Abstract
The kinetics of sodium-stimulated phosphate flux and phosphate-stimulated sodium flux in human red cells have been previously described (Shoemaker, D.G., C.A. Bender, and R.B. Gunn. 1988. J. Gen. Physiol. 92:449–474). However, despite the identification of multiple isoforms in three gene families (Timmer, R.T., and R.B. Gunn. 1998. Am. J. Physiol. Cell Physiol. 274:C757–C769), the molecular basis for the sodium-phosphate cotransporter in erythrocytes is unknown. Most cells express multiple isoforms, thus disallowing explication of isoform-specific kinetics and function. We have found that erythrocyte membranes express one dominant isoform, hBNP-1, to which the kinetics can thus be ascribed. In addition, because the erythrocyte Na-PO4 cotransporter can also mediate Li-PO4 cotransport, it has been suggested that this transporter functions as the erythrocyte Na–Li exchanger whose activity is systematically altered in patients with bipolar disease and patients with essential hypertension. To determine the molecular basis for the sodium-phosphate cotransporter, we reasoned that if the kinetics of phosphate transport in a nucleated erythroid-like cell paralleled those of the Na-activated pathway in anucleated erythrocytes and yet were distinct from those known for other Na-PO4 cotransporters, then the expressed genes may be the same in both cell types. In this study, we show that the kinetics of sodium phosphate cotransport were similar in anuclear human erythrocytes and K562 cells, a human erythroleukemic cell line. Although the erythrocyte fluxes were 750-fold smaller, the half-activation concentrations for phosphate and sodium and the relative cation specificities for activation of 32PO4 influx were similar. Na-activation curves for both cell types showed cooperativity consistent with the reported stoichiometry of more than one Na cotransported per PO4. In K562 cells, external lithium activation of phosphate influx was also cooperative. Inhibition by arsenate, K I = 2.6–2.7 mM, and relative inhibition by amiloride, amiloride analogs, phosphonoformate, and phloretin were similar. These characteristics were different from those reported for hNaPi-3 and hPiT-1 in other systems. PCR analysis of sodium-phosphate cotransporter isoforms in K562 cells demonstrated the presence of mRNAs for hPiT-1, hPiT-2, and hBNP-1. The mRNAs for hNaPi-10 and hNaPi-3, the other two known isoforms, were absent. Western analysis of erythrocytes and K562 cells with isoform-specific antibodies detected the presence of only hBNP-1, an isoform expressed in brain neurons and glia. The similarities in the kinetics and the expression of only hBNP-1 protein in the two cell types is strong evidence that hBNP-1 is the erythrocyte and K562 cell sodium-phosphate cotransporter.
Keywords: cotransport, phosphate, sodium, lithium, BNP-1
INTRODUCTION
The kinetics of sodium-stimulated phosphate flux and phosphate-stimulated sodium flux in human red cells were initially described by Shoemaker et al. 1988. Recently, we have shown that the Na-phosphate cotransporter can also mediate Li-phosphate cotransport with similar kinetics and pharmacology (S. Elmariah and R.B. Gunn, manuscript in preparation). It is known that the rabbit kidney Na-phosphate cotransporter (NaPi-2) can mediate Na–Na exchange (Forster et al. 1998) and no other examined isoform transports lithium. Thus, the erythrocyte Na-phosphate cotransporter may mediate both the ouabain-insensitive Na–Na exchange and Na–Li exchange activities. The molecular identification of the Na–Li exchanger in erythrocytes and neurons has important clinical implications for understanding the treatment of bipolar disease with lithium and the development of essential hypertension. Although more than 24 Na-phosphate cotransporters in three gene families (Fig. 1) have been cloned and some are expressed in oocytes (Werner et al. 1991; Magagnin et al. 1993) and cultured mammalian cells (Quabius et al. 1996; Timmer and Gunn 1998), the molecular basis for the sodium–phosphate cotransporter in erythrocytes is unknown. Human representatives of each type have been cloned: hNaPi-10, a type Ia; hBNP-1, a type Ib; hNaPi-3, a type IIa; hPiT-1, a type IIIa; and hPiT-2, a type IIIb. Some of these isoforms show very restricted expression; e.g., type II cotransporters are principally found in the kidney and small intestine. In contrast, the type III isoforms are found in nearly every tissue in the body and thus are felt to be the constitutive housekeeping forms of Na-phosphate cotransporters that every cell seems to require to maintain intracellular phosphate above electrochemical equilibrium with respect to plasma phosphate. Because most cells express several isoforms of Na-phosphate cotransporters, the kinetics of individual isoforms have been evaluated principally in brush border vesicles, oocytes, and mammalian expression systems that may lack native regulatory mechanisms. The type Ia isoform was originally identified in proximal tubules of the kidney (Werner et al. 1991). A type Ib Na-phosphate cotransporter was recently cloned by subtractive hybridization of N-methyl-d-aspartate (NMDA)–treated primary cultures of cerebellar granule cells isolated from rat pups (Ni et al. 1994). This Na-phosphate cotransporter was originally reported to be a brain specific Na-PO4–inorganic cotransporter (BNPI or BNP-1), but the human isoform was also detected by Northern analysis in intestine, colon, and testes (Ni et al. 1996). Expression of BNP-1 in oocytes increased Na-dependent 32PO4 uptake (Ni et al. 1994). The amino acid sequences of hBNP-1 (Ni et al. 1996) and hNaPi-10 (Chong et al. 1993) share 22% identity, as determined using the ClustalX algorithm for sequence alignment (Higgins et al. 1996). Any of these or an unknown gene's product could be responsible for erythrocyte Na-PO4 cotransport.
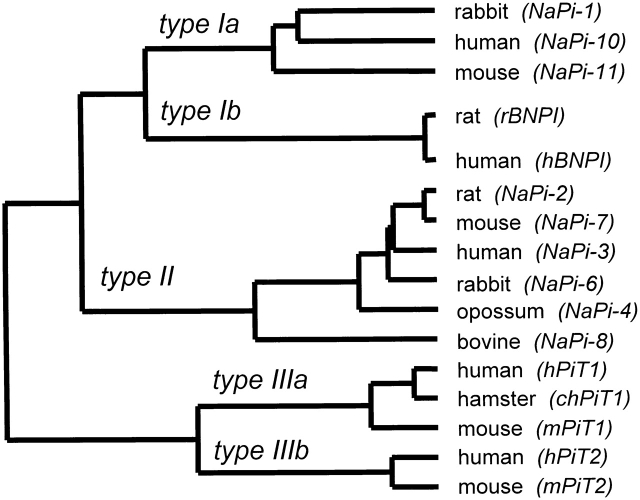
Figure 1.
Dendrogram showing the three major families of Na-phosphate cotransporters identified in mammalian cells. The multiple sequence alignment was carried out using ClustalX on the derived amino acid sequences for the indicated isoforms (protein weight matrix used was Dayhoff PAM 250; gap penalty, 10; and floating gap penalty, 10; see Thompson et al. 1994). The dendrogram was constructed from the dendrogram output file of ClustalX. The commonly used designation for the given isoform is given in parentheses next to the species type from which the cDNA was isolated. The corresponding gene names for these isoforms as designated by the Human Gene Nomenclature Committee (HGNC) are as follows: (a) SLC17A1 for NaPi-10; (b) SLC20A1 for hPiT1; (c) SLCS20A2 for hPiT2; and (d) SLC34A1 for NaPi-3. There is no name designated by HGNC for hBNP-1.
K562 cells were first isolated by Lozzio and Lozzio in 1971 from a patient with chronic mylogenous leukemia in blast crisis (Lozzio and Lozzio 1975). The cells have many characteristics of a red cell precursor; e.g., they can be stimulated to produce embryonic and fetal hemoglobin after treatment with dimethylsulfoxide, hemin, butarate, thymidine, etc. In addition, they constitutively express many red cell proteins, including spectrin and glycophorin A (Yurchenco and Furthmayr 1980) and L-type pyruvate kinase (Jansen et al. 1983). However, there are no previous reports regarding the existence of a Na-phosphate cotransport system in these cells or the identity of specific isoforms that may be expressed. K562 cells may express any combination of Na-PO4 cotransporter genes to feed their high metabolic demand for phosphate. It is known that they do not express AE1, which is the dominant phosphate pathway in mature erythrocytes (Horton et al. 1981; Law et al. 1983; Dissing et al. 1984), and thus the Na-PO4 cotransporter present in erythrocytes (Shoemaker et al. 1988) may be overexpressed to meet these needs.
Since mature human red blood cells are anucleated and have no DNA or RNA, the identification of the gene of origin of a given protein is not always straightforward. We reasoned that if the kinetics of phosphate transport in K562 cells paralleled those of the Na-activated pathway in erythrocytes and yet were distinct from those known for other Na-PO4 cotransporters, then the expressed genes may be the same in K562 cells and mature erythrocytes. Furthermore, the identification of the mRNAs for Na-PO4 cotransporters in K562 cells would restrict the likely gene candidates, and concordance of Western analysis on the two cell types could specifically identify the transporter, provided its known kinetic characteristics did not conflict with those of the two cell types. In this paper, we show that the transporter that mediates the Na-phosphate cotransporter in K562 cells has the same kinetic and pharmacological characteristics as the transporter responsible for Na-phosphate cotransport in mature human red cells. The characteristics of Na-dependent 32PO4 influx into K562 cells and red blood cells are distinct from those of the renal sodium-phosphate cotransporter, hNaPi-3, or a widely expressed Na-PO4 cotransporter, hPiT-1. We demonstrate by reverse transcription–PCR (RT-PCR) that the K562 cells contain mRNA for hPiT-1, hPiT-2, and hBNP-1, but not hPiT-10 or hNaPi-3. We show by Western analysis using specific antibodies to the individual cotransporters that hBNP-1 is the only Na-phosphate cotransporter polypeptide detectable in K562 cells and human red cells. We conclude that the K562 cells have a Na-phosphate cotransporter that is the gene product of hBNP-1, and that this transporter protein is the same as, or closely related to, the one in mature human red cells.
No native cells have been used previously for the separate study of PiT-1, PiT-2, or BNP-1. The transport characteristics of these isoforms are wholly derived from their expression in Xenopus oocytes (Kavanaugh et al. 1994; Ni et al. 1994; Kavanaugh and Kabat 1996), although the expression of one isoform may dominate, such as PiT-2 in rat fibroblasts (Kavanaugh et al. 1994). We propose here that erythrocytes and K562 cells are model systems for the behavior of BNP-1 which is also expressed in neuronal and glial cells, particularly the amygdala and hippocampus. Erythrocytes and K562 cells are the only cells known to express a single sodium-phosphate cotransporter isoform. The homologs of all three isoforms are widely expressed in rat brain, possibly in the same cells. The expression of rBNP-1 is selectively reduced in CA1 pyramidal neurons of the hippocampus, after ischemia (Ni et al. 1997) and significantly upregulated in cerebellar granule neurons after subtoxic doses of the excitatory amino acid NMDA (Ni et al. 1994). The only Na-PO4 cotransport measurements in neurons have been in cells whose compliment of sodium phosphate isoforms were not determined (Glinn et al. 1995).
Another possible reason for the importance of identifying hBNP-1 as the erythrocyte Na-PO4 cotransporter is the observation that lithium can substitute for Na on the cotransporter. There are no other good molecular candidates for the Na–Li exchanger in erythrocytes. The strong arguments against the sodium–hydrogen exchanger isoform (NHE1–NHE4) as the Na/Li exchanger are summarized by West et al. 1998. It has been shown that the activity of the erythrocyte Na–Li exchanger correlates with the therapeutic responsiveness of patients with bipolar (manic depressive) disease to lithium therapy (Ostrow et al. 1978; Pandey et al. 1984; Zaremba and Rybakowski 1986), but this remains controversial since it is not found in all patient populations (Werstiuk et al. 1984). Similarly, the activity has been shown to correlate with the development of essential hypertension (Canessa et al. 1980; Adragna et al. 1982; Cooper et al. 1983; West et al. 1998). Consequently, the activity of BNP-1 in erythrocytes may be a marker for its activity in the brain and other tissues inaccessible to diagnostic assays.
MATERIALS AND METHODS
Materials
K562 cells (CCL 243) were obtained from American Type Culture Collection. Fetal calf serum was obtained from Atlanta Biologicals; RPMI 1640, l-glutamine, and other media components were obtained from Life Technologies, Inc. Disposable plastic culture flasks and dishes were obtained from Corning, Inc. All chemicals were reagent grade or better, and were obtained from either Fisher Scientific or Sigma-Aldrich. The sodium salt of 4,4′-dinitro-2,2′-stilbenedisulfonic acid (DNDS) was obtained from Pfaltz and Bauer, Inc. Reagents used in PCR and RT-PCR were obtained from CLONTECH Laboratories, Inc. or PE Biosystems. Isotopes were purchased from New England Nuclear. Optifluor scintillation fluor was obtained from Packard Instrument Co.
K562 Cells
Cells were maintained and grown in suspension in RPMI 1640 media supplemented with 10% heat-inactivated fetal calf serum containing penicillin (50 U/ml) and streptomycin (50 μg/ml). The cells were grown and incubated at 37°C in a 5% CO2 atmosphere. The cells in all experiments were harvested and used from suspensions in logarithmic growth phase after being seeded at a density of 105 cells/ml.
Flux in K562 Cells
Cells (6–7 × 108) were harvested by decanting the cell suspension into several 50-ml tubes and centrifuging them at 3,000 g for 5 min at 4–10ºC. The clear media was aspirated, the cell pellets resuspended in wash media to a total volume of ∼100 ml, and the cell suspensions were combined. Wash media contained (mM): 0.81 MgCl2, 5.55 d-glucose, 0.3 KH2PO4, 25 HEPES, pH 7.64 at 20ºC. The cell suspensions were centrifuged briefly at 3,000 g, the supernatant was aspirated, and the cell pellets were resuspended in wash media. This cell washing procedure was repeated three times. After the last centrifugation step, the cells were resuspended in a minimal volume of wash medium (8–10 ml) by gentle vortexing to form a markedly concentrated cell suspension (∼7 × 107 cells/ml) that was held at 20°C until use (<15 min).
The sodium-dependent 32PO4 influx was calculated as the difference between the influx measured in the presence and absence of sodium. The influx media contained (mM): 4.36 KCl, 0.81 MgCl2, 5.55 d-glucose, 25 HEPES, and either 143.5 NaCl titrated with NaOH or 143.5 NMDG titrated with HCl to pH 7.64 at 20–21°C. Stoppered Erlenmeyer flasks (50 ml) contained 10 or 15 ml of a medium with a prescribed composition and 1–2 μCi/ml of 32PO4. The flasks were placed in a shaking water bath at 37°C. After temperature equilibration of the media, the 32PO4 influx measurements were initiated by adding ∼1 ml of the concentrated cell suspension prepared as described above. This addition dilutes the medium Na to ∼130 mM during the flux measurement. Duplicate samples were taken at known times (∼1, 2, 4, 7, 20, and 30 min) by removing 1-ml samples from the flasks and transferring them to labeled, 13-ml polystyrene test tubes containing 5 ml of ice-cold stop solution. Stop solution contained (mM): 144.5 NaCl, 6 KH2AsO4, 1 KH2PO4, and 20 HEPES, pH 7.64 at room temperature with KOH. These tubes were placed in a refrigerated centrifuge and spun at 2,000–3,000 g for 3–4 min. The supernatant was aspirated, and the pellets were immediately washed a further two times with 5 ml of ice-cold stop solution. The K562 cell pellets were lysed by addition of 250 μl of lysis solution A (1 mM MgCl2, 20 HEPES, 0.5 Triton X-100, pH 7.6 at room temperature), followed by incubation overnight at room temperature or for 2 h at 37°C. Next, 500 μl of lysis solution B (1% sodium deoxycholate) was added, mixed by vortexing, and diluted by addition of 750 μl of glass distilled water to yield 1.5 ml lysate for each sample. After the last timed sample was taken, the remaining cell suspension in each Erlenmeyer flask was transferred to a test tube, centrifuged, and the clear supernatant was saved for analysis of 32PO4, inorganic phosphate, and sodium. The lysed pellets of K562 cells were each analyzed for 32PO4 and protein. The radioactivity of the lysate (0.5 ml) was counted in duplicate in Optifluor (3 ml). The protein was measured in duplicate using 100-μl samples in a standard BCA protein assay (Smith et al. 1985) using bovine serum albumin as a standard. The reaction was performed in microtiter plates and measured in an MRX plate reader (Dynex Technologies) at 570 nm, as previously described (Timmer and Gunn 1998). The radioactivity of the influx solution was measured by counting 10 μl of the flux suspension supernatant together with 490 μl of a mixture of the Triton X-100 and deoxycholate lysis solutions. The phosphate concentration of the influx solution was measured using a standard malachite green method (Henkel et al. 1988; Timmer and Gunn 1998). The samples for influx solution–specific activity and cell lysis samples were counted contemporaneously and no correction (<0.5%) for radioactive decay was made.
Flux into Red Blood Cells
Human red blood cells were taken by venepuncture from one of us (R.B. Gunn) into a heparinized graduated cylinder. The whole blood was centrifuged in a 50-ml centrifuge tube at 12,000 g for 5 min at 4–10°C. The plasma and buffy coat were aspirated before the red cell pellet was dispersed with a glass rod into three volumes of ice-cold isotonic MgCl2 (110 mM) and again centrifuged briefly at 12,000 g. This wash procedure was repeated two to three more times and the mixed cells held on ice at 90% hematocrit until the flux measurement was initiated (<2 h).
The sodium-dependent 32PO4 flux into erythrocytes was measured in the same way as into K562 cells using the same flux medium with the following exceptions: (a) the influx media contained 0.25 mM DNDS, to block AE1-mediated phosphate influx; (b) samples were usually taken at ∼15, 45, and 75 min; and (c) at the end of the washings, the red cells were lysed by adding 1 ml of glass-distilled H2O and vortexing. The lysed red blood cell pellets were each analyzed by: (a) transferring 50 μl into 5-ml test tubes with 2 ml of modified Drabkin's solution, vortexing the test tube, and reading the concentration of cyanmethemoglobin at 540 nM in a Novaspec II spectrophotometer (Amersham Pharmacia Biotech) (the product of the OD540, the dilution in Drabkin's solution, and the factor 1.465 was used to calculate the milligrams of hemoglobin per milliliter in the lysate); and (b) taking 500 μl of each lysate into a 2-ml microcentrifuge tube with 250 μl of 7% (vol/vol) perchloric acid (PCA). After vortexing and a brief centrifugation of the tubes, 350 μl of the clear supernatant was counted with 3 ml of Optifluor in a Tricarb-4000 liquid scintillation counter (Packard Instrument Co.). The radioactivity of the influx solution was measured by counting either 10 μl of the flux suspension supernatant together with 340 μl of 2% PCA or measuring 350 μl of a PCA extract of the supernatant prepared in the same way as the red cell lysate samples. In Na+-free media, choline2-DNDS was used. It was prepared by ion exchange of Na2DNDS on a choline column of Rexyn 101 (H) (Fisher Scientific).
Flux Analysis
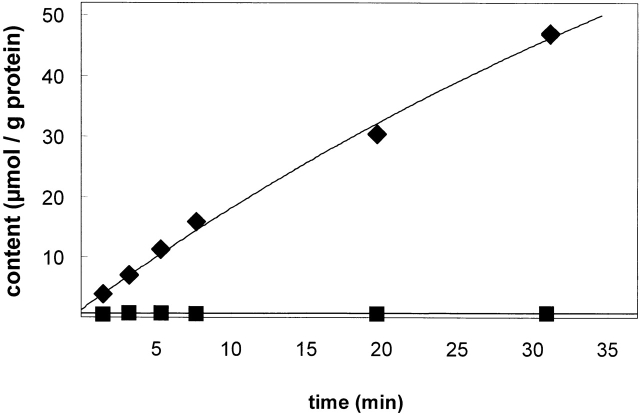
The values of picomoles per microgram protein for each of the six or more samples were plotted against the time elapsed between initiation of the influx and the time at which the samples were added to the ice-cold arsenate (K562 cells) or MgCl2 (red cells) solution to stop the influx reaction. The time course of 32PO4 content of K562 cells in a typical experiment is shown in Fig. 2. The 12–16 data points were fit to an exponential equation: y(t) = y infinity *[1 − exp(−kt)], where y is the cell content in picomoles per microgram protein at time t. The cell content of phosphate was determined from the ratio of the tracer content of the cells and the specific activity of the medium. The steady state content of the cells was estimated by the best-fit value to y infinity. The initial influx rate is given by the initial slope of the graph and is equal to the product of the two best-fit parameters: y infinity and k. In Fig. 2, the flux in a high sodium medium was 1,800 nmol/(g protein · min). The activation curves for phosphate were fit by minimization of the sum of squares for the Michaelis-Menten equation: v = V max PO4/(K 12 PO4+ PO4), where V is the flux, PO4 is the extracellular total phosphate concentration (mM), and K 12 PO4 is the concentration of total extracellular phosphate that causes half-maximal 32PO4 influx. The minimization algorithm used was the Solver module in the Excel spreadsheet (Microsoft Corp.). The activation curves for sodium were fit using the Hill equation: Flux = h[Na]o n/(1 + h[Na]o N), where h is the Hill constant and N is the Hill coefficient.
Figure 2.
Typical time course of phosphate influx in K562 cells in the presence and absence of Nao. Time course of traced phosphate content into K562 cells. (Top) 32PO4 influx into K562 cells in 128 mM NaCl medium. (Bottom) 32PO4 influx into K562 cells in the absence of Na (150 N-methyl-d-glucamine medium), pH 7.4, 37ºC, 0.3 mM PO4. The cell content of traced phosphate as a function of time was a single exponential y(t) − y o = (y ∞ − y o) · (1 − e − kt) where y o is the extrapolated initial traced content, y ∞ is the steady state traced content, and the initial slope of the curve is the influx: (y ∞ − y o) · (k) = 1,800 ± 570 nmol/(g protein · min). Duplicate values are obscured by the symbols.
Water Content
The water content of the red cells and K562 cells were measured. Red cells or K562 cells washed in appropriate media were loaded in nylon tubes at ∼50–70% cytocrit and packed under standard centrifugation conditions (Gunn et al. 1973). The grams of cell water content was measured by drying to constant weight at 95°C, a known weight of wet packed cells and correcting for 2.5% (wt/wt) trapped extracellular space in red cells and 8–20% (wt/wt) trapped extracellular space between the K562 cells. The trapped space was measured using 14C-polyethylene glycol or H3-inulin under the same packing conditions.
Ion Content
The ion contents of K562 cells were determined by making extracts of known weights of wet packed cells in 7% (vol/vol) PCA and measuring the Na and K concentration by atomic emission spectroscopy, the chloride concentration by coulombometric titration, and the phosphate concentration as described above. The ion contents of K562 cells were measured after washing in either RPMI, 110 mM MgCl2, the NMDG-Cl or NaCl solutions given above.
Antibody Production
Peptides were synthesized and purified by reversed-phase HPLC (Emory University Microchemical Facility). The peptides that were synthesized are as follows: (a) the hPiT-1–specific peptide was CEERTVSFKLGDLEEAPERER, (b) the hPiT-2-specific peptide was CNAVAEAEIEAEEGGVEMKLASE, and (c) the hBNP-1–specific peptide was CYESPALHPSISEEERKYIED. The N-terminal cysteine residue is not present in the target isoform polypeptide sequence and was added to facilitate conjugation to keyhole limpet hemocyanin (KLH; Pierce Chemical Co.) with sulfosuccinimidyl 4-[N-maleimidomethyl]cyclohexane-1-carboyxlate (sulfo-SMCC; Pierce Chemical Co.). In brief, 2.0 ml KLH (10 mg/ml in water) was first activated by adding 1.0 ml of sulfo-SMCC (4.6 mM or 2 mg/ml) dissolved in conjugation buffer (83 mM Na2HPO4, 900 mM NaCl, and 100 mM EDTA). The activation reaction of KLH and sulfo-SMCC was allowed to proceed for 1 h at room temperature (18–22ºC). The unreacted sulfo-SMCC is removed from KLH and sulfo-SMCC-activated KLH by gel filtration column chromatography. The reaction mixture was applied to a 10-ml column (EconoPac 10DG column; BioRad, Inc.) equilibrated with conjugation buffer and fractions (2.5 ml) collected. The protein concentration in each fraction was determined and the fractions containing the protein peak were pooled. The activated protein (sulfo-SMCC–activated KLH) was incubated with 10 mg of the peptide dissolved in 2.5 ml of conjugation buffer. The conjugation reaction was allowed to proceed for 2 h at room temperature, and then dialyzed overnight at 4ºC against 500 ml phosphate-buffered saline (1.76 mM KH2PO4, 8.05 mM Na2HPO4, 136 mM NaCl, and 2.68 mM KCl, pH 7.2). The final solution contained the peptide-KLH in PBS (∼0.4 mg peptide/ml and 1.7 mg KLH/ml). The peptide conjugate (100 μl at 0.8–0.9 mg/ml in PBS) was emulsified with Titermax (250 μl; CytRx Corp.) using a syringe according to the manufacturer's instructions. The emulsified conjugate adjuvant (50 μl) was injected in each hind quadricep of New Zealand White rabbits. At week five, a second injection of conjugate adjuvant was carried out as described above. Positive antisera were obtained by week three. Injections were carried out and antisera were obtained according to standard protocols at Lampire Biological Laboratories. Control sera were obtained from the same rabbits before the first injection.
Western Analysis
We followed the detailed protocol previously published (Timmer and Gunn 1998), except that protein was transferred to PVDF (polyvinylidene difluoride) membranes instead of nitrocellulose membranes. The concentration of protein in the cell lysates was determined by the BCA method, as described above. Blots were probed with the primary antibody directed against the indicated Na-PO4 cotransporter protein isoforms, then developed using goat anti–rabbit immunoglobulin conjugated to horseradish peroxidase and chemiluminescence according to the manufacturer's protocol (ECL Western Blot kit; Amersham Pharmacia Biotech). The primary antibodies used in these studies were prepared as described above (anti–PiT-1, anti–PiT-2, and anti–BNP-1) or as previously described (anti–NaPi-3; Timmer and Gunn 1998).
RT-PCR Reaction and Analysis
PCR reactions were carried out on cDNA using the Advantage 2 PCR enzyme system (CLONTECH Laboratories, Inc.). This enzyme system contains a mixture of a recombinant engineered Taq polymerase, a proofreading thermophilic DNA polymerase, and an antibody to the Taq polymerase to enable “hot start” PCR reactions. Thermocycling conditions were as follows: 1 cycle at 95ºC for 1 min; 30 cycles of 30 s at 95ºC and 3 min at 68ºC; and then the reaction was held at 25ºC for 1–8 h. The primer concentration used in these reactions was 0.4 μM, and all other reaction conditions were as suggested by the manufacturer. The cDNA used in the reactions was obtained from CLONTECH Laboratories, Inc. Control PCR reactions used primers specific for GAPDH. The GAPDH primer sequences used are as follows: (a) GAPDH-F, (5′) ACC ACA GTC CAT GCC ATC AC (3′); and, (b) GAPDH-R, (5′) TCC ACC ACC CTG TTG CTG TA (3′). The expected product from the GAPDH primers is 452 bp. The isoform-specific primers were designed with the aid of computer software (Primer Designer; Scientific and Educational Software). The primer pairs with sequences, target isoform, and expected PCR product size are shown in Table .
Table 1.
Primers Used for RT-PCR Analysis of Isoform Expression
| Target isoform* | Forward primer | Reverse primer | Forward primer sequence‡ | Reverse primer sequence‡ | Expected product |
|---|---|---|---|---|---|
| bp | |||||
| hPiT-1 | Pit1-1451F | Pit1-2275R | CAG TTC AGT CCA GCC GTC AG | CCA GCC AAC AGA CAC AAC AG | 825 |
| hPiT-1 | Pit1-1882F | Pit1-2191R | TGA CCA GGA TAA GCC TGA AG | GGC AGA TGC CAG TTC AAT AC | 310 |
| hPiT-2 | Pit2-482F | Pit2-1416R | ACA ACG AGA CGG TGC TCA TC | CAC ACT TCT CCT CGC TCA TC | 935 |
| hBNP-1 | BNP-1410F | BNP-1875R | CGT TCT ACC TGC TGC TCA TC | CAC ACT TCT CCT CGC TCA TC | 466 |
| hBNP-1 | BNP-1410F | BNP-1985R | CGT TCT ACC TGC TGC TCA TC | AAC ACG TAC TGC CAC TCC TC | 576 |
| NaPi-10 | NP10-512F | NP10-970R | GAG GCC GAC TTA CTT CTA TGA G | GAC CTG CTA GGT TAC CAC AGA T | 459 |
| NaPi-10 | NP10-267F | NP10-971R | TTC CAC CTC CTA TGG TGT CAT C | TGA CCT GCT AGG TTA CCA CAG A | 705 |
| NaPi-3 | NP03-952F | NP03-1430R | AGT CTC ATY CRG ATY TGG TG | GGC CAG KGC WGC CAG GAT | 479 |
RESULTS
Phosphate Activation
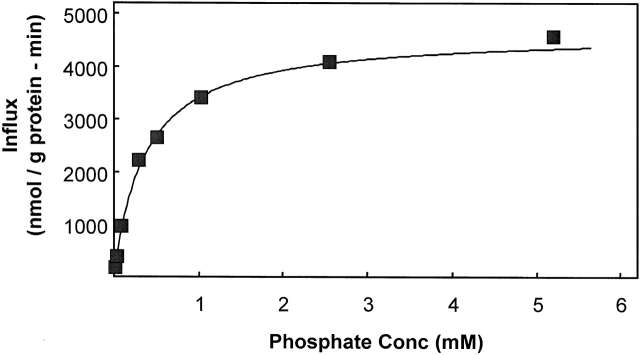
The K 12 PO4 for phosphate activation of 32PO4 influx into K562 cells was determined from flux measurements at several different total phosphate concentrations in sodium media at pH 7.4 at 37°C (Fig. 3). The K 1/2 = 0.36 mM when fit to the Michaelis-Menten equation by weighted least squares analysis. For comparison, we previously reported that the K 1/2 for 32PO4 flux into human erythrocytes was 0.30 ± 0.02 mM under similar conditions (Shoemaker et al. 1988).
Figure 3.
Determination of the K 1/2 for phosphate activation of 32PO4 influx. Extracellular phosphate activation of 32PO4 flux into K562 cells at Naout = 130 mM. The data were fit to the Michaelis-Menten equation, v = Vmax · [PO4]out/(K 1/2 + [PO4]out), by nonlinear least squares. For the data shown, Vmax = 4,630 nmol PO4/(g protein · min) and K 1/2 = 0.36 mM. The phosphate influx was almost entirely Na dependent: the flux carried out at 0.3 mM PO4 in the absence of Na+ was only ∼75 nmol PO4/(g protein · min). Each point is a single flux calculated from the time course of six values at the given extracellular phosphate concentration. One of five similar experiments.
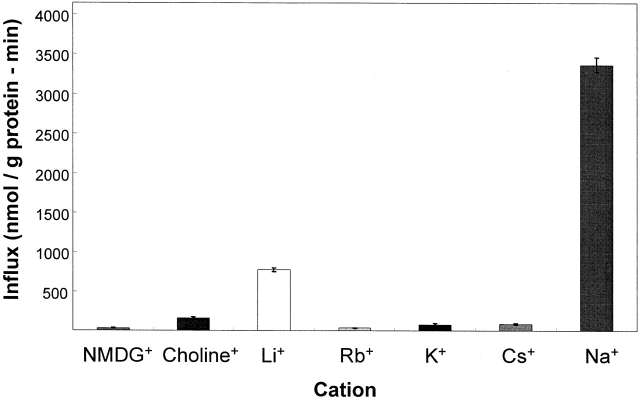
Selectivity of Monovalent Cation Activation
Activation of the 32PO4 influx by monovalent cations is shown in Fig. 4 for K562 cells. Chloride media with the only monovalent cation being 143 mM N-methyl-d-glucamine, choline, rubidium, potassium, or cesium gave small or negligible 32PO4 influxes into K562 cells. Only lithium and sodium were able to activate the flux relative to the other cations. The activation of the 32PO4 influx in K562 cells (Fig. 4) by 143 mM external lithium was 19% (±6%, n = 15) of the activation by 143 mM external sodium (100%). This relative activation by these two alkali cations was the same in erythrocytes (S. Elmariah and R.B. Gunn, manuscript in preparation) and was much greater than the relative activation (<2%) reported in HEK-293 cells expressing hNaPi-3 (Timmer and Gunn 1998) or seen in oocytes expressing hPiT-1 (Olah et al. 1994).
Figure 4.
Activation of 32PO4 influx by monovalent cations in erythrocytes and K562 cells. Monovalent cation activation of 32PO4 influx in the presence of 1.0 mM external phosphate, pH 7.4, 37ºC, and fluxes were carried out as described with each indicated cation present at 143 mM using the chloride salt. The standard errors of six determinations in one of four similar experiments.
Sodium Activation
The concentration dependence for external sodium activation of 32PO4 influx in both K562 cells and human erythrocytes is shown in Fig. 5. In both cases, the sodium concentration was adjusted by isomolar substitution with NMDG. In both cell types, the cooperative activation by Na is evident by the sigmoidicity of the activation graph. This is consistent with more than one Na+ ion being required for phosphate influx. Although this cooperativity has not been previously reported in erythrocytes, it is consistent with the previous measurements of the stoichiometry showing that more than one sodium is transported for each phosphate transported (Shoemaker et al. 1988). In K562 cells the K 1/2 Na was 34 mM with Hill coefficient n = 1.5 and the constant h = 0.0047. However, these data can be equally well fit [nearly the same sum of squares of residuals: 886,797 (Hill) vs. 968,855 (M–M)] by a Michaelis-Menten equation (n = 1.0) with best fit parameters for K 1/2 Na = 80 mM and for Vmax = 6,000 nmol/(g protein · min). In erythrocytes, the K 1/2 Na was 46 mM, n = 1.9, and h = 0.00058.
Figure 5.
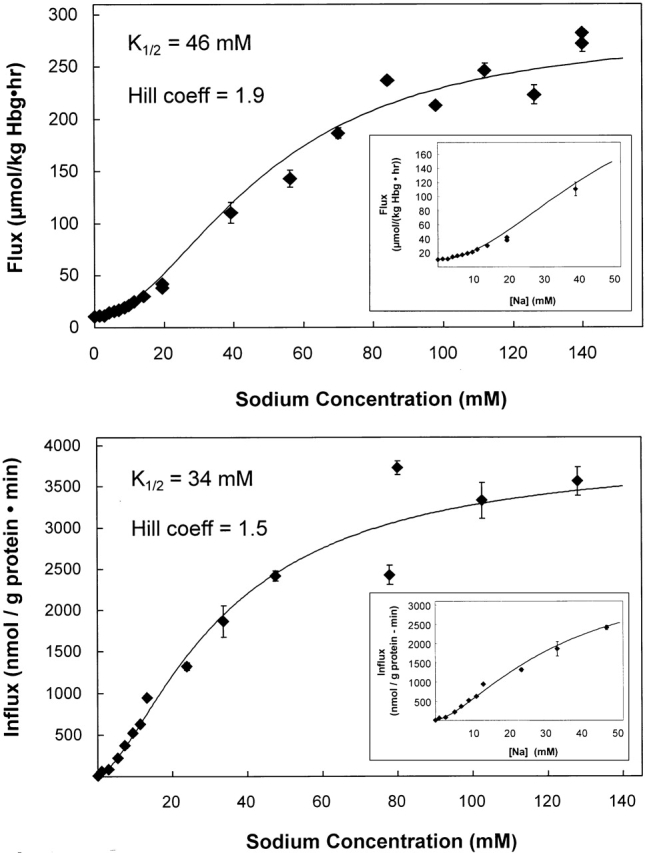
Determination of the K 1/2 for sodium activation of 32PO4 influx. Sodium activation of 32PO4 flux into erythrocytes (top) and K562 cells (bottom) was determined. In these experiments, the total phosphate concentration was 0.3 mM. Both curves appear sigmoidal, although the data from K562 cells is equally well fit by a single site (Michaelis-Menten) equation. Using the Hill equation, the concentration of sodium required for half maximal activation of phosphate influx was 46 mM for erythrocytes and 34 mM for K562 cells. The extrapolated maximum flux was 277 μmol/[kg hemoglobin (Hgb) · h] in erythrocytes and 3,900 nmol/(g protein · min) in K562 cells. The Hill coefficient, N, was 1.9 for erythrocytes and 1.5 for K562 cells. When the Michaelis-Menten equation was used to fit the data from K562 cells, the Vmax was 6,000 nmol/(g protein · min) and K 1/2 = 80 mM.
Lithium Activation
The graph of the activation of 32PO4 influx by external lithium was also cooperative in K562 cells (Fig. 6) and in erythrocytes (S. Elmariah and R.B. Gunn, manuscript in preparation). This suggests that there are at least two sites for lithium on the extracellular domain of the transporter that must be occupied by lithium for transport of phosphate in the absence of extracellular Na. Although the K 1/2 Li was >140 mM Li in sodium-free media (Fig. 6, K 1/2 Li ∼ 400–500 mM), the extrapolated maximum flux was about the same for Li as for Na activation under the same conditions [3,900 nmol PO4/(g protein · min)]. If the two cation binding sites for Na and Li are the same two sites, then the apparent affinity of the lower affinity site is greater for sodium than for lithium. This follows from using the reciprocal of the K 1/2 values as a measure of the apparent affinity of the second (lower affinity) cation activation site. Namely, (34 mM; Na,K562)−1 ≅ (46 mM; Na,RBC)−1 >> (∼450 mM; Li,K562)−1. This lower affinity for Li in the context of comparable Vmax values for the phosphate flux activated by either cation appears to be the sole reason why 143 mM Li only activates 19% as well as 143 mM Na+.
Figure 6.
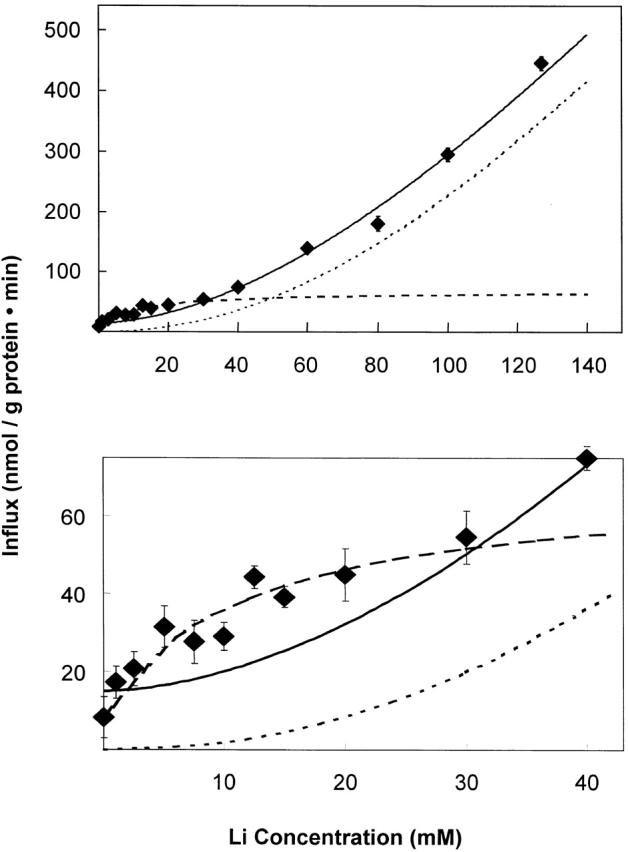
Determination of the K 1/2 for lithium activation of 32PO4 influx in K562 cells. In these experiments, the total external phosphate concentration was 0.3 mM. When the data over the entire concentration range (0–143.5 mM Li+) are fit to the Hill equation (top and bottom, solid line), the activation was cooperative with K 1/2 Li = 430 mM and Hill coefficient, N = 1.7 for the best fit line as shown assuming Vmax = 3,900 nmol PO4/(g protein · min). However, when the data at low lithium concentration (0–30 mM) are fit to the Hill equation (bottom, dashed line), the activation was not cooperative with K 1/2 Li = 7 mM and Hill coefficient, N = 1.0 for the best fit line, as shown assuming Vmax = 90 nmol PO4/(g protein · min). When the values from this line are subtracted from the original data, and the resulting values fit to the Hill equation (top and bottom, dotted line), the activation was cooperative with K 1/2 Li = 270 mM and Hill coefficient, N = 2.1 for the best fit line as shown assuming Vmax = 2,100 nmol PO4/(g protein · min). It appears that the activation of 32PO4 influx has two components: a hyperbolic component with a Hill coefficient of 1 (cotransport of 1 Li+ and 1 PO4) and a sigmoidal component with a Hill coefficient of 2 (cotransport of two Li+ and one PO4).
The activation of 32PO4 influx at low concentrations (0–30 mM) of lithium (Fig. 6, bottom) did not appear to be cooperative upon careful inspection in two experiments, but rather could be fit with a Michaelis-Menten equation using K 1/2 Li ≅ 7 mM and Vmax ≅ 90 nmol PO4/(g protein · min). This component could be subtracted from the total flux to reveal the cooperative component as shown by the dotted curves in Fig. 6.
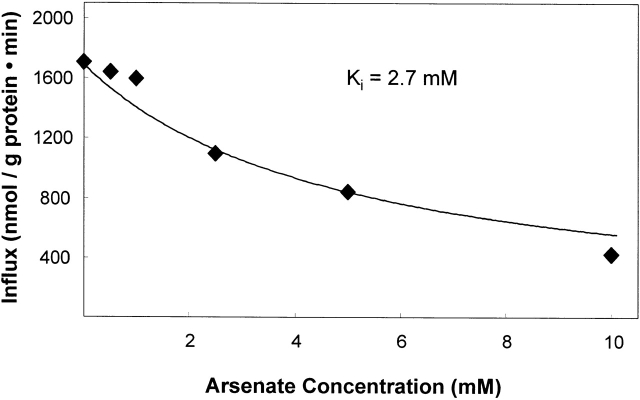
Arsenate Inhibition
Phosphate transport in the dog kidney (Ginsburg and Lotspeich 1962), rat kidney brush border membrane vesicles (Hoffmann et al. 1976), and gut (Berner et al. 1976) is inhibited by arsenate, which behaves as a phosphate congener (Gmaj and Murer 1986; Kenney and Kaplan 1988). Arsenate inhibits sodium-activated 32PO4 influx into erythrocytes (S. Elmariah and R.B. Gunn, manuscript in preparation) with a K i = 2.6 mM assuming complete competitive inhibition. Arsenate inhibits 32PO4 influx into K562 cells (Fig. 7) with a K i = 2.7 mM. These values are in contrast to the K i = 0.4 mM reported for the inhibition of hNaPi-3 expressed in HEK-293 cells (Timmer and Gunn 1998).
Figure 7.
Determination of the K I for arsenate inhibition of the sodium-activated 32PO4 influx in K562 cells. The data were fit to the equation for a competitive inhibitor: influx = Vmax · [PO4]/{K 1/2 · (1 + [arsenate]/K i + [PO4])} by varying K i. The constants were used from Fig. 2: Vmax = 3,712, K 1/2 = 0.36 mM, [PO4] = 0.3 mM. The K i value is 2.7 mM. The error bars are obscured by the symbols.
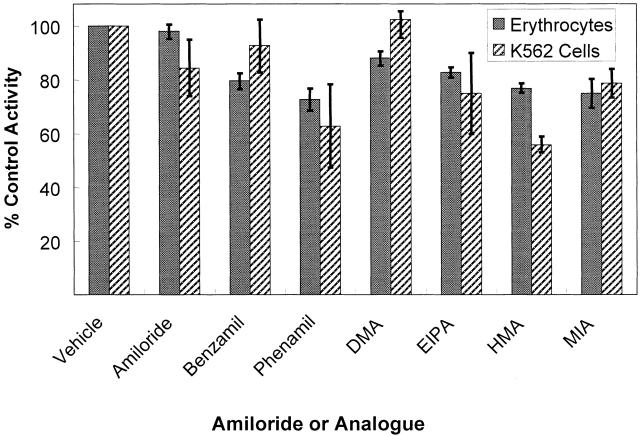
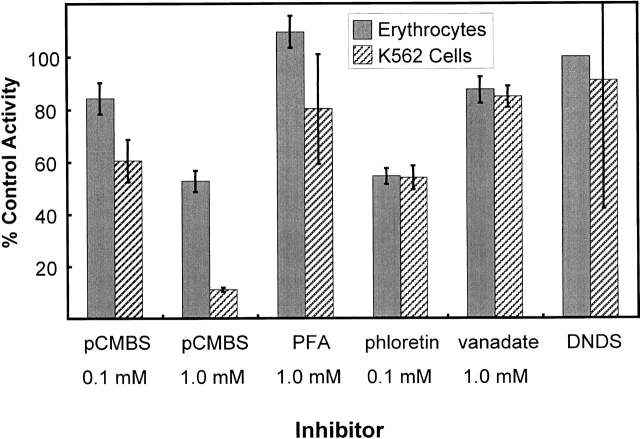
Pharmacology of Inhibition
In addition to the kinetics of activation, the pharmacology of inhibition, or lack thereof, is the same in mature erythrocytes and K562 cells measured under the same conditions. Shown in Fig. 8 is a comparison of the inhibition of 32PO4 influx by amiloride and six amiloride analogs in the two cell types. There are some minor variations between the two cell types, but, in general, amiloride and its analogs are poor inhibitors of the Na-dependent flux in these two cell types. Thus, the amiloride binding sites or its ability to inhibit this Na-PO4 cotransporter are different from that on the Na/H and Na/Ca exchangers (Kleyman and Cragoe 1988), epithelial Na channels of the principal cells of the distal nephron (Kleyman and Cragoe 1988), the Na-hexose (Cook et al. 1987), or the Na-alanine (Harris et al. 1985) cotransporters, which are all amiloride sensitive and over 90% inhibited at these concentrations. Results in Fig. 9 show the percent of the control Na-phosphate flux into K562 cells and erythrocytes after treatment with p-chloromercuri phenylsulfonate (pCMBS), phosphonoformate (PFA), phloretin, vanadate, and DNDS. The effect of these drugs on NaPi-3–mediated Na-phosphate cotransport has been reported (Timmer and Gunn 1998). There is only an apparent difference in the inhibition by pCMBS in erythrocytes and K562 cells. This is probably due to the high concentration of glutathione (1 mM) in red cells but not K562 cells since glutathione reacts with pCMBS and reduces the effective concentration in the erythrocyte experiments. In erythrocytes, the sensitivity of the cotransporter to DNDS cannot be directly determined since it was always present in the assays to block the AE1-mediated phosphate flux. However, since the Na-activated portion of the phosphate flux was always the same in the absence or presence of DNDS, the Na-phosphate cotransporter is not believed to be inhibited by DNDS. PFA has little effect on the Na-activated phosphate transport into K562 cells or erythrocytes, but is a strong inhibitor of NaPi-3 (Timmer and Gunn 1998).
Figure 8.
Comparison of the sensitivity of erythrocytes and K562 cells to inhibition by amiloride and its analogs. The flux assays were carried out as described, except that the media contained 0.3 mM total phosphate and 32.5 mM DMSO. The control (100%) flux values for erythrocytes were 0.26–0.30 mmol PO4/kg Hgb · h in different experiments. The control flux values for K562 cells were 850–960 nmol/g protein · min. The standard deviation of two to four determinations is shown by the error bars. Each of the indicated compounds was used at 0.1 mM final concentration. The compounds used are indicated using their abbreviated names, which are used for the following chemical names: Phenamil is phenamil methanesulfonate; DMA is 5-(N,N-dimethyl)-amiloride; EIPA is 5-(N-ethyl-N-isopropyl)-amiloride; HMA is 5-(N,N-hexamethylene)-amiloride; and, MIA is 5-(N-methyl-N-isobutyl)-amiloride.
Figure 9.
Pharmacology of inhibition of phosphate influx in erythrocytes and K562 cells. Various compounds were tested for their ability to inhibit the Na-dependent phosphate influx in human erythrocytes (shaded bars) or K562 cells (cross-hatched bars). All erythrocyte suspensions also contained 0.25 mM DNDS to block AE1. The control (100%) flux values for erythrocytes were 0.20–0.41 mmol PO4/kg Hgb · h in different experiments. The control flux values for K562 cells were 750–1,940 nmol/g protein · min. The standard deviation of two to four measurements is shown by the error bars. Each of the indicated compounds was used at the indicated concentrations. The following compounds were tested: pCMBS, PFA, phloretin, sodium vanadate (vanadate), and DNDS.
These data show that the kinetic and pharmacological characteristics of the Na- and Li-activated 32PO4 influx is the same in K562 cells and mature erythrocytes. These kinetics are very different from the renal transport mechanisms mediated by hNaPi-3 (NaPi-2, rat). This is most easily explained by a common molecular mechanism in K562 cells and erythrocytes and one that is different from that in renal cells. We have, therefore, determined which genes of known sodium-phosphate cotransporter proteins are expressing mRNA in the K562 cells, and thus which protein was probably responsible for these flux characteristics.
PCR Analysis of Sodium-Phosphate Cotransporter Isoforms
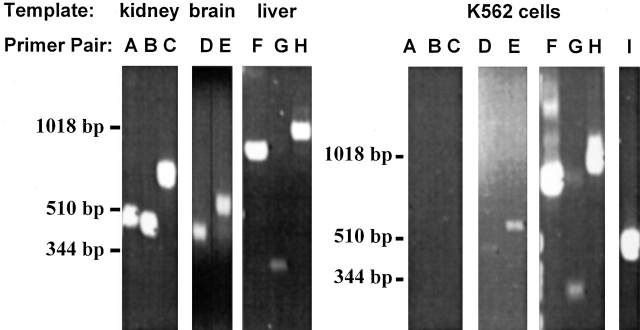
We used primer design software to design several sets of primers specific for the human forms of the major known Na-dependent phosphate transporters shown in the dendrogram in Fig. 1. The primers used in these experiments are described in Table . PCR reactions were carried out using cDNA templates (Fig. 10) derived from specific human tissues (brain, liver, and kidney) and K562 cells. The cDNA from human tissues was used as a positive control for these primer sets; e.g., liver cDNA was a positive control for the hPiT-1 and hPiT-2 primer sets, brain cDNA was a positive control for the BNP-1 primer sets, and kidney cDNA was a positive control for the NaPi-3 and NaPi-10 primer sets. It has been previously reported that PiT-1 and PiT-2 mRNAs are ubiquitously expressed as determined by Northern analysis (Kavanaugh et al. 1994). The expression of the two isoforms is approximately equal in liver (Kavanaugh and Kabat 1996), where there is Na-dependent, arsenate-sensitive phosphate transport (Escoubet et al. 1993; Ghishan et al. 1993; Younus and Butterworth 1993). The expression of BNP-1 mRNA was originally described as being restricted to the brain (Ni et al. 1994). The highest level of expression of BNP-1 in human is in brain, although there is low-level expression in intestine, colon, and testes (Ni et al. 1996). The expression of NaPi-3, a type IIa isoform, is restricted to the renal proximal tubule (Magagnin et al. 1993). The principal tissue of expression for the type Ia isoform (e.g., NaPi-10) is also the kidney (Chong et al. 1993). PCR carried out using these primer sets specifically amplifies the predicted products from the cDNA prepared from tissues that are known to express these isoforms (compare predicted product sizes in Table and the results in Fig. 10, left, A–H). When cDNA prepared from K562 cells is used as the template in the PCR reactions, only the primer sets for PiT-1, PiT-2, and BNP-1 yield the expected products (Fig. 10, right, D–H). In contrast, the primer sets for NaPi-3 and NaPi-10 do not yield any products from the same template cDNA. Thus, the mRNAs for PiT-1, PiT-2, and BNP-1 appear to be expressed in K562 cells. Similar results were obtained using total RNA isolated from the K562 cells used in these experiments and a single-tube RT–PCR system (data not shown). All experiments were carried out using a minus template negative control (water substitution for cDNA template). In these samples, there was no visible bands produced, indicating the absence of contaminating DNA in the PCR reagents (data not shown). GAPDH primers spanning a known intron indicate the samples were not contaminated by genomic DNA; i.e., products of distinct size were realized with amplification from cDNA versus genomic DNA (data not shown).
Figure 10.
Expression of sodium-phosphate cotransporter isoforms in human tissues and K562 cells as determined by PCR. cDNA was obtained from CLONTECH Laboratories, Inc. for several human tissues, including kidney, brain, liver, and K562 cells. The templates used for the given various PCR primers are indicated above the gel image. Primer pairs specific for the various sodium-phosphate cotransporter isoforms or GAPDH are indicated above the gel image. The primer pairs and their expected products were as follows (see Table for description): (A) NaPi-3 (479 bp), (B) NaPi-10 (459 bp), (C) NaPi-10 (705 bp), (D) hBNP-1 (466 bp), (E) hBNP-1 (576 bp), (F) hPiT-1 (825 bp), (G) hPiT-1 (310 bp), (H) hPiT-2 (935 bp), and (I) GAPDH (452 bp).
Western Analysis of Sodium-Phosphate Cotransporter Isoforms
The expression of a particular mRNA does not always mean that the corresponding protein is also expressed. To determine whether the polypeptides for PiT-1, PiT-2, and BNP-1 were expressed in K562 cells, it was necessary to carry out Western analysis on total cell lysates. We had previously prepared antibodies specific for NaPi-3, which is a type II sodium-phosphate cotransporter (Fig. 1) that is localized exclusively to the kidney (Timmer and Gunn 1998). The other known human sodium-phosphate cotransporters are: (a) NaPi-10, the human type Ia sodium-phosphate cotransporter, which is found predominantly in the kidney; (b) hBNP-1, the human type Ib cotransporter that was originally localized to brain tissue; (c) the hPiT-1 cotransporter; and (d) hPiT-2, which is homologous to hPiT-1, but is a distinct isoform.
Based on the isoforms detected by RT-PCR, we prepared isoform-specific antipeptide polyclonal antibodies to the hBNP-1, hPiT-1, and hPiT-2 cotransporter isoforms. To prepare these polyclonal antibodies, we followed a five-step procedure to construct the peptide antigens for the several sodium-phosphate cotransporters. First, the sequence for each isoform was analyzed using three different algorithms for prediction of membrane protein topology (Kyte and Doolittle 1982; Eisenberg et al. 1984; Mohana Rao and Argos 1986) to develop an approximate picture of the number and location of membrane-spanning segments for each. Second, the sequence was compared against the PROSITE database (Bairoch et al. 1997) of protein motifs to determine the location of potential sites for post-translational modification (e.g., glycosylation or phosphorylation). Third, the known sequences for each type from several species were aligned using the Clustal algorithm (Higgins et al. 1992) to define regions of homology between several species to maximize the utility of the antisera. Fourth, the sequences for hNaPi-10, hBNP-1, hPiT-1, and hPiT-2 using the Clustal algorithm (Higgins et al. 1992) to define regions that are distinct for each of these cotransporters. Finally, we analyzed the sequence using the algorithm of Hopp and Woods 1981 to identify regions with a high probability for eliciting an immune response. The peptides that we used to make polyclonal antibodies based upon these analyses are given in Table . Each immunizing peptide has a unique sequence with minimal potential for post-translational modification; e.g., glycosylation or phosphorylation. We were unable to identify a single peptide sequence for either hBNP-1 or hPiT-2 that would both be highly antigenic and recognize human and rat isoforms. Therefore, we used a peptide sequence that had lower predicted antigenicity but was common to both rat and human.
Table 2.
Peptides Used for Preparation of Isoform-specific Antibodies
| Target isoform | Genbank accession No. | Peptide sequence* | Position in human protein | Predicted to cross-react with rat isoform‡ |
|---|---|---|---|---|
| hPiT-1 | 4885601 | CEERTVSFKLGDLEEAPERER | E313-R332 | yes |
| hPiT2 | 5830173 | CNAVAEAEIEAEEGGVEMKLASE | N438-E459 | yes§ |
| hBNP-1 | I59302 | CYESPALHPSISEEERKYIED | Y257-D276 | yes |
| hNaPi-3‖ | 4506985 | CKLALEEEQKPESRLVPK | K77-K93 | yes¶ |
*The amino-terminal cysteine is not part of the native protein sequence and is added to the peptide only for conjugation to the carrier protein. ‡Unless otherwise noted, the sequence is identical in rat or mouse. §The sequence “MKL” in the human sequence is “MRL” in the rat sequence. ‖Preparation of this antibody was previously described (Timmer and Gunn 1998). ¶The comparable sequence in rat is (bold indicates mismatches): KLAQEEEQKPEPRLSPK. We have previously shown that this antibody specifically detects the appropriate polypeptide in rat kidney (Timmer and Gunn 1998).
The antigens were prepared by conjugating the isoform-specific peptide to KLH, and the resulting antisera assayed in Western analysis using the isoform-specific peptide conjugated to BSA. None of the antisera had any reactivity to BSA at the dilutions used (data not shown). The specificity of the antibodies for the target peptide sequence is shown in Fig. 11 A. The antisera prepared using the PiT-1 peptide does not cross-react with the peptide sequences used for PiT-2 or BNP-1. This lack of cross-reactivity is also true for the other antisera. We used the antisera to assay the expression of the three isoforms in rat brain and liver; these isoforms have been previously demonstrated in these tissues by Northern analysis (Kavanaugh et al. 1994) or RT-PCR (see above). The antisera for PiT-1 detects an ∼70-kD polypeptide in rat brain and liver (Fig. 11B). However, the antisera fails to detect the appropriate polypeptide in either human erythrocytes or K562 cells. The predicted molecular weight of this protein is 74 kD. Competition with the immunizing peptide shows that the band detected is specific (data not shown). The antisera for PiT-2 detects an ∼40-kD polypeptide in rat liver (Fig. 11B) and the reactivity for this polypeptide is specifically blocked with immunizing peptide (data not shown). However, the PiT-2 antisera does not detect any proteins in either K562 cells or human erythrocytes. The predicted molecular weight for PiT-2 is 70 kD, which is significantly larger than the protein detected in liver. We believe that this may be due to PiT-2 sensitivity to proteolysis, despite the inclusion of protease inhibitors in the extraction buffer used in these experiments. The discrepancy in molecular weight remains to be resolved. Finally, the antisera for BNP-1 detects an ∼116-kD polypeptide in brain, liver, human erythrocytes, and K562 cells. This apparent molecular weight exceeds the predicted 61 kD without glycosylation. Similarly, NaPi-3 has an apparent molecular weight of 125 kD by SDS-PAGE that exceeds the predicted 69 kD without glycosylation (Timmer and Gunn 1998). Of the three isoforms that were detected at the mRNA level by RT-PCR, we can only detect the expression of the BNP-1 polypeptide. We tentatively conclude that the Na-dependent phosphate activity measured in K562 cells arises principally from the BNP-1 isoform of the sodium-phosphate cotransporter, despite the apparent expression of the mRNA for PiT-1, PiT-2, and BNP-1. Whether or not the PiT-1 or PiT-2 polypeptides can be expressed in K562 cells remains to be explored. It is possible that the polypeptides for these proteins are synthesized in response to certain environmental conditions (low or high phosphate) or upon induction to the erythrocyte or leukocyte phenotype.
Figure 11.
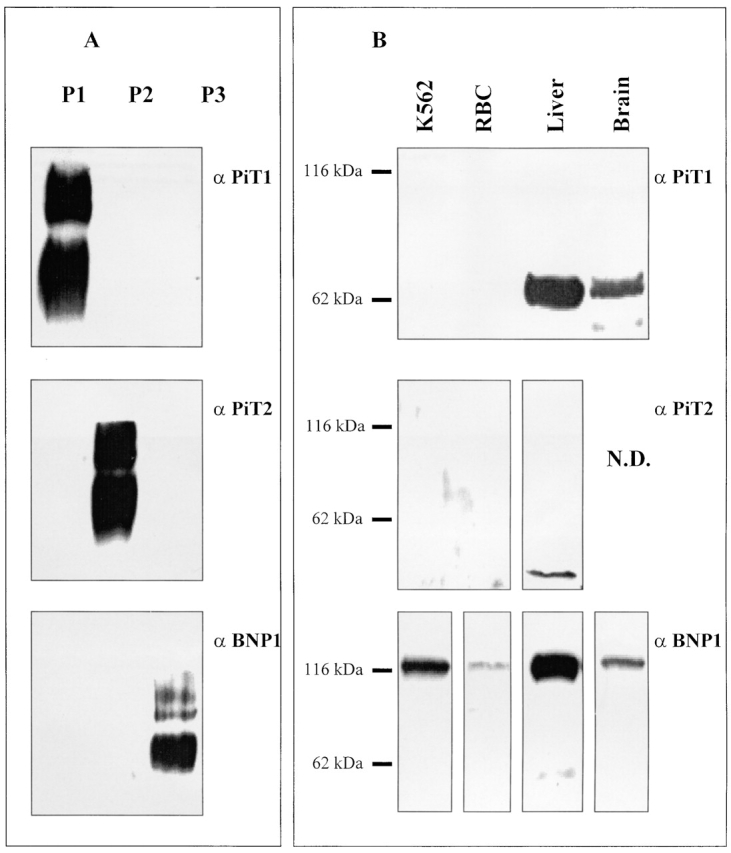
Expression of sodium-phosphate cotransporter isoforms in human tissues and K562 cells as determined by Western analysis. (A) The specificity of the antibodies for the antigenic peptides was tested by Western analysis of BSA-peptide conjugates. The lanes are labeled as follows: P1 (BSA-PiT-1 peptide conjugate), P2 (BSA-PiT-2 peptide conjugate), and P3 (BSA-BNP-1 peptide conjugate). Each lane was loaded with 1 ng of total BSA-peptide conjugate. The blot in α PiT-1 shows reactivity of these BSA-peptide conjugates with the anti–PiT-1 polyclonal antibody prepared using a KLH-PiT-1 peptide conjugate. The other blots were developed using the anti-PiT-2 polyclonal antibody (α PiT-2) or the anti–BNP-1 polyclonal antibody (α BNP-1). The antibody dilution of the primary antibody used in all cases was 1/1,000 of the crude sera. (B) Whole cell lysates were prepared from rat tissues, K562 cells and human erythrocytes as described in materials and methods. Total cell protein from these samples were used for Western analysis. The samples are indicated at the top and are as follows (left to right): K562, which is K562 total cell protein (10 μg); RBC, which is total human erythrocyte protein (10 μg); liver, which is total rat liver protein (10 μg); and brain, which is total rat brain protein (10 μg). The blots were developed using the indicated polyclonal antibodies: α PiT-1, the anti–PiT-1 polyclonal antibody; α PiT-2, the anti–PiT-2 polyclonal antibody; and α BNP-1, the anti–BNP-1 polyclonal antibody.
Ion Composition
The ion composition of erythrocytes is well known (Funder and Wieth 1966a,Funder and Wieth 1966b). Although K562 cells have been the subject of many papers, there are no measurements of ionic composition reported. In Table , there are three experiments showing the composition of the supernatants and cells after being washed in different media, packed by centrifugation, and separately analyzed. K562 cells were washed in their culture medium (RPMI) with fetal calf serum, isotonic MgCl2 (110 mM), the NaCl flux medium, or the N-methyl-glucamine Cl preflux wash medium. After washing in the RPMI culture medium, the contents should reflect the steady state values. However, the supernatant includes both the composition of the wash medium and any ions lost by the cells between the last wash and separating the supernatant and packed cells. The loss of K from the cells is apparent in the values measured in the supernatants after the NaCl wash medium, where the supernatant K all comes from the cells and was variable (Table , 4.5 mM in experiment 2 and 23 mM in experiment 3) due to different lengths of time between the last wash and centrifugation. This variability is reciprocally displayed in the cell K contents from these experiments (690 mmol/kg dry cell solid in experiment 2 and 650 in experiment 3). Some of the variability between experiments may arise from differences in the time of harvest relative to the last split of the cultured cells (i.e., whether they were in log or stationary phase of growth). In general, K562 cells are high K, low Na cells with intracellular chloride at 50–60 mM, which is above electrochemical equilibrium if the membrane potential is more hyperpolarized than −15 mV. The apparent water fraction (0.H2O) was constant (0.796–0.833), though after correction for the concurrently measured 14C-inulin or 14C-polyethylene trapped space, the water content, D gram of intracellular water/(gram dry cell solids), varied by 28%. The preflux wash reduced intracellular sodium content and concentration by half, but intracellular K, Cl, and PO4 remained relatively constant. This reduction would increase the initial driving for Na-phosphate influx measured in NaCl medium.
Table 3.
Composition of K562 Cells and the Supernatant after Washing and Packing
| Supernatants (mM) Water Content and Trapped Space | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Experiment | Wash medium | Na | K | Cl | PO4 | 0.H20 | D | Trapped space | |
| 1 | RPMI | 118 | 8.6 | 100 | 32.4 | 0.833 | 5.00 | 0.097 | |
| MgCl2 | 2.6 | 7.0 | 205 | 33.5 | 0.829 | 4.83 | 0.084 | ||
| 2 | NaCl | 135 | 4.5 | 146 | 41.8 | 0.796 | 3.91 | 0.117 | |
| NMDG Cl | 3.6 | 3.2 | 131 | 41.7 | 0.799 | 3.98 | 0.118 | ||
| 3 | NaCl | 132 | 23 | 145 | 34.9 | 0.804 | 4.09 | 0.095 | |
| NMDG Cl | 2.5 | 6.7 | 128 | 35.5 | 0.804 | 4.11 | 0.106 | ||
| Packed Cell Intracellular Concentration and Content | |||||||||
| Experiment | Wash medium | Na mM | Na content | K mM | K content | Cl mM | Cl content | PO4 mM | PO4 content |
| 1 | RPMI | 27 | 135 | 133 | 660 | 56 | 280 | 32.4 | 162 |
| MgCl2 | 26 | 125 | 133 | 640 | 58 | 280 | 33.5 | 162 | |
| 2 | NaCl | 28 | 111 | 176 | 690 | 63 | 250 | 41.8 | 164 |
| NMDG Cl | 17 | 67 | 167 | 660 | 54 | 220 | 41.7 | 166 | |
| 3 | NaCl | 31 | 125 | 159 | 650 | 60 | 250 | 34.9 | 143 |
| NMDG Cl | 12 | 51 | 152 | 630 | 51 | 210 | 35.5 | 146 | |
0.H20 is the water fraction corrected for trapped space. D is the grams of intracellular water/grams of dry cell solids. Trapped space is the grams of plasma space/grams wet packed cells, the average of duplicate measurements on packed cells centrifuged concurrently with the other cells analyzed for water and ions. K562 cells were washed in the indicated medium (see materials and methods) three times and packed in duplicate tubes. The RPMI and MgCl2 experiments were performed on paired aliquots of the same cell suspension. Each NaCl and NMDG Cl experiment below it were performed on aliquots of the same cell suspension on the same day. The values are the means of the duplicates that differed by no more than two in the least significant figure. Ion concentrations in millimoles per kilogram cell water, contents in millimoles per kilogram dry cell solids.
DISCUSSION
All cells of the body, except erythrocytes, maintain intracellular phosphate above electrochemical equilibrium using the inward sodium flux down its gradient to actively transport phosphate against its electrochemical potential gradient. The influx of 32PO4 into red cells is normally by three pathways and, in plasma-like media at 37°C, the contributions of these three pathways are as follows: (a) 70% is mediated by anion exchanger AE1 (band 3) and can be blocked by DNDS or other stilbene disulfonates; (b) 25% is sodium-dependent Na-phosphate cotransport; and (c) 5% is by a pathway linear in extracellular phosphate concentration (up to 1 mM) (Gunn et al. 1989). In erythrocytes, only the monovalent phosphate species is transported by AE1. The phosphate is passively transported 2.8× faster by AE1 than by the Na-PO4 cotransporter under physiological conditions (Gunn et al. 1989). Thus, in erythrocytes, the steady state intracellular phosphate is close to equilibrium with the plasma phosphate.
The molecular basis for the sodium-dependent phosphate flux in erythrocytes has heretofore been unknown and provided the rationale for this study. For better or worse, erythrocytes are anucleated and thus do not have RNA or DNA. This attribute helps to make them ideally suited for detailed tracer transport measurements, but poorly suited for many molecular biological manipulations. Although reticulocytes are a commonly used nucleated model for erythrocytes, it is very possible that the cotransport in reticulocytes is not mediated by the same molecule as cotransport in mature erythrocytes. However, to determine whether a cotransport activity in reticulocytes is the same as mature erythrocytes requires tracer influx experiments, and it is difficult to obtain large numbers of pure reticulocytes required for such studies. Thus, we chose to examine a model cell that has similar cotransporter kinetics and selectivity, is nucleated, and is easily obtained as a homogeneous population. The model cell used in these studies is the erythroleukemic cell line K562.
The results described above show that this cell line has the kinetics of Na-PO4 cotransport that are the same as in mature erythrocytes. We found, in K562 cells, that the Na activation kinetics were sigmoidal (Fig. 5, bottom), which led us to the careful reexamination of the Na activation kinetics in erythrocytes (Fig. 5, top). The clear sigmoidal Na-activation kinetics in erythrocytes now supports a mechanism with two (or more) Na sites and agrees with the previously observed stoichiometry of >1 Na per phosphate transported. There is no conflict between the data presented here and our previously reported data (Shoemaker et al. 1988). In the previous erythrocyte studies we reported that the ratio of the [PO4]o-activated 22Na influx and the [Na]o-activated 32PO4 influx was 1.72 ± 0.02 at three different pH values and consequently a majority of the Na-PO4 cotransport coupled 2 Na per phosphate (Shoemaker et al. 1988). However, we also reported that Na-activation kinetics were described by the Michaelis-Menten equation for a single Na site. If the earlier data (Figure 4 in Shoemaker et al. 1988) were graphed after being corrected for the phosphate concentration used in the experiments shown in Fig. 5, top, the data points would scatter along the curve drawn in this figure. The sigmoidicity was not previously noticed because there were too few data points at low Na concentrations.
We show above that phosphate transport in both K562 cells is activated not only by sodium, but also by lithium (Fig. 4). There is no significant activation of phosphate transport by any other cation tested (Fig. 4). This pattern of cation activation is identical to that observed for erythrocytes (S. Elmariah and R.B. Gunn, manuscript in preparation). The lithium activation of 32PO4 influx in K562 cells appears to have two separable components: a cooperative component similar to the Na-activated 32PO4 influx and a hyperbolic, saturating component. This second component of lithium activation may arise either from a 1:1 cotransport mode of the normally 2:1 cotransporter or from a different cotransporter isoform. Alternatively, K562 cells may have an additional lithium-phosphate cotransporter besides hBNP-1, but it must be lithium specific since the evidence is against any 1:1 Na-phosphate cotransport (Fig. 5, bottom, inset). This additional cotransporter must not be in erythrocytes since 1:1 cotransport of Li or Na with phosphate is not apparent in the phosphate kinetics measured in erythrocytes (Fig. 5, top, inset).
The pharmacological properties of the Na-dependent phosphate transport activity in K562 cells and erythrocytes are strikingly similar (Fig. 8, and Fig. 9). Previously, we reported that arsenate had a variable inhibitory effect on erythrocyte Na-PO4 cotransport (Shoemaker et al. 1988), but this may have been due to uncompensated pH changes in those experiments. It was the marked arsenate inhibition of phosphate transport by K562 cells (Fig. 7) that motivated a more careful study in erythrocytes that confirmed the similarity of arsenate inhibition in these two systems (S. Elmariah and R.B. Gunn, manuscript in preparation). In erythrocytes, the Na-activated 32PO4 influx is the same in the absence and presence of 0.25 mM DNDS (Shoemaker et al. 1988) thus the cotransporter is not blocked by this compound, although >99% of the chloride and phosphate transport on AE1 is blocked. Other compounds tested (pCMBS at 0.1 mM, phloretin at 0.1 mM, vanadate at 1 mM, and PFA at 1 mM; Fig. 9) showed very similar effects on the Na-dependent phosphate transport in both K562 cells and erythrocytes.
The above data suggests that the Na-dependent phosphate transport activities in K562 cells and erythrocytes are kinetically similar, thus supporting a common molecular identity for the cotransporter in these two cell types. We then determined which specific cotransporter isoform mRNAs were expressed in K562 cells and found that these cells express mRNA for three human Na-phosphate cotransporters: hBNP-1, hPiT-1, and hPiT-2 (Fig. 10). It may also express the mRNA from an unknown Na-PO4 cotransporter gene. Antibodies to hBNP-1, hPiT-1, and hPiT-2 react only with BSA conjugated to the peptide sequence used as the antigen in their preparation (Fig. 11). Both erythrocyte and K562 cells only react with the anti–hBNP-1 antibody. Although the positive controls for all three antibodies demonstrated they could identify their appropriate substrates in other tissues, the absence of any detectable hPiT-1 or hPiT-2 in erythrocytes or K562 cells does not prove they are absent, but does suggest they are of much lower abundance than hBNP-1.
In summary, we made the following observations on K562 cells: (a) the kinetics and pharmacology of Na-PO4 cotransport in these cells are the same as in mature erythrocytes; (b) K562 cells and erythrocytes have significant Li-activated 32PO4 influx, whereas the (Type II) renal isoforms and hPiT-1 do not; (c) RT-PCR shows that the mRNA for hBNP-1, hPiT-1, and hPiT-2 are the only sodium-phosphate cotransporters expressed in these cells; and (d) Western analysis shows that only hBNP-1 is the only sodium-phosphate cotransporter isoform polypeptide that can be detected in erythrocyte membranes and K562 cell membranes. Since both erythrocytes and K562 cells are shown here to have only hBNP-1 protein by Western analysis and the two cell types have similar transport kinetics, it seems likely that these similarities are caused by the presence of hBNP-1–mediated transport kinetics. Thus, we tentatively conclude that the erythrocyte sodium-phosphate cotransporter is the same as hBNP-1. But this conclusion requires additional experiments; e.g., heterologous expression and kinetic characterization of hBNP-1.
The other supportive evidence for this conclusion is the exclusion of both hNaPi-3 and hPiT-1 based upon their previously reported functional properties. For example, the expression of hPiT-1 in oocytes induces external Na-activated inward current in the presence of external phosphate but no external Li-activated inward current (Olah et al. 1994). This suggests that Li is not a substrate for hPiT-1, unlike the cotransporter in erythrocytes and K562 cells. Therefore, although mRNA for hPiT-1 was found in K562 cells, the apparent absence of hPiT-1 protein in K562 cells and erythrocytes is consistent with some other isoform mediating the Li-PO4 cotransport in these cells. Although Na-PO4 cotransport in K562 cells (Fig. 10) and in HEK 293 cells expressing NaPi-3 share some similar pharmacology (e.g., both are not inhibited by DNDS, equally inhibited by phloretin (50% at 0.1 mM) and are equally inhibited by pCMBS (>90% at 1 mM) (Timmer and Gunn 1998), these cotransport activities have some distinct pharmacology. For example, PFA and vanadate had little effect on either erythrocytes or K562 cells, but are significant inhibitors of NaPi-3 expressed in HEK 293 cells (Timmer and Gunn 1998) and NaPi-3 (or the rat homolog) in cortical brush border membranes (PFA: K i rat = 4.6 × 10−4 M, Szczepanska-Konkel et al. 1986; K i rat = 2.1 × 10−4 M and K i human = 3.1 × 10−5 M, Yusufi et al. 1986). These differences rule out NaPi-3 as a candidate for the K562 and erythrocyte cotransporter. As shown in Table , the characteristics of the different isoforms when expressed in different systems do not always agree. The differences between hNaPi-3 and the form in erythrocytes as described above are apparent. But the kinetic differences between hPiT-1 and rPiT-2 expressed in oocytes and that of erythrocytes are probably insufficient to exclude them as candidates in those cells, except the apparent absence of Li as a substrate in hPiT-1. Unfortunately, there are no kinetic or pharmacological studies of native or expressed hPiT-2 or hBNP-1. A side-by-side study of their kinetics and pharmacology in a common expression system could strengthen the identification of hBNP-1 as the K562 and erythrocyte Na-PO4 cotransporter.
Table 4.
Functional Properties of Na-PO4 Cotransporters
| hNaPi-3 | |||||||
|---|---|---|---|---|---|---|---|
| B-B vesicles | Oocytes | Cultured cells | hPiT-1 | rPiT-2 | Erythrocytes | K562 cells | |
| K 1/2-out Na (mM) | 63* | 57‡ | 128§ | 40–50‖ | 46 | 34 | |
| K 12-out PO4(mM) | 0.19* | 0.17‡ | 0.089§ | 0.024 ± 0.005 | 0.025 ± 0.006 | 0.30 | 0.36 |
| Hill coefficient (Na) | 1.5* | 2.1‡ | 1§ | 1.5–2¶ | 1.9 | 1.5 | |
| Arsenate, K I (mM) | 1.1** | 0.4§ | 2.6 | 2.7 | |||
| VLi 32PO4VNa 32PO4 | <0.02§ | <0.06‡‡ | 0.19 | 0.19 | |||
| PFA inhibition | +§§ | +§‖ ‖ | − | − | |||
| Stilbene disulfonate inhibition | +¶¶*** | −§ | − | − | |||
The functional properties of hNaPi-3 in brush border vesicles (B-B vesicles) and expressed in Xenopus oocytes or cultured mammalian cells (HEK-293) are different from those found in erythrocytes and K562 cells in this report. There are no reports of functional parameters of expressed hPiT-2 or hBNP-1 or any other known native cells that express only these isoforms. The presence or absence of inhibition of transport is indicated by + or +, respectively. *Werner et al. 1990; ‡Magagnin et al., 1993; §Timmer and Gunn, 1998; ‖Kavanaugh and Kabat, 1996; ¶Kavanaugh et al., 1994; **Kinne et al. 1977; ‡‡Olah et al., 1994; §§ Loghman-Adham and Dousa 1992; ‖ ‖Yusufi et al., 1986; ¶¶ Azzarolo et al. 1991; ***Tenenhouse and Martel 1993.
The results of this study are significant for two reasons. First, the apparent expression of a single Na-PO4 cotransporter, hBNP-1, in erythrocytes and K562 cells potentially provides a native cell system in which to characterize the isoform-specific kinetics as reported here. In addition, this suggests a native cell system to study the isoform-specific regulation of phosphate transport in future studies. The predominance of rPiT-2 expression in rat fibroblasts provides another system for the study of isoform-specific characteristics (Kavanaugh et al. 1994). However, many cells express multiple isoforms of Na-phosphate cotransporters. The mechanisms by which cells control their intracellular phosphate concentration are poorly understood, though changes in plasma phosphate concentration alter cell metabolism, presumably through altering cytoplasmic phosphate activity and cytoplasmic phosphoryl potential. Since the Na-phosphate cotransport mechanisms use the energy in the Na gradient, they are probably closely regulated. Second, there is the suggestion that the erythrocyte Na-phosphate cotransporter is the molecular basis for Na–Li exchange in red cells. This suggestion is based in part upon the observation that the Na-phosphate cotransporter mediates Li-phosphate cotransport (Fig. 4). In addition, a Na-dependent cotransporter can mediate cation–cation exchange through a slippage mechanism when only a cation, but no phosphate, is bound. For example, the rat homolog of NaPi-3 can mediate Na–Na exchange (Forster et al. 1998), and the Na-PO4 cotransporter (hBNP-1) in erythrocytes can mediate Na–Li exchange (S. Elmariah and R.B. Gunn, manuscript in preparation). Thus, hBNP-1 function may extend beyond the transport of phosphate to include lithium transport and its therapeutic efficacy in neurons and glia that express this Na-phosphate cotransporter. Proof that hBNP-1 is also the molecular mechanism of Na–Li exchange in red cells is beyond the scope of this paper and is the subject of current studies in progress.
Acknowledgments
We thank P.M. Smith for maintaining the K562 cells, B. Stockman for performing SDS-PAGE and Western analysis, and B.M. Medley for preparing the manuscript.
This work was supported in part by the National Heart, Lung, and Blood Institute grant R37-HL-28674 to R.B. Gunn.
Footnotes
Abbreviations used in this paper: DNDS, 4,4′-dinitro-2,2′-stilbenedisulfonic acid; Hgb, hemoglobin; PCA, perchloric acid; pCMBS, p-chloromercuri phenylsulfonate; PFA, phosphonoformate; RT-PCR, reverse transcription–PCR.
References
- Adragna N.C., Canessa M.L., Solomon H., Slater E., Tosteson D.C. Red cell lithium-sodium countertransport and sodium–potassium cotransport in patients with essential hypertension. Hypertension (Dallas). 1982;4:795–804. doi: 10.1161/01.hyp.4.6.795. [DOI] [PubMed] [Google Scholar]
- Azzarolo A.M., Ritchie G., Quamme G. Inhibition of sodium-phosphate cotransport in renal brush-border membranes with the stilbenedisulfonate, H2-DIDS. Biochim. Biophys. Acta. 1991;1069:70–76. doi: 10.1016/0005-2736(91)90105-h. [DOI] [PubMed] [Google Scholar]
- Bairoch A., Bucher P., Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berner W., Kinne R., Murer H. Phosphate transport into brush-border membrane vesicles isolated from rat small intestine. Biochem. J. 1976;160:467–474. doi: 10.1042/bj1600467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canessa M., Adragna N., Solomon H.S., Connolly T.M., Tosteson D.C. Increased sodium–lithium countertransport in red cells of patients with essential hypertension. N. Engl. J. Med. 1980;302:772–776. doi: 10.1056/NEJM198004033021403. [DOI] [PubMed] [Google Scholar]
- Chong S.S., Kristjansson K., Zoghbi H.Y., Hughes M.R. Molecular cloning of the cDNA encoding a human renal sodium phosphate transport protein and its assignment to chromosome 6p21.3-p23. Genomics. 1993;18:355–359. doi: 10.1006/geno.1993.1476. [DOI] [PubMed] [Google Scholar]
- Cook J.S., Shaffer C., Cragoe E.J., Jr. Inhibition by amiloride analogues of Na+-dependent hexose uptake in LLC-PK1/C14 cells. Am. J. Physiol. Cell Physiol. 1987;253:C199–C204. doi: 10.1152/ajpcell.1987.253.2.C199. [DOI] [PubMed] [Google Scholar]
- Cooper R., LeGrady D., Nanas S., Trevisan M., Mansour M., Histand P., Ostrow D., Stamler J. Increased sodium–lithium countertransport in college students with elevated blood pressure. J. Am. Med. Assoc. 1983;249:1030–1034. [PubMed] [Google Scholar]
- Dissing S., Hoffman R., Murnane M.J., Hoffman J.F. Chloride transport properties of human leukemic cell lines K562 and HL60. Am. J. Physiol. Cell Physiol. 1984;247:C53–C60. doi: 10.1152/ajpcell.1984.247.1.C53. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Escoubet B., Garestier M.-C., Le Grimellec C., Amiel C. Multiple modulation of Na-dependent P i uptake by cellular Ca in MDCK cells. Am. J. Physiol. Cell Physiol. 1993;265:C19–C27. doi: 10.1152/ajpcell.1993.265.1.C19. [DOI] [PubMed] [Google Scholar]
- Forster I., Hernando N., Biber J., Murer H. The voltage dependence of a cloned mammalian renal type II Na+/Pi cotransporter (NaPi-2) J. Gen. Physiol. 1998;112:1–18. doi: 10.1085/jgp.112.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funder J., Wieth J.O. Determination of sodium, potassium, and water in human red blood cells Scand. J. Clin. Lab. Invest. 18 1966. 151 166a [DOI] [PubMed] [Google Scholar]
- Funder J., Wieth J.O. Potassium, sodium, and water in normal human red blood cells Scand. J. Clin. Lab. Invest. 18 1966. 167 180b [DOI] [PubMed] [Google Scholar]
- Ghishan F.K., Rebeiz R., Honda T., Nakagawa N. Characterization and expression of a novel Na+-inorganic phosphate transporter at the liver plasma membrane of the rat. Gastroenterology. 1993;105:519–526. doi: 10.1016/0016-5085(93)90729-v. [DOI] [PubMed] [Google Scholar]
- Ginsburg J.M., Lotspeich W.D. Interrelations of arsenate and phosphate transport in the dog kidney. Am. J. Physiol. 1962;205:707–714. doi: 10.1152/ajplegacy.1963.205.4.707. [DOI] [PubMed] [Google Scholar]
- Glinn M., Ni B., Paul S.M. Characterization of Na+-dependent phosphate uptake in cultured fetal rat cortical neurons. J. Neurochem. 1995;65:2358–2365. doi: 10.1046/j.1471-4159.1995.65052358.x. [DOI] [PubMed] [Google Scholar]
- Gmaj P., Murer H. Cellular mechanisms of inorganic phosphate transport in kidney. Physiol. Rev. 1986;66:36–66. doi: 10.1152/physrev.1986.66.1.36. [DOI] [PubMed] [Google Scholar]
- Gunn R.B., Dalmark M., Tosteson D.C., Wieth J.O. Characteristics of chloride transport in human red blood cells. J. Gen. Physiol. 1973;61:185–206. doi: 10.1085/jgp.61.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn R.B., Fröhlich O., King P.A., Shoemaker D.G. Anion transport. In: Agre P., Parker J.C., editors. Red blood Cell MembranesStructure, Function and Clinical Implications. Marcel Dekker, Inc.; New York, NY: 1989. pp. 563–596. [Google Scholar]
- Harris R.C., Lufburrow R.A., Cragoe E.J., Seifter J.L. Amiloride analogs inhibit Na-glucose and alanine cotransport in renal brush border membrane vesicles (BBMV) Kidney Int. 27 1985. 310(Abstr.) [Google Scholar]
- Henkel R.D., Vandeberg J.L., Walsh R.A. A microassay for ATPase. Anal. Biochem. 1988;169:312–318. doi: 10.1016/0003-2697(88)90290-4. [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Bleasby A.J., Fuchs R. CLUSTAL Vimproved software for multiple sequence alignment. Comput. Appl. Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Thompson J.D., Gibson T.J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hoffmann N., Thees M., Kinne R. Phosphate transport by isolated renal brush border vesicles. Pflügers Arch. 1976;362:147–156. doi: 10.1007/BF00583641. [DOI] [PubMed] [Google Scholar]
- Hopp T.P., Woods K.R. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton M.A., Cedar S.H., Edwards P.A.W. Expression of red cell specific determinants during differentiation in the K562 erythroleukaemia cell line. Scand. J. Haematol. 1981;27:231–240. doi: 10.1111/j.1600-0609.1981.tb00478.x. [DOI] [PubMed] [Google Scholar]
- Jansen G., Rijksin G., De Gast G.C., Staal G.E.J. Glycolytic enzymes of an erythroleukemic cell line, K562, before and after hemoglobin induction. Exp. Hematol. 1983;11:626–638. [PubMed] [Google Scholar]
- Kavanaugh M.P., Kabat D. Identification and characterization of a widely expressed phosphate transporter/retrovirus receptor family. Kidney Int. 1996;49:959–963. doi: 10.1038/ki.1996.135. [DOI] [PubMed] [Google Scholar]
- Kavanaugh M.P., Miller D.G., Zhang W., Law W., Kozak S.L., Kabat D., Miller A.D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc. Natl. Acad. Sci. USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney L.J., Kaplan J.H. Arsenate substitutes for phosphate in the human red cell sodium pump and anion exchanger. J. Biol. Chem. 1988;263:7954–7960. [PubMed] [Google Scholar]
- Kinne R., Berner W., Hoffman N., Murer H. Phosphate transport by isolated renal and intestinal plasma membranes. Adv. Exp. Med. Biol. 1977;81:265–277. doi: 10.1007/978-1-4613-4217-5_26. [DOI] [PubMed] [Google Scholar]
- Kleyman T.R., Cragoe E.J. Amiloride and its analogs as tools in the study of ion transport. J. Membr. Biol. 1988;105:1–21. doi: 10.1007/BF01871102. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Law F.-Y., Steinfeld R., Knauf P.A. K562 cell anion exchange differs markedly from that of mature red blood cells. Am. J. Physiol. Cell Physiol. 1983;244:C68–C74. doi: 10.1152/ajpcell.1983.244.1.C68. [DOI] [PubMed] [Google Scholar]
- Loghman-Adham M., Dousa T.P. Dual action of phosphonoformic acid on Na+-phosphate cotransport in opossum kidney cells. Am. J. Physiol. Renal Physiol. 1992;263:F301–F310. doi: 10.1152/ajprenal.1992.263.2.F301. [DOI] [PubMed] [Google Scholar]
- Lozzio C.B., Lozzio B.B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- Magagnin S., Werner A., Markovich D., Sorribas V., Stange G., Biber J., Murer H. Expression cloning of human and rat renal cortex Na/Pi cotransport. Proc. Natl. Acad. Sci. USA. 1993;90:5979–5983. doi: 10.1073/pnas.90.13.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohana Rao J.K., Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim. Biophys. Acta. 1986;869:197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- Ni B., Du Y., Wu X., DeHoff B.S., Rosteck P.R., Jr., Paul S.M. Molecular cloning, expression, and chromosomal localization of a human brain-specific Na+-dependent inorganic phosphate cotransporter. J. Neurochem. 1996;66:2227–2238. doi: 10.1046/j.1471-4159.1996.66062227.x. [DOI] [PubMed] [Google Scholar]
- Ni B., Rosteck P.R., Jr., Nadi N.S., Paul S.M. Cloning and expression of a cDNA encoding a brain-specific Na+-dependent inorganic phosphate cotransporter. Proc. Natl. Acad. Sci. USA. 1994;91:5607–5611. doi: 10.1073/pnas.91.12.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B., Stephenson D., Wu X., Smalstig E.B., Clemens J., Paul S.M. Selective loss of neuronal Na+-dependent phosphate cotransporter mRNA in CA1 pyramidal neuron following global ischemia. Brain Res. 1997;48:132–139. doi: 10.1016/s0169-328x(97)00090-9. [DOI] [PubMed] [Google Scholar]
- Olah Z., Lehel C., Anderson W.B., Eiden M.V., Wilson C.A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J. Biol. Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- Ostrow D.G., Pandey G.N., Davis J.M., Hurt S.W., Tosteson D.C. A heritable disorder of lithium transport in erythrocytes of a subpopulation of manic-depressive patients. Am. J. Psychiatry. 1978;135:1070–1078. doi: 10.1176/ajp.135.9.1070. [DOI] [PubMed] [Google Scholar]
- Pandey G.N., Dorus E., Casper R.C., Janicek P., Davis J.M. Lithium transport in red cells of patients with affective disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 1984;8:547–555. doi: 10.1016/0278-5846(84)90014-9. [DOI] [PubMed] [Google Scholar]
- Quabius E.S., Murer H., Biber J. Expression of proximal tubular Na-Pi and Na-SO4 cotransporters in MDCK and LLC-PK1 cells by transfection. Am. J. Physiol. Renal Physiol. 1996;270:F220–F228. doi: 10.1152/ajprenal.1996.270.1.F220. [DOI] [PubMed] [Google Scholar]
- Shoemaker D.G., Bender C.A., Gunn R.B. Sodium-phosphate cotransport in human red blood cells. J. Gen. Physiol. 1988;92:449–474. doi: 10.1085/jgp.92.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.K., Krohn R.I., Germanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Szczepanska-Konkel M., Yusufi A.N.K., VanScoy M., Webster S.K., Dousa T.P. Phosphonocarboxylic acids as specific inhibitors of Na+-dependent transport of phosphate across renal brush border membrane. J. Biol. Chem. 1986;261:6375–6383. [PubMed] [Google Scholar]
- Tenenhouse H.S., Martel J. Na+-dependent sulfate transport in opossum kidney cells is DIDS sensitive. Am. J. Physiol. Cell Physiol. 1993;265:C54–C61. doi: 10.1152/ajpcell.1993.265.1.C54. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL Wimproving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer R.T., Gunn R.B. Phosphate transport by the human renal cotransporter NaPi-3 expressed in HEK-293 cells. Am. J. Physiol. Cell Physiol. 1998;274:C757–C769. doi: 10.1152/ajpcell.1998.274.3.C757. [DOI] [PubMed] [Google Scholar]
- Werner A., Biber J., Forgo J., Palacin M., Murer H. Expression of renal transport systems for inorganic phosphate and sulfate in Xenopus laevis oocytes. J. Biol. Chem. 1990;265:12331–12336. [PubMed] [Google Scholar]
- Werner A., Moore M.L., Mantei N., Semenza G., Biber J., Murer H. Cloning and expression of cDNA for a Na/Pi cotransport system of kidney cortex. Proc. Natl. Acad. Sci. USA. 1991;88:9608–9612. doi: 10.1073/pnas.88.21.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werstiuk E.S., Rathbone M.P., Grof P. Erythrocyte lithium transport in bipolar affective disorders. The effect of membrane transport inhibitors. Neuropsychobiology. 1984;12:86–92. doi: 10.1159/000118115. [DOI] [PubMed] [Google Scholar]
- West I.C., Rutherford P.A., Thomas T.H. Sodium–lithium countertransportphysiology and function. J. Hypertens. 1998;16:3–13. doi: 10.1097/00004872-199816010-00002. [DOI] [PubMed] [Google Scholar]
- Younus M.J., Butterworth P.J. Sodium-dependent transport of phosphate by rat liver plasma membrane vesicles. Biochim. Biophys. Acta. 1993;1143:158–162. doi: 10.1016/0005-2728(93)90138-6. [DOI] [PubMed] [Google Scholar]
- Yurchenco P.D., Furthmayr H. Expression of red cell membrane proteins in erythroid precursor cells. J. Supramol. Struct. 1980;13:255–269. doi: 10.1002/jss.400130213. [DOI] [PubMed] [Google Scholar]
- Yusufi A.N.K., Szczepanska-Konkel M., Kempson S.A., McAteer J.A., Dousa T.P. Inhibition of human renal epithelial Na+-Pi cotransport by phosphonoformic acid. Biochem. Biophys. Res. Commun. 1986;139:679–686. doi: 10.1016/s0006-291x(86)80044-4. [DOI] [PubMed] [Google Scholar]
- Zaremba D., Rybakowski J. Erythrocyte lithium transport during lithium treatment in patients with affective disorders. Pharmacopsychiatry. 1986;19:63–67. doi: 10.1055/s-2007-1017156. [DOI] [PubMed] [Google Scholar]