoffken W, Huber R. The ornithodorin-thrombin crystal structure, a key to the TAP enigma? Embo J. 1996;15:6011–6017. [PMC free article] [PubMed] [Google Scholar]
- Visintin A, Iliev DB, Monks BG, Halmen KA, Golenbock DT. Md-2. Immunobiology. 2006;211:437–47. doi: 10.1016/j.imbio.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao Q, Christensen BM. Identification and characterization of the fibrinogen-like domain of fibrinogen-related proteins in the mosquito, Anopheles gambiae, and the fruitfly, Drosophila melanogaster, genomes. BMC Genomics. 2005;6:114. doi: 10.1186/1471-2164-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman L, Connolly TM. Isolation of an inhibitor selective for collagen-stimulated platelet aggregation from the soft tick Ornithodorus moubata. J Biol Chem. 1993;268:5445–5449. [PubMed] [Google Scholar]
- Waxman L, Smith DE, Arcuri KE, Vlasuk GP. Tick anticoagulant peptide (TAP) is a novel inhibitor of blood coagulation Factor Xa. Science. 1990;248:593–596. doi: 10.1126/science.2333510. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Umeda F, Masakado M, Isaji M, Mizushima S, Nawata H. Purification and molecular cloning of prostacyclin-stimulating factor from serum-free conditioned medium of human diploid fibroblast cells. Biochem J. 1994;303(Pt 2):591–8. doi: 10.1042/bj3030591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Umeda F, Masakado M, Ono Y, Nawata H. Serum-free conditioned medium of human diploid fibroblast cells contains an activity that stimulates prostacyclin production by cultured bovine aortic endothelial cells. Biochem Mol Biol Int. 1993;31:65–71. [PubMed] [Google Scholar]
- Zhang SM, Loker ES. The FREP gene family in the snail Biomphalaria glabrata: additional members, and evidence consistent with alternative splicing and FREP retrosequences. Fibrinogen-related proteins. Dev Comp Immunol. 2003;27:175–87. doi: 10.1016/s0145-305x(02)00091-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ribeiro JM, Guimaraes JA, Walsh PN. Nitrophorin-2: a novel mixed-type reversible specific inhibitor of the intrinsic factor-X activating complex. Biochemistry. 1998;37:10681–90. doi: 10.1021/bi973050y. [DOI] [PubMed] [Google Scholar]
- Zhou J, Ueda M, Umemiya R, Battsetseg B, Boldbaatar D, Xuan X, Fujisaki K. A secreted cystatin from the tick Haemaphysalis longicornis and its distinct expression patterns in relation to innate immunity. Insect Biochem Mol Biol. 2006;36:527–35. doi: 10.1016/j.ibmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Zhu K, Bowman AS, Dillwith JW, Sauer JR. Phospholipase A2 activity in salivary glands and saliva of the lone star tick (Acari: Ixodidae) during tick feeding. J Med Entomol. 1998;35:500–4. doi: 10.1093/jmedent/35.4.500. [DOI] [PubMed] [Google Scholar]
- Zhu K, Dillwith JW, Bowman AS, Sauer JR. Identification of hemolytic activity in saliva of the lone star tick (Acari:Ixodidae) J Med Entomol. 1997;34:160–6. doi: 10.1093/jmedent/34.2.160. [DOI] [PubMed] [Google Scholar]
- Zimlichman S, Danon A, Nathan I, Mozes G, Shainkin-Kestenbaum R. Serum amyloid A, an acute phase protein, inhibits platelet activation. J Lab Clin Med. 1990;116:180–6. [PubMed] [Google Scholar]
- Zudaire E, Portal-Nunez S, Cuttitta F. The central role of adrenomedullin in host defense. J Leukoc Biol. 2006;80:237–44. doi: 10.1189/jlb.0206123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


