Abstract
Ligand-specific molecular switches composed of RNA were created by coupling preexisting catalytic and receptor domains via structural bridges. Binding of ligand to the receptor triggers a conformational change within the bridge, and this structural reorganization dictates the activity of the adjoining ribozyme. The modular nature of these tripartite constructs makes possible the rapid construction of precision RNA molecular switches that trigger only in the presence of their corresponding ligand. By using similar enzyme engineering strategies, new RNA switches can be made to operate as designer molecular sensors or as a new class of genetic control elements.
Mastery of the molecular forces that dictate biopolymer folding and function would allow molecular engineers to participate in the design of enzymes—a task that to date has been managed largely by the random processes of evolution. The reward for acquiring this capability is substantial, considering that many applications in medicine, industry, and biotechnology demand high-speed enzymes with precisely tailored catalytic functions. “Modular rational design” has proven to be an effective means for conferring additional chemical and kinetic complexity on existing protein (1–4) and RNA enzymes (5–9). This engineering strategy takes advantage of the modular nature of many protein (10) and RNA subdomains (11–13), which can be judiciously integrated to form new multifunctional constructs. The recent discoveries of new catalytic RNA motifs (14, 15) and new ligand-binding motifs (16, 17) have considerably expanded the opportunities for ribozyme engineering.
We have used modular rational design to create several artificial ribozymes that are activated or deactivated by the binding of specific small organic molecules such as ATP (5, 8) and flavin mononucleotide (FMN) (unpublished data). Each of these allosteric† ribozymes is composed of two independent structural domains: one an RNA-cleaving ribozyme and the other a receptor (or “aptamer”) for a specific ligand. The conformational changes that occur within an aptamer domain on introduction of the ligand, termed “adaptive binding” (22–25), can trigger kinetic modulation of the adjoining catalytic domain by several different mechanisms that ultimately influence ribozyme folding (8, 9). In this report, we describe the combined application of modular rational design and in vitro selection techniques that provide an effective strategy for the rapid generation of precision molecular switches made of RNA.
MATERIALS AND METHODS
Oligonucleotides.
Synthetic DNA and the 14-nt substrate RNA were prepared by standard solid phase methods (Keck Biotechnology Resource Laboratory, Yale University) and purified by denaturing (8 M urea) PAGE as described (5). RNA substrate was 5′ 32P-labeled with T4 polynucleotide kinase and [γ-32P]ATP, and repurified by PAGE. Double-stranded DNA templates for in vitro transcription using T7 RNA polymerase were generated by extension of primer A (5′-TAATACGACTCACTATAGGGCGACCCTGATGAG) on a DNA template complementary to the desired RNA. Extension reaction were conducted with reverse transcriptase as described (8).
In Vitro Selection.
Selection for allosteric activation was performed by first preselecting each successive population (1 μM internally 32P-labeled RNA; ref. 5) for self-cleavage without FMN in 10 μl reaction buffer [50 mM Tris⋅HCl (pH 7.5 at 23°C) and 20 mM MgCl2] for 20 hr at 23°C. Preselections for G4–G6 were punctuated at 5-hr intervals by heating to 65°C for 1 min to denature and refold any misfolded molecules. Uncleaved RNA was purified by denaturing (8 M urea) 10% PAGE, eluted from excised gel, and precipitated with ethanol. The resulting RNA was selected by incubation in the reaction buffer in the presence of 200 μM FMN for the times indicated. Reaction times for positive selections during subsequent iterations of the selective-amplification process were decreased to favor allosteric ribozymes with the fastest rates of self-cleavage. Products separated by 10% PAGE were imaged and quantitated by using a PhosphorImager and imagequant software (Molecular Dynamics). The 5′ cleavage fragments produced in the presence of FMN were isolated as described above, amplified by RT-PCR (primer A and primer B: 5′-GGGCAACCTACGGCTTTCACCGTTTCG; ref. 5), and the resulting double-stranded DNA was transcribed in vitro (5) to generate the next RNA population. Selection for FMN inhibition was conducted in an identical fashion, except that FMN was included in both the transcription and the preselection, but was excluded in the selection reaction. Individual molecules from G6 populations of both selections were isolated by cloning (TA Cloning Kit, Invitrogen) and analyzed by sequencing (ThermalSequenase Kit, Amersham).
Allosteric Ribozyme Assays.
Reactions containing internally 32P-labeled self-cleaving ribozyme (100–500 nM) and either 200 μM FMN, 1 mM theophylline, or 1 mM ATP were initiated by the addition of reaction buffer and incubated through several half lives with periodic sampling. Products were separated by denaturing PAGE, and yields were quantitated as described above. Rate constants were derived by plotting the natural logarithm of the fraction of uncleaved RNA versus time and establishing the negative slope of the resulting line. The values for each rate constant given are the average of a minimum of three replicate assays, each that differed by less than 2-fold. Ribozymes carrying the class I induction element and the class II inhibition element were arbitrarily chosen for detailed analysis.
Bimolecular assays were conducted under single-turnover conditions with ribozyme (500 nM) in excess over trace amounts (≈5 nM) of 5′ 32P-labeled substrate. Reactions were initiated by combining ribozyme and substrate that were preincubated separately for 10 min at 23°C in reaction buffer. Kinetic parameters were generated as described above. Product yields were corrected for the amount of substrate that remained uncleaved after exhaustive incubation with the unmodified hammerhead ribozyme (5). The values for each rate constant given are the average of a minimum of two replicate assays that differed by less than 2-fold.
RESULTS AND DISCUSSION
In Vitro Selection of Allosteric Ribozymes.
We generated a population of >65,000 variant RNAs composed of separate FMN-binding aptamer (26) and hammerhead ribozyme (27, 28) domains that are joined by a random-sequence bridge (Fig. 1A). The bridge replaces a majority of the natural “stem II” portion of the hammerhead motif—a structural element that is a critical determinant of ribozyme activity (29, 30). The randomized domain within the resulting tripartite construct will provide a sampling of alternative stem II elements that might respond to FMN binding in the adjacent aptamer domain and confer either positive or negative allosteric control on the adjoining ribozyme domain. Two identical RNA pools (≈6 × 1012 molecules each) were subjected to in vitro selection (14, 15) either for FMN-dependent allosteric induction (Fig. 1B) or allosteric inhibition (Fig. 1C). To isolate bridges that direct the allosteric induction of ribozymes, a “negative selection” for self-cleavage in the absence of FMN was applied to the first pool. RNAs that remained uncleaved during this reaction were isolated and subsequently subjected to a “positive selection” for self-cleavage in the presence of FMN. This method is expected to favor the isolation of ribozymes that activate only when FMN is detected. In contrast, the second pool was both transcribed and preselected in the presence of FMN. The remaining RNA precursors were then subjected to positive selection in the absence of ligand, which favors the isolation of bridges that direct ribozymes to undergo allosteric inhibition.
Figure 1.
Combined modular rational design and in vitro selection for FMN-sensitive allosteric ribozymes. (A) Tripartite construct consisting of a hammerhead ribozyme joined to an FMN-binding aptamer (boxed) via a random-sequence bridge composed of eight nucleotides (N). The three stems that form the unmodified ribozyme are designated I, II and III, and the site of RNA cleavage is indicated by the arrowhead. The randomized bridge serves both as a partial replacement for stem II of the ribozyme and as a flanking stem for the aptamer. The G-C base pair immediately adjacent to the catalytic core is needed for the hammerhead ribozyme to achieve maximal catalytic activity (9, 42). Selection for FMN-inducible (B) and FMN-inhibited (C) allosteric ribozymes gave rise to RNA populations that respond either positively or negatively to the presence of FMN, respectively. The initial RNA pool (G0) and successive RNA populations (G1–G6) are identified.
Both RNA populations isolated after six rounds of selection (G6) display high sensitivity to FMN, demonstrating that the combined engineering approach is an effective means to generate ribozymes that function as highly specific molecular switches. The in vitro selection process could have produced RNA structures that cleave by some other means under the permissive reaction conditions. For example, isoalloxazine rings like that found in FMN have been shown to promote photocleavage of RNA molecules (31) and could conceivably serve as a cofactor for a novel FMN-dependent ribozyme. However, the RNAs isolated by selection appear to cleave in a reaction that is solely mediated by the original hammerhead ribozyme domain that was integrated into each construct as determined by gel mobility of RNA cleavage fragments.
Sequence and Functional Characteristics of Isolated Bridge Elements.
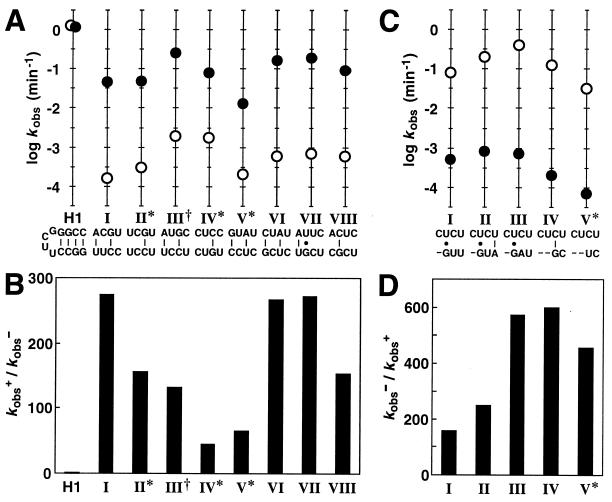
The G6 populations from both selections were cloned, sequenced, and assayed for allosteric function (Fig. 2). Eight distinct classes of bridges, designated as “induction elements” I–VIII, were identified in the FMN-inducible RNA population. Ribozymes with these different classes of induction elements show unique rate constants for self-cleavage in the absence (kobs−) or presence (kobs+) of ligand (Fig. 2A). Most classes exhibit greater than 100-fold allosteric activation (kobs+/kobs−), with classes I, III, and VII exhibiting FMN-dependent rate enhancements of ≈270-fold (Fig. 2B). This allosteric induction is similar in magnitude to the kinetic modulation seen with some natural allosteric proteins (32). Furthermore, the kobs+ values attained by nearly all classes approach the maximum kobs (1.1 min−1) measured for an unmodified hammerhead ribozyme (Fig. 2A).
Figure 2.
Bridge sequences and kinetic parameters for individual allosteric ribozymes. (A) Sequences and corresponding ribozyme rate constants for eight classes of induction elements isolated from G6. Plotted for each class is the logarithm of the observed rate constant for self-cleavage in the absence (○) or presence (●) of FMN. The base-pairing schemes depicted for each bridge were generated by assuming that no base-pair shift relative to the G-C base pair remaining in stem II had occurred. Indicated are classes that display greater than 20% misfolding (∗) and a class wherein an extraneous mutation exists in the stem-loop region of the aptamer domain (†). H1 is an unmodified hammerhead ribozyme (5, 8) that displays maximum catalytic activity and that remains unaffected by the presence of FMN. (B) Fold-activation of catalytic activity (kobs+/kobs−) achieved in the presence of ligand for each class of FMN-inducible ribozyme. (C) Sequences and corresponding ribozyme rate constants for five classes of inhibition elements isolated from G6. Nucleotide deletions are represented as dashes. (D) Fold-inhibition of catalytic activity (kobs−/kobs+) achieved in the presence of ligand for each class of FMN-inhibited ribozyme.
Likewise, five distinct classes of bridges were identified and were designated as “inhibition elements” I–V (Fig. 2C). Unlike the FMN-inducible populations which showed an immediate response to in vitro selection, ligand-dependent inhibition of ribozyme function was not detected until G3 of this parallel selection. Interestingly, each of the five classes carries a 1- or 2-nt deletion within the randomized bridge domain, suggesting that none of the sequence variants comprising the original RNA pool formed an adequate ligand-responsive element that could confer allosteric inhibition. The relative delay in deriving an FMN-inhibited RNA population may have been caused by the necessary emergence of specific nucleotide deletions within the bridge domain, an occurrence that depends on the frequency of deletion events during the selective-amplification process. Consistent with this hypothesis is the fact that sequences of the inhibition elements are highly homologous, indicating that the emergence and diversification of a single responsive bridge domain may have given rise to all classes examined. All five classes demonstrate substantial allosteric inhibition (200- to 600-fold) in the presence of FMN (Fig. 2D).
Many of the bridge elements isolated by selection display maximum rate enhancements that are at least 10-fold lower than that measured for the unmodified hammerhead ribozyme H1 (Fig. 2). The allosteric ribozymes that display the largest rate constants for RNA cleavage carry the class III induction element (kobs+ = 0.25 min−1) or the class III inhibition element (kobs− = 0.45 min−1). The maximum rate constants for these two ribozymes are, respectively, only 4- and 2-fold slower than H1. By using similar in vitro selection methods, we have recently isolated a population of theophylline-dependent ribozymes that use a tripartite configuration like that described for the FMN-sensitive RNAs. Individual theophylline-sensitive ribozymes from this population display rate constants that exceed 1 min−1 (unpublished data), thereby confirming that allosteric hammerhead ribozymes indeed can be made to operate as efficiently as the unmodified ribozyme.
Rapid Interconversion Between Active and Inactive Ribozyme Structures.
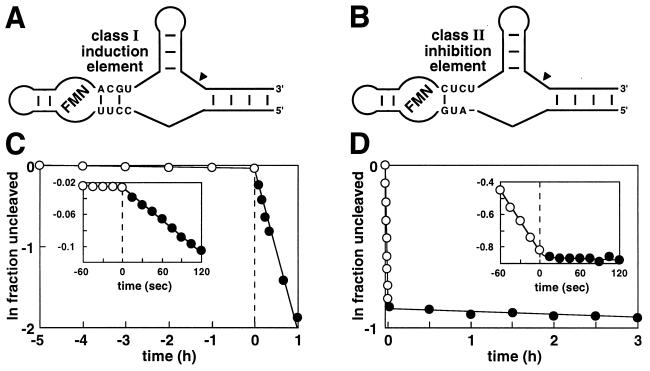
The inactive state for ribozymes that carry the class I induction element (Fig. 3A) is maintained for long periods of time in the absence of FMN, yielding only ≈1% self-cleavage per hour (Fig. 3C). However, self-cleavage is triggered almost instantaneously on the addition of ligand (Fig. 3C, Inset), in this case bringing about a 270-fold increase in catalytic rate. Possibly the “off” state maintained by induction elements in the absence of FMN lacks the ability to form the stable stem II structure that is necessary for ribozyme activity. Alternatively, each element forms a distinct structure that prevents formation of this essential stem. FMN binding establishes the “on” state by inducing a conformational change in the aptamer that rapidly converts the induction element into a structure that is compatible with ribozyme function. In contrast, ribozymes that carry the class II inhibition element (Fig. 3B) rapidly self-cleave in the absence of FMN, but quickly convert to an inactive state on addition of ligand (Fig. 3D, Inset). Here, inhibition elements maintain the off state by binding FMN and stabilizing specific bridge structures that preclude ribozyme function. Release of the ligand results in structural reorganization of the bridge and establishes the on state of the adjoining ribozyme. However, it remains unclear what structural state is responsible for the slow rate of cleavage seen with the class II inhibition element when FMN is present. Further experimentation is needed to determine whether the FMN–ribozyme complex remains weakly active, or whether the small number of FMN-free RNAs present under equilibrium binding conditions solely contribute to the RNA cleavage rate that is observed.
Figure 3.
Rapid ligand-dependent modulation of allosteric ribozymes. Tripartite ribozyme constructs carrying either a class I induction element (A) or a class II inhibition element (B) are depicted. Sequences for the aptamer and ribozyme domains are as shown in Fig. 1. The performance of these ribozymes in the presence and absence of FMN are evident from plots C and D, which show the natural logarithm of the fraction of ribozyme remaining uncleaved versus time relative to FMN addition. (Insets) Plots provide an expanded view of ribozyme responses to FMN addition.
Mechanism for Allosteric Function.
The rapid ligand-dependent activation or inhibition of ribozyme function indicates that the conformational changes required to modulate activity must be highly responsive to ligand binding. We propose that for some bridge elements this allosteric transition is achieved through localized base pairing changes within each element, and that binding energy derived from ligand–aptamer complex formation is used to create this shift in structural configuration.
A critical component of the proposed mechanism for both allosteric induction and inhibition is a single sheared A⋅G base pair, located within the aptamer domain immediately adjacent to the bridge, which forms only when FMN is bound (33, 34). With class I induction elements, the presence of FMN stabilizes the A⋅G base pair which in turn establishes a specific register for base pairing within the bridge (Fig. 4A). In the absence of this FMN-dependent structural constraint, base pairing throughout the bridge may “slip” 1 bp relative to the A⋅G interaction, thereby displacing the G-C base pair needed for ribozyme function. This inactive conformation would be maintained if no single nucleotide is bulged from the top strand of the bridge. Symmetric internal bulges are known to be more stable than asymmetric or single-nucleotide bulges (35). Therefore, the register that is set by the sheared A⋅G base pair may be faithfully propagated along the bridge element if the presence of symmetric internal bulges favor a continuously stacked stem II domain. Interestingly, all inhibition modules acquired deletions that appear to be essential for their function. This corresponds well with a “slip-structure” mechanism, as a continuously stacked bridge in this case would disrupt the critical G-C base pair of the ribozyme when FMN was bound, whereas the absence of FMN would allow proper ribozyme folding (Fig. 4B).
Figure 4.
The proposed slip-structure mechanism for allosteric regulation mediated by the class I induction element (A) and class II inhibition element (B) is illustrated. Shown are the proposed stem II secondary structures of the ligand-bound and unbound states of the FMN-modulated ribozymes. Not depicted are the left- and right-flanking sequences which comprise the aptamer and ribozyme domains, respectively. Asterisks denote the G and C residues of the hammerhead ribozyme that must pair to support catalysis, and the A and G residues of the FMN aptamer that become paired on ligand binding. Also shown are bimolecular ribozyme constructs containing stem II elements designed to simulate the active or inactive slip structures proposed for the class I induction module (C; I-1 through I-3) or the class II inhibition module (D; II-1 and II-2). Thick lines identify nucleotides that form the bridge elements. Mutations made within I-3 to reinforce the desired base pairing conformation are encircled.
To further investigate this slip-structure mechanism for allosteric regulation, several ribozyme constructs were created by using stable stem–loop structures in place of the FMN-binding domain (Fig. 4C). In its occupied state, the FMN aptamer forms a compact, approximately A-form RNA structure (34). Therefore, the stem–loop structures integrated into the test constructs should simulate the FMN-bound aptamer and enforce the putative slip structures necessary to either induce or inhibit ribozyme function. For example, construct I-1 is designed to simulate the structure of a class I induction element bound to FMN by enforcing the formation of the sheared A⋅G pair. Indeed, the kobs for I-1 in the absence of FMN is identical to the rate constant for the FMN-induced form of the parent allosteric ribozyme (Table 1). Two additional constructs (I-2 and I-3) were used to determine the rate constants when the opposing “slipped” version is enforced with progressively stronger base pairing. Construct I-2 is not significantly inhibited when the aptamer is replaced by structures that should favor the inactive conformation. Perhaps in this context, a single bulged nucleotide along the top strand of the bridge may occur which would restore proper ribozyme folding. However, the activity of the adjoining ribozyme is substantially diminished when potential bulge formation is precluded by the introduction of additional base pairs in the bridge that forms construct I-3, consistent with the proposed mechanism for allosteric function.
Table 1.
Catalytic rate constants for the on and off states of class I (induction) and class II (inhibition) ribozymes compared to constructs designed to simulate these states
| Allosteric ribozyme |
kobs (×10−1 min−1)
|
Simulant construct |
kobs (×10−1 min−1)
|
||
|---|---|---|---|---|---|
| On | Off | On | Off | ||
| Class I (induction) | 0.46 | 0.0017 | I-1 | 0.46 | — |
| I-2 | — | 0.21 | |||
| I-3 | — | 0.04 | |||
| Class II (inhibition) | 2.0 | 0.0080 | II-1 | 0.47 | — |
| II-2 | — | 0.0020 | |||
Further evidence for a slip-structure mechanism was provided by examining the class II inhibition element. Here, FMN binding enforces a base pairing pattern that precludes formation of the active ribozyme conformation (Fig. 4D). In the absence of FMN, the loss of the A⋅G base pair may permit the remaining base pairs to slip by 1 nt, thereby forming the active ribozyme conformation. Constructs II-1 and II-2, designed with stem–loop structures that enforce the two different base pairing conformers, display rate constants that correspond closely with the values for the active and inactive states of the parent allosteric ribozyme, respectively (Table 1). In all examples, the bridge elements contain unpaired bases that presumably destabilize the stem structures and allow rapid interconversion between different structural states. A similar RNA switch mechanism may serve an important role in the structure and function of 16S ribosomal RNA (36, 37), a finding which indicates that this mechanism for allosteric function may not be unprecedented. Although alternative mechanisms for allosteric function may be in operation, these striking correlations all are consistent with the proposed slip-structure mechanism. Similar studies with the remaining classes of bridge elements might reveal whether this mechanism is also more general in occurrence.
Engineering Allosteric Ribozymes with New Ligand Specificities.
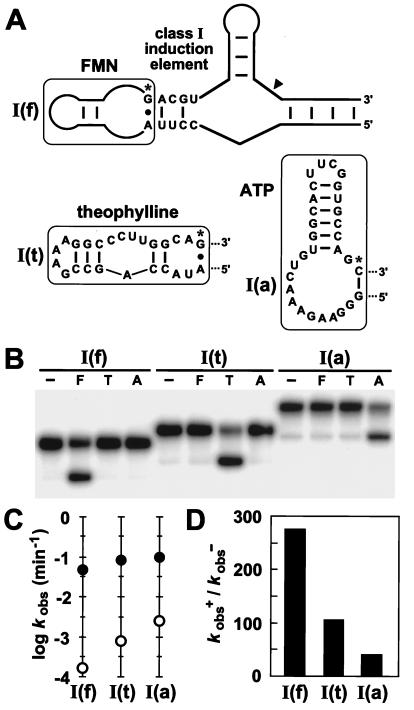
We reasoned that if binding energy derived from the ligand–aptamer complex is used to shift the thermodynamic balance between two slip-structure conformations, then each bridge may act as a generic reporter of the occupation state of the adjoining aptamer domain in a manner that is independent of the sequence and ligand specificity of the aptamer. To examine this possibility, the FMN aptamer was removed from the class I induction element of an FMN-sensitive ribozyme and replaced with either an aptamer that binds theophylline (38) or an aptamer that binds ATP (39) (Fig. 5A). In each case, ligand binding is known to stabilize base pairing of the terminal nucleotides of the appended aptamer (33, 38, 39). Therefore, adaptive binding of ligand by the aptamer may trigger the allosteric transition necessary for class I function. Indeed, each ribozyme construct undergoes self-cleavage only in the presence of its cognate ligand (Fig. 5B). Kinetic analyses (Fig. 5 C and D) show that the activity of the FMN-inducible ribozyme increases 270-fold in the presence of FMN, whereas the theophylline- and ATP-inducible ribozymes are activated 110- and 40-fold, respectively, only by their corresponding ligands. These findings indicate that the task of regulating ribozyme activity rests mainly on the bridge element, which relays information concerning the binding state of the aptamer to the adjoining ribozyme domain.
Figure 5.
Modular characteristics of the class I induction element. (A) Sequence and secondary structures of allosteric ribozyme constructs containing either an FMN, theophylline, or ATP aptamer [constructs I(f), I(t), and I(a), respectively]. The terminal A⋅G or G-C base pairs of each aptamer (denoted by asterisks) are interactions stabilized by ligand binding. (B) Qualitative assessment of the specificity of ligand-induced ribozyme self-cleavage. Internally 32P-labeled constructs were incubated at 23°C for 15 min in the absence (−) or presence of FMN (F; 200 μM), theophylline (T; 1 mM), or ATP (A; 1 mM). (C) Kinetic parameters kobs− (○) and kobs+ (●) determined for each allosteric ribozyme construct in the absence or presence of its cognate ligand, respectively. (D) Allosteric activation of ribozyme function (kobs+/kobs−) is depicted for each construct.
Although the class I induction element can be engineered to respond to several unrelated effector molecules, this characteristic is not universally applicable. For example, appending an aptamer for arginine (40) to the class I induction element failed to produce a significant allosteric effect. We did find that two of three other classes of induction elements tested (classes VI and VII) also display modularity when engineered to carry the theophylline aptamer. However, class III induction elements and class III inhibition elements showed no response to the addition of effector when similarly appended to the same aptamer. These findings indicate that the successful design of an allosteric ribozyme using this modular approach requires the fusion of compatible “matched pairs” of aptamer and bridge domains.
Conclusions.
The simultaneous use of rational and combinatorial approaches to enzyme engineering (41) provides a powerful approach to the design of new ribozymes with enhanced kinetic characteristics. The tripartite ribozyme constructs generated by using this strategy of enzyme engineering function as highly specific sensors for various small organic compounds. A critical component of these constructs are the ligand-responsive bridge elements. These dynamic structural domains act as simple “communication modules” that can be used to rapidly engineer new RNA molecular switches simply by swapping domains within the context of the tripartite construct. In addition, the introduction of mutations into the receptor domain of the construct should make possible the in vitro selection of new ligand-binding domains based on the modulation of a catalytic report. In a similar manner, new RNA molecular switches could be made that serve as new precision biosensors, or that function in vivo as genetic control elements that regulate gene expression in response to the presence of almost any effector molecule.
Acknowledgments
We thank G. A. Emilsson, A. Roth, G. F. Joyce, and S. Altman for helpful comments on the manuscript. G.A.S. was supported by a Seessel Postdoctoral Fellowship from Yale University. Additional funding was provided by a Hellman Family Fellowship and a Young Investigator Award from the Arnold and Mabel Beckman Foundation to R.R.B.
ABBREVIATION
- FMN
flavin mononucleotide
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
The catalytic rate of an allosteric enzyme is regulated by the binding of an effector molecule to an allosteric binding site that is topographically distinct from the active site of the enzyme. Conformational changes brought about by the binding of a specific effector molecule result in modulation of the activity of the adjacent active site (18–21).
References
- 1.McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Nature (London) 1995;375:549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 2.Marsden A F A, Wilkinson B, Cortés J, Dunster N J, Staunton J, Leadlay P F. Science. 1998;279:199–202. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- 3.Hellinga H W, Marvin J S. Trends Biotechnol. 1998;16:183–189. doi: 10.1016/s0167-7799(98)01174-3. [DOI] [PubMed] [Google Scholar]
- 4.Crane D E, Walsh C T, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 5.Tang J, Breaker R R. Chem Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 6.Tang J, Breaker R R. RNA. 1997;3:914–925. [PMC free article] [PubMed] [Google Scholar]
- 7.Araki M, Okuno Y, Hara Y, Sugiura Y. Nucleic Acids Res. 1998;26:3379–3384. doi: 10.1093/nar/26.14.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang J, Breaker R R. Nucleic Acids Res. 1998;26:4214–4221. doi: 10.1093/nar/26.18.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberston M P, Ellington A D. Nat Biotechnol. 1999;17:62–66. doi: 10.1038/5236. [DOI] [PubMed] [Google Scholar]
- 10.Gerstein M, Lesk A M, Chothia C. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 11.Pyle A M, Green J B. Curr Biol. 1995;5:303–310. doi: 10.1016/0959-440x(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 12.Draper D E. Trends Biochem Sci. 1996;21:145–149. [PubMed] [Google Scholar]
- 13.Westhof E, Masquida B, Jaeger L. Fold Des. 1996;1:78–88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 14.Willians K P, Bartel D P. Nucleic Acids Mol Biol. 1996;10:367–381. [Google Scholar]
- 15.Breaker R R. Chem Rev. 1997;97:371–390. doi: 10.1021/cr960008k. [DOI] [PubMed] [Google Scholar]
- 16.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 17.Osborne S E, Ellington A D. Chem Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 18.Monod J, Jacob F. Cold Spring Harbor Symp Quant Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 19.Monod J, Changeux J-P, Jacob F. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 20.Kurganov B I. Allosteric Enzymes. New York: Wiley; 1978. [Google Scholar]
- 21.Perutz M. Mechanisms of Cooperativity and Allosteric Regulation in Proteins. New York: Cambridge Univ. Press; 1994. [DOI] [PubMed] [Google Scholar]
- 22.Ye X, Gorin A, Ellington A D, Patel D J. Nat Struct Biol. 1996;3:1026–1033. doi: 10.1038/nsb1296-1026. [DOI] [PubMed] [Google Scholar]
- 23.Hsiung C, Patel D J. Nat Struct Biol. 1996;3:1046–1050. doi: 10.1038/nsb1296-1046. [DOI] [PubMed] [Google Scholar]
- 24.Lin C H, Patel D J. Chem Biol. 1997;4:817–832. doi: 10.1016/s1074-5521(97)90115-0. [DOI] [PubMed] [Google Scholar]
- 25.Patel D J, Suri A K, Jiang F, Jiang L, Fan P, Kumar R A, Nonin S. J Mol Biol. 1997;272:645–664. doi: 10.1006/jmbi.1997.1281. [DOI] [PubMed] [Google Scholar]
- 26.Burgstaller P, Famulok M. Angew Chem Int Ed Engl. 1994;33:1084–1087. [Google Scholar]
- 27.Forster A C, Symons R H. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 28.Fedor M J, Uhlenbeck O C. Biochemistry. 1992;31:12042–12054. doi: 10.1021/bi00163a012. [DOI] [PubMed] [Google Scholar]
- 29.Tuschl T, Eckstein F. Proc Natl Acad Sci USA. 1993;90:6991–6994. doi: 10.1073/pnas.90.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long D M, Uhlenbeck O C. Proc Natl Acad Sci USA. 1994;91:6977–6981. doi: 10.1073/pnas.91.15.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgstaller P, Hermann T, Huber C, Westhof E, Famulok M. Nucleic Acids Res. 1997;25:4018–4027. doi: 10.1093/nar/25.20.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carey J. Proc Natl Acad Sci USA. 1988;85:975–979. doi: 10.1073/pnas.85.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgstaller P, Famulok M. Bioorg Med Chem Lett. 1996;6:1157–1162. [Google Scholar]
- 34.Fan P, Suri A K, Fiala R, Live D, Patel D J. J Mol Biol. 1996;258:480–500. doi: 10.1006/jmbi.1996.0263. [DOI] [PubMed] [Google Scholar]
- 35.Serra M J, Turner D H. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 36.Lodmell J S, Dahlberg A E. Science. 1997;277:1262–1267. doi: 10.1126/science.277.5330.1262. [DOI] [PubMed] [Google Scholar]
- 37.von Ahsen U. Chem Biol. 1998;5:R3–R6. doi: 10.1016/s1074-5521(98)90080-1. [DOI] [PubMed] [Google Scholar]
- 38.Jenison R D, Gill S C, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 39.Sassanfar M, Szostak J W. Nature (London) 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 40.Famulok M. J Am Chem Soc. 1994;116:1698–1706. [Google Scholar]
- 41.Breaker R R, Joyce G F. Trends Biotechnol. 1994;12:268–275. doi: 10.1016/0167-7799(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 42.Ruffner D E, Stormo G D, Uhlenbeck O C. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]