Abstract
Multiple DNA polymerases participate in replicating the leading and lagging strands of the eukaryotic nuclear genome. Although 50 years have passed since the first DNA polymerase was discovered, the identity of the major polymerase used for leading-strand replication is uncertain. We constructed a derivative of yeast DNA polymerase ε that retains high replication activity but has strongly reduced replication fidelity, particularly for thymine-deoxythymidine 5′-monophosphate (T-dTMP) but not adenine-deoxyadenosine 5′-monophosphate (A-dAMP) mismatches. Yeast strains with this DNA polymerase ε allele have elevated rates of T to A substitution mutations. The position and rate of these substitutions depend on the orientation of the mutational reporter and its location relative to origins of DNA replication and reveal a pattern indicating that DNA polymerase ε participates in leading-strand DNA replication.
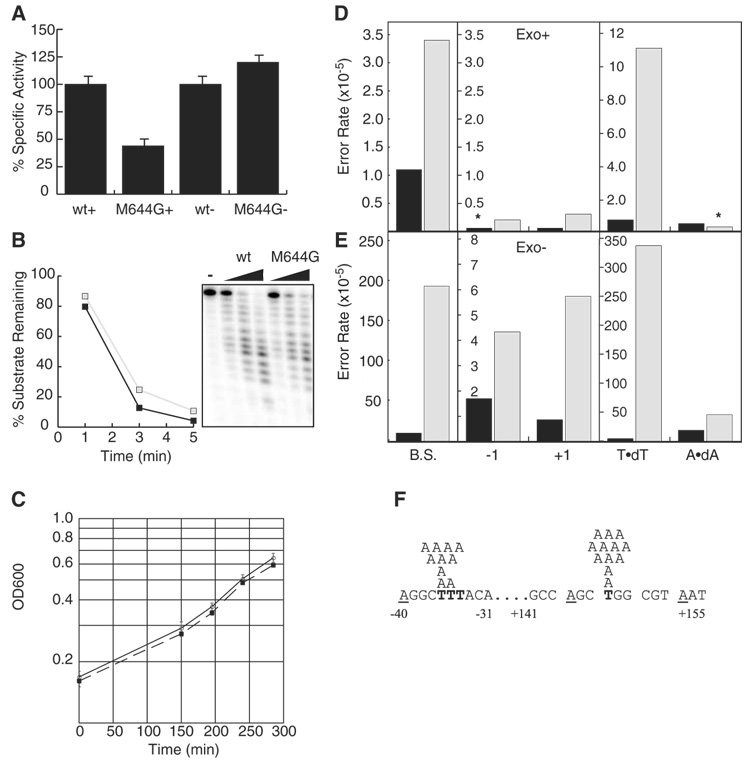
Replication of the eukaryotic nuclear genome requires DNA polymerase α to initiate synthesis at origins and to initiate synthesis of Okazaki fragments on the lagging strand, allowing DNA polymerases δ (pol δ) and ε (pol ε) to then perform the bulk of chain elongation (1, 2). Pol δ is implicated in lagging-strand replication (1), but the identity of the polymerase(s) that replicates the leading strand is unknown (1, 2). Null alleles of the POL2 (pol ε) and POL3 (pol δ) genes are uninformative for identifying the leading-strand polymerase, because both genes are essential for normal replication. To retain replication activity while generating a distinct mutational signature in vivo that allows assignment of pol ε to leading- and/or lagging-strand replication in yeast cells, we substituted glycine for Met644 at the Saccharomyces cerevisiae pol ε active site. Yeast pol ε with the Met644Gly change retains 44% of wild-type polymerase activity (Fig. 1A) and retains full 3′ exonuclease activity (Fig. 1B). A haploid pol2-M644G yeast strain grows at a rate similar to a POL2 strain (Fig. 1C), indicating that M644G pol ε retains substantial replicative capacity. In both its exonuclease-proficient (Fig. 1D) and exonuclease-deficient forms (Fig. 1E), M644G pol ε synthesizes DNA in vitro with reduced fidelity in comparison with wild-type (i.e., Met644) pol ε (Fig. 1 and table S1) (3), i.e., it is defective in discriminating against deoxynucleotide triphosphate (dNTP) misinsertion. Even the exonuclease-proficient polymerase has an elevated base-substitution error rate (Fig. 1D), indicating that despite retaining proofreading potential (Fig. 1B), M644G pol ε does not efficiently proofread certain mismatches, for example, T-dTMP mismatches. This is more obvious in some sequence contexts than others. Among 16 positions in the lacZ template where T to A substitutions can be detected (fig. S1), errors are particularly prevalent at template T+147 and T−36 (Fig. 1F), resulting in a high error rate of exonuclease-proficient M644G pol ε for T-dTMP mismatches (11 × 10−5). In contrast, the lowest error rate for exonuclease-proficient M644G pol ε is ≤0.28 × 10−5 for the A-dAMP mismatch, a difference of at least 39-fold (Fig. 1D). This large difference in the rate of stable misincorporation of dTMP opposite T compared to dAMP opposite A is critical for interpreting M644G pol ε’s distinctive mutational signature in yeast, because these mismatches are the two possible intermediates that could result in an A–T to T–A substitution in vivo.
Fig. 1.
Specific activity, growth, and fidelity analyses of M644G pol ε. (A) Relative specific activity of pol ε derivatives with activated DNA. (B) Exonuclease activity of wild-type (black boxes) and M644G pol ε (gray boxes). (C) Growth curves for wild-type (solid line) and pol2-M644G (dashed line) strains. (D) Average error rates for exonuclease-proficient wild-type (black bars) and M644G pol ε (gray bars) for base substitutions (B.S.), single-base deletions (−1), single-base insertions (+1), T to A transversions (T•dT), and A to T transversions (A•dA). Asterisks denote error rates that are ≤ the indicated value. (E) As in (D) but for exonuclease-deficient pol ε. (F) T to A transversions at −36 and +147 positions in lacZ using exonuclease-proficient M644G pol ε. Sites where T to A and A to T transversions are detectable are in bold and underlined, respectively.
In haploid yeast containing the exonuclease-proficient pol2-M644G allele, the spontaneous mutation rate at the CAN1 locus was elevated by a factor of 3.9 compared with a wild-type strain (table S3) (3). When repair of single-base mismatches was inactivated by disrupting MSH6, the mutation rate at CAN1 in the pol2-M644G strain was 58 times as high as that for the Δmsh6 strain with wild-type POL2 (table S3), consistent with inaccurate DNA replication in vivo by exonuclease-proficient M644G pol ε. When 11 independent Canr mutants from the pol2-M644G strain were sequenced, six contained T–A to A–T substitutions predicted by the high rate of T-dTMP mismatch formation by M644G pol ε (Fig. 1D).
Based on these results, we investigated whether M644G pol ε participates in leading-and/or lagging-strand replication using a strategy previously employed to follow replicative mutagenesis on the two strands in strains encoding wild-type or exonuclease-deficient DNA polymerases (4–6). In those studies, the template strand for replication errors was assigned by monitoring incorporation of 6-hydroxylaminopurine monophosphate opposite template cytosine on one strand, or incorporation of dAMP opposite template 8-oxo-G on the other strand. In the present study, we assigned the template strand based on the strong preference of M644G pol ε for stable misincorporation of dTMP opposite T rather than dAMP opposite template A (Fig. 1D). We compared spontaneous mutation rates in strains containing the pol2-M644G allele or the wild-type POL2 gene (Table 1). Rates were measured using the URA3 reporter gene, which was inserted in each of the two possible orientations, and on opposite sides of, but close to, ARS306 (Fig. 2), an origin of replication on the left arm of chromosome III that fires in early S phase with >90% efficiency (7).
Table 1.
Mutation rates at URA3. Mutation rates were measured as described in (15). Parentheses contain the numbers of A–T to T–A plus T–A to A–T substitutions observed among the total number of ura3 mutants sequenced. CI, confidence interval.
| Yeast strain | Location and origin | URA3 orient. | Mut. rate (× 10−8) | 95% CI | A–T to T–A plus T–A to A–T (× 10−8) | Occurrences A–T to T–A: T–A to A–T |
|---|---|---|---|---|---|---|
| Wild type | R-ARS306 | 1 | 6.6 | 3.5–12 | 1.6 (3/12) | 2:1 |
| 2 | 5.9 | 1.6–17 | 1.5 (2/8) | 1:1 | ||
| pol2-M644G | L-ARS306 | 1 | 16 | 11–36 | 5.3 (7/21) | 1:6 |
| 2 | 45 | 23–60 | 29 (20/31) | 18:2 | ||
| R-ARS306 | 1 | 28 | 13–40 | 19 (45/61) | 43:2 | |
| 2 | 16 | 9.7–36 | 2.1 (5/38) | 1:4 | ||
| Wild type | R-ARS501 | 1 | 3.9 | 3.2–7.2 | 0.16 (1/24) | 0:1 |
| 2 | 3.1 | 2.2–6.4 | 0.25 (2/25) | 1:1 | ||
| pol2-M644G | R-ARS501 | 1 | 26 | 17–40 | 17 (22/34) | 22:0 |
| 2 | 21 | 15–44 | 5.9 (9/32) | 2:7 |
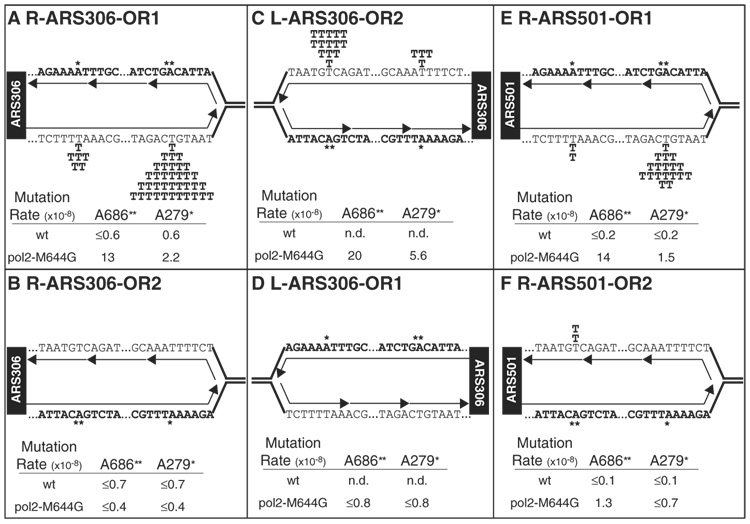
Fig. 2.
Variation in the rates of T to A transversions by location and gene orientation. Six haploid strains were constructed containing the pol2-M644G mutant allele. URA3 was to the right (A, B, E, and F) or left (C and D) of the indicated ARS, with the coding sequence in the Watson (A, D, and E) or Crick (B, C, and F) strand. A replication fork is shown moving away from the ARS (black box) and replicating the URA3 gene. The nascent leading strand is depicted as a single, unbroken arrow, whereas nascent Okazaki fragments on the lagging strand are depicted as broken arrows. The A–T to T–A hot spots (* for A279 and ** for A686) are represented as the inferred T-dTMP mispair generated during replication.
URA3 mutation rates in the wild-type POL2 strains ranged from 3.1 × 10−8 to 6.6 × 10−8, whereas rates in the pol2-M644G strains ranged from 16 × 10−8 to 45 × 10−8 (Table 1). When independent ura3 mutants from these strains were sequenced, a variety of mutations were observed in the wild-type (M644) strains, consistent with many sources of mutagenesis in wild-type yeast. Among the mutations, A–T to T–A substitutions were rare, resulting in low spontaneous mutation rates for these two events in wild-type strains (Table 1). In contrast, and as predicted by the high rate of T-dTMP mismatch formation by M644G pol ε in vitro (Fig. 1D), the rates of A–T to T–A substitutions were higher in the pol2-M644G strains (Table 1). Just as misincorporation in vitro was prevalent at certain template locations in the lacZ template (e.g., T+147 and T−36, Fig. 1F), the majority of these transversions generated in the pol2-M644G strains occurred at two specific base pairs (hot spots) in the URA3 coding sequence, numbers 279 and 686 (Fig. 2).
The rate of these substitutions depended strongly on the orientation and location of the URA3 gene. When URA3 was placed to the right of ARS306 in the pol2-M644G strain, 43 of 61 mutants recovered contained an A to T transversion, defined relative to the URA3 coding strand (Table 1), and 36 of these 43 were at base pair number 686 (Fig. 2A). This yields a rate of A to T transversion at base pair 686 in orientation 1 of 13 × 10−8 (Fig. 2A). This rate is higher than that observed in the corresponding wild-type strain (≤0.6 × 10−8) (Fig. 2A), which indicates that the A to T mutations are dependent on replication by M644G pol ε. Because ARS306 is only ~1700 base pairs distant from the URA3 gene, the replication fork emanating from ARS306 reaches base pair 686 long before the fork emanating from ARS307, which is over 32,000 base pairs to the right of URA3. In this case, if we assume from the error specificity in vitro (Fig. 1D) that these events resulted from T-dTMP mismatches generated by M644G pol ε rather than by A-dAMP mismatches, pol ε is inferred to replicate the leading-strand template, as depicted in Fig. 2A.
In contrast to the two A to T hot spots, only two T to A events were detected among 61 mutations recovered from this strain (Table 1, R-ARS306, orientation 1), even though there are many sites where such substitutions can be scored. Such T to A events would be inferred to result from T-dTMP mismatches generated if M644G pol ε was replicating the lagging-strand DNA template. This paucity of T to A substitutions could be due to the fact that pol ε participates much more in leading-strand replication than in lagging-strand replication. Alternatively, a hot spot sequence context may not be present in the lagging-strand template. These two possibilities were distinguished by comparing results in a second strain in which URA3 was again placed to the right of ARS306, but now in the opposite orientation. In orientation 2, where a T-dTMP mismatch at base pair 686 would be a lagging-strand error, the rate of A to T transversions at base pair 686 was observed to be lower by a factor of at least 32 (≤0.4 × 10−8, Fig. 2B) as that in orientation 1. Thus, the lack of T to A substitutions in orientation 1 is not simply due to the absence of the appropriate hot spot sequence context in the lagging-strand template, but is most simply explained by prominent participation of pol ε in leading-strand replication as compared with lagging-strand replication.
This interpretation is reinforced by the fact that the opposite mutational asymmetry holds when URA3 is located to the left of ARS306. Here, the rate of A to T transversions at base pair 686 was high (20 × 10−8) in orientation 2 (Fig. 2C), where a T-dTMP mismatch by M644G pol ε would again be a leading-strand error, and the rate is lower by at least a factor of 25 (≤0.5 × 10−8) in orientation 1 (Fig. 2D), where a T-dTMP mismatch would be a lagging-strand error. These effects are not confined to the A–T base pair at position 686, because a similar pattern is observed at position 279 in the URA3 gene (Fig. 2). A to T substitutions at position 279 are only seen in two of the four strains (Fig. 2, A and C), and in both cases the pattern is consistent with T-dTMP mismatches generated by M644G pol ε during replication of the leading-strand template. Finally, a similar pattern is observed at a second genomic location examined by inserting URA3 in opposite orientations 850 base pairs to the right of ARS501, an origin of replication on the right arm of chromosome V that is used late in S phase (8). The results at ARS501 are similar to those at ARS306, in that the rate of A to T transversion at base pair 686 is 11 times as high in orientation 1 (Table 1 and Fig. 2E) compared with orientation 2 (Table 1 and Fig. 2F). Given that the nearest flanking origin is at least 10,000 base pairs to the right of ARS501, this pattern is consistent with T-dTMP mismatches generated by M644G pol ε during replication of the leading-strand template.
From the M644G pol ε bias for T-dTMP as opposed to A-dAMP errors (Fig. 1) and the patterns of mutagenesis in vivo (Fig. 2), we infer that M644G pol ε, and therefore likely wild-type pol ε, participates in leading-strand replication. This interpretation is consistent with evidence for pol δ participation in lagging-strand replication (1) and with evidence that the exonuclease activities of pol δ and pol ε edit 6-hydroxylaminopurine–induced mismatches on opposite strands (6). The interpretation that pol ε replicates the leading strand does not exclude its participation in lagging-strand replication under certain circumstances. The interpretation also does not exclude the possibility that pol δ might contribute to leading-strand replication. Both models for the participation of pol δ and pol ε in leading-and lagging-strand replication (1, 2) may be correct, with the choice of polymerase dependent on such variables as replication timing (9), DNA sequence context, chromosomal organization and/or chromatin status, or various types of replicative stress. Given that pol ε is a known checkpoint sensor (10–12), our evidence that pol ε has an important role in replicating the leading strand is consistent with the idea that the status of leading-strand synthesis of replication fork progression determines whether the S phase checkpoint is activated (13, 14).
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/317/5834/127/DC1 Materials and Methods, SOM Text, Fig. S1, Tables S1 to S3, References
References and Notes
- 1.Garg P, Burgers PM. Crit. Rev. Biochem. Mol. Biol. 2005;40:115. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 2.Johnson A, O'Donnell M. Annu. Rev. Biochem. 2005;74:283. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 3.Materials and methods are available as supporting material on Science Online.
- 4.Pavlov YI, Mian IM, Kunkel TA. Curr. Biol. 2003;13:744. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- 5.Pavlov YI, Newlon CS, Kunkel TA. Mol. Cell. 2002;10:207. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- 6.Shcherbakova PV, Pavlov YI. Genetics. 1996;142:717. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poloumienko A, Dershowitz A, De J, Newlon CS. Mol. Biol. Cell. 2001;12:3317. doi: 10.1091/mbc.12.11.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson BM, Brewer BJ, Reynolds AE, Fangman WL. Cell. 1991;65:507. doi: 10.1016/0092-8674(91)90468-e. [DOI] [PubMed] [Google Scholar]
- 9.Fuss J, Linn S. J. Biol. Chem. 2002;277:8658. doi: 10.1074/jbc.M110615200. [DOI] [PubMed] [Google Scholar]
- 10.Dua R, Edwards S, Levy DL, Campbell JL. J. Biol. Chem. 2000;275:28816. doi: 10.1074/jbc.M002376200. [DOI] [PubMed] [Google Scholar]
- 11.Dua R, Levy DL, Campbell JL. J. Biol. Chem. 1999;274:22283. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 12.Navas TA, Zhou Z, Elledge SJ. Cell. 1995;80:29. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 13.Harrison JC, Haber JE. Annu. Rev. Genet. 2006;40:209. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 14.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Annu. Rev. Biochem. 2004;73:39. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 15.Shcherbakova PV, Kunkel TA. Mol. Cell. Biol. 1999;19:3177. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.We thank D. Nguyen for DNA substrate preparation and sequence analysis of lacZ mutants, and R. Schaaper and D. Gordenin for critically reading this manuscript. This work was funded in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (T.A.K.) and partly by the Swedish Research Council, The Swedish Cancer Society, and the Fund for Basic Science-Oriented Biotechnology and the Medical Faculty at Umeå University (E.J.).