Summary
Recombinant adenoviruses are frequently used as gene transfer vehicles for therapeutic gene delivery. Strategies to amend their tropism include the incorporation of polypeptides with high affinity for cellular receptors. Single-chain antibodies have a great potential to achieve such cell type-specificity. In this study we evaluated the efficiency of incorporation of a single-chain antibody fused with the adenovirus minor capsid protein IX in the capsid of adenovirus type 5 vectors. To this end the codons for the single-chain antibody scFv 13R4 were fused with those encoding the C-terminus of the pIX via a 75-Angstrom spacer sequence. The 13R4 is a hyper-stable single-chain antibody directed against β-galactosidase, which was selected for its capacity to fold correctly in a reducing environment such as the cytoplasm. A lentiviral vector was used to stably express the pIX.flag.75.13R4.MYC.HIS fusion gene in 911 helper cells. Upon propagation of pIX-gene deleted HAdV-5 vectors on these cells the pIX-fusion protein was efficiently incorporated in the capsid. Here, the 13R4 scFv was functional as was evident from its capacity to bind its ligand β-galactosidase. These data demonstrate that the minor capsid protein IX can be used as an anchor for incorporation of single-chain antibodies in the capsids of adenovirus vectors.
Keywords: Adenoviridae; genetics; ultrastructure; Blotting, Western; methods; Capsid Proteins; genetics; Cell Line, Transformed; Gene Expression; Gene Therapy; Genetic Engineering; Genetic Vectors; analysis; genetics; Humans; Immunoglobulin Fragments; genetics; Immunohistochemistry; Lentivirus; genetics; Microscopy, Immunoelectron; Recombinant Fusion Proteins; genetics; Transduction, Genetic; methods; beta-Galactosidase; genetics
Keywords: hyper-stable scFv, adenovirus, targeting, protein IX, capsid
Introduction
Human Adenovirus (HAdV)-derived vectors are among the most frequently used gene delivery vehicles for human gene therapy and vaccination.1 Much effort has been devoted to improve the cell-type specificity of gene delivery. Whereas the use of bispecific antibodies has been employed with considerable success, retargeting of adenovirus vectors by genetic incorporation of cell specific-ligands and single-chain antibodies proved more difficult. Many attractive ligands for insertion into the virus capsid elements for retargeting purposes are molecules that are normally excreted from the cells, such as epidermal growth factor (EGF), antibodies, and their derivatives e.g. single-chain antibody fragments (scFv).2, 3 An important hurdle is that many of such polypeptide ligands are normally routed via the protein secretory pathway, while the adenovirus particles assemble in the nucleus. Hence, the ligands fused with capsid proteins lack the post-translational modifications that may be essential for their function.2, 4 The reducing environment in the cytoplasm, which prevents the formation of disulphide bridges, and the absence of accessory factors to help these proteins to fold correctly, avert their maturation to functional proteins.5 Indeed, studies in which scFv were fused with capsid proteins were not very successful.2, 6, 7 However, some antibodies can be produced in a soluble form in the cytoplasm and retain their activity. These are the called hyper-stable single-chain antibodies.8–12 Recently, such hyper-stable scFv have been incorporated in HAdV particles on de-knobbed, fibritin-foldon trimerized fibers.4 Although the exact location of pIX in the adenovirus capsid is under revision13–16, we and others have shown that this minor protein is an efficient platform for retargeting and imaging moieties.17–21 Here we demonstrate efficient and functional incorporation of the hyper-stable scFv 13R4 that was fused with the minor capsid protein pIX via a 75-Angstrom spacer. The scFv 13R4 originates from a naïve human phage display library22 and was isolated after random mutagenesis by error-prone polymerase chain reaction, and selection for increased cytoplasmic solubility.9 We show that 13R4 fused with pIX via the 75-Angstrom spacer is accessible on the surface of purified viruses. Moreover the 13R4 is functional in the capsid as evidenced from its capacity to bind E.coli β-galactosidase.
Results
For incorporation of the 13R4 scFv in the adenovirus capsid a fusion gene was constructed in which the coding region of pIX was fused via the flag epitope with the codons for a 75Angstrom spacer, and with the codons for the 13R4 scFv (Figure 1a). The resulting fusion gene coding for pIX.flag.75.13R4.MYC.HIS was inserted into the lentiviral expression vector pLV-CMV-IRES-NPTII.23 A schematic outline of the vector is provided in Figure 1b. To test the pIX.flag.75.13R4.MYC.HIS production after lentivirus transduction, 911 cells were exposed to LV-CMV-pIX.flag.75.13R4.MYC.HIS-IRES-NPTII at 40 ng p24 per 105 cells (911-pIX.flag.75.13R4.MYC.HIS). Forty-eight hours post transduction, the 911-pIX.flag.75.13R4.MYC.HIS cells were fed fresh medium with 200 μg/ml G418. No clonal cells were isolated, since the lentiviral transduction leads to a polyclonal cell line that produce homogenous levels pIX, sufficient to fully restore the heat-stable phenotype of the HAdV particle.23 Immunohistochemistry analysis showed homogeneous amounts of the fusion proteins in the transduced cells (Figure 2a). The pIX.flag.75.13R4.MYC.HIS amounts produced by the transduced 911 cells are similar to the pIX level produced by 911 cells during infection with a wt HAdV-5 (Figure 2b).23
Figure 1.
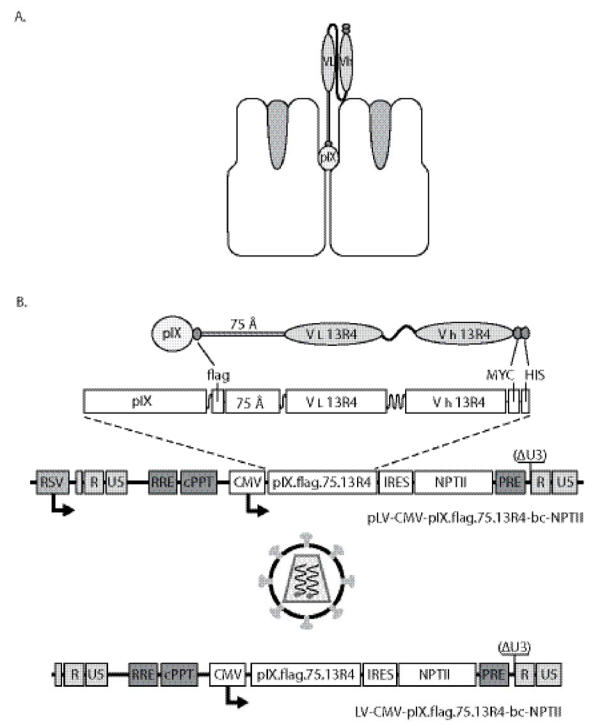
A. Schematic representation of the pIX.flag.75.13R4.MYC.HIS fusion protein exposing the 13R4 scFv above the hexon capsomers. B. Schematic representation of the lentiviral system. The lentiviral vectors used in this study are so-called SIN vectors35, and contain the Rev-responsive element sequence36, the central poly-purine tract (cPPT)33, 37, 38, and the human hepatitis B virus-derived posttranscriptional regulatory element. The encephalomyocarditis virus internal ribosomal entry site (IRES) was obtained from pTM339, the NPTII coding region was isolated from peGFPn2 (Clontech, BD Biosciences, The Netherlands).
Figure 2.
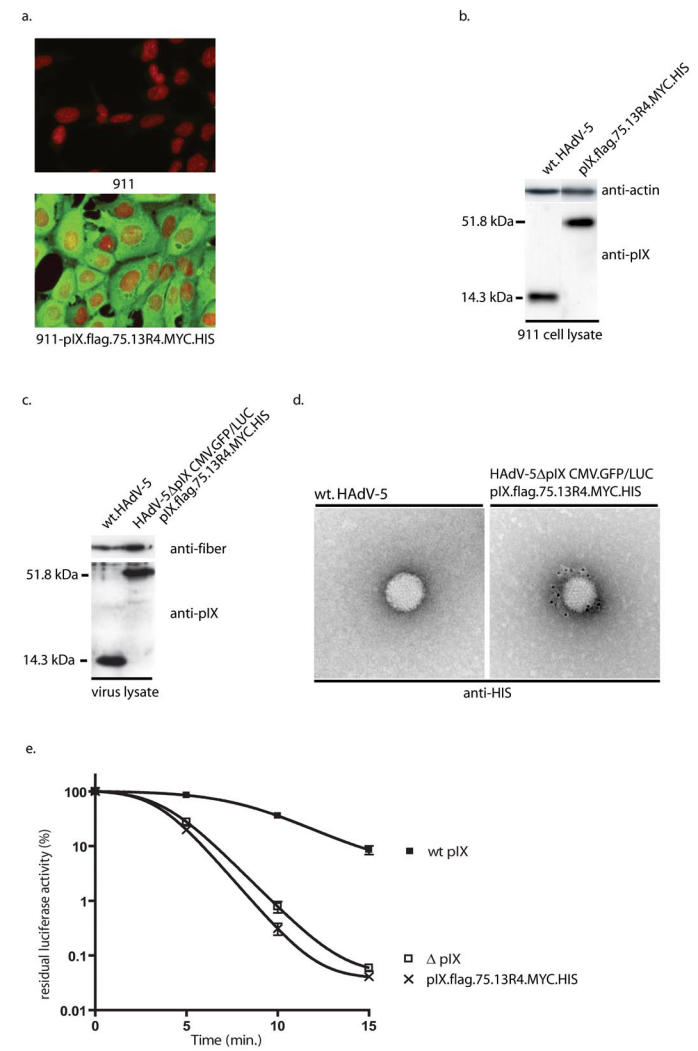
A. Immunohistochemistry assay for detection of the pIX.flag.75.13R4.MYC.HIS produced by the 911-pIX.flag.75.13R4.MYC.HIS cells. The production of pIX.flag.75.13R4.MYC.HIS was visualized using mouse anti-flag and FITC-labeled goat-anti-mouse antibodies. The nuclei were stained using propidium iodide. B. Western analysis of pIX.flag.75.13R4.MYC.HIS levels in the 911-pIX.flag.75.13R4.MYC.HIS cells. The pIX.flag.75.13R4.MYC.HIS amount in the complementing cell line 911-pIX.flag.75.13R4.MYC.HIS was compared with the pIX amounts during wt HAdV-5 infection. The Western analysis was performed using anti pIX serum23. C. Western analysis of the incorporation efficiency of pIX.flag.75.13R4.MYC.HIS. To test the incorporation efficiency of pIX.flag.75.13R4.MYC.HIS produced by the 911-pIX.flag.75.13R4.MYC.HIS cells, HAdV-5ΔpIX.CMV.GFP/LUC was propagated on the cell line, purified by CsCl centrifugation, and protein lysates of the purified viruses sample were made for Western analysis. The amount of the pIX fusion proteins in HAdV-5ΔpIX.CMV.GFP/LUC propagated on 911-pIX.flag.75.13R4.MYC.HIS was compared with wt HAdV-5, with anti-pIX serum and, as a virus-particle loading control, the 4D2 antibody directed against the fiber protein. D. Immunoelectron microscopic analysis of HAdV-5ΔpIX.CMV.GFP/LUC loaded with pIX.flag.75.13R4.MYC.HIS. To test the accessibility of the HIS epitope on viruses loaded with pIX.flag.75.13R4.MYC.HIS were bound on copper grids. The HIS epitope was detected with penta-HIS antibody, followed by rabbit anti-mouse immunoglobulin and gold-labeled Prot.A. E. Heat stability of HAdV-5ΔpIX.CMV.GFP/LUC with pIX.flag.75.13R4.MYC.HIS in their capsid. HAdV-5ΔpIX.CMV.GFP/LUC was propagated on 911-pIX.flag.75.13R4.MYC.HIS cells. Similarly, HAdV-5ΔpIX.CMV.GFP/LUC and HAdV-5.CMV.GFP/LUC were propagated on 911 cells as negative and positive control, respectively. Freeze-thaw lysates were incubated at 45°C for various times. Residual infectious virus titers were estimated by determining the capacity of the virus to induce luciferase activity in U2OS cell 24 h after infection. The results are presented as percentages of residual luciferase activity. Each bar represents the cumulative mean ± standard deviation of triplicate analyses.
Next, we tested the incorporation of pIX.flag.75.13R4.MYC.HIS into the capsid of the HAdV-5 vector HAdV-5ΔpIX.CMV.GFP/LUC.23 This vector lacks a functional pIX gene and carries the eGFP and the firefly luciferase reporter genes under control of two separate CMV-promoters. HAdV-5ΔpIX.CMV.GFP/LUC viruses were propagated on the 911-pIX.flag.75.13R4.MYC.HIS cell line, harvested and purified via the conventional CsCl purification method. During purification, particle-associated pIX molecules were separated from the non-associated pIX, since variants of pIX that cannot be incorporated into the capsid do not co-purify with the particles of CsCl gradients.19, 24 To examine the amount of pIX.flag.75.13R4.MYC.HIS in the particles, 5×109 CsCl-gradient purified particles were analyzed by Western analysis (Figure 2c). The amount of pIX.flag.75.13R4.MYC.HIS in the pIX.flag.75.13R4.MYC.HIS-loaded HAdV-5ΔpIX.CMV.GFP/LUC particles is similar to the amounts in wt HAdV-5 particles.
To test if the 13R4.MYC.HIS fusion protein was accessible on the outside of the viral capsid, the purified pIX.flag.75.13R4.MYC.HIS-loaded HAdV-5ΔpIX.CMV.GFP/LUC particles were subjected to immunoelectron microscopy. The presence of the 13R4.MYC.HIS fusion protein was visualized using penta-HIS antibodies and gold-conjugated prot.A (Figure 2d). Wt HAdV-5 particles were used as negative control. There was no gold label found on the negative control whereas viruses loaded with the 13R4.MYC.HIS show specific binding of the gold-conjugated prot.A.
To study if incorporation of the pIX.flag.75.13R4.MYC.HIS could restore the heat stability of the pIX-gene deleted viruses, HAdV-5ΔpIX.CMV.GFP/LUC was propagated on 911-pIX.flag.75.13R4.MYC.HIS cells. Similarly, HAdV-5ΔpIX.CMV.GFP/LUC and HAdV-5.CMV.GFP/LUC were propagated on 911 cells as negative and positive control, respectively. Heat-inactivation analysis demonstrated that although the particles were fully loaded, the pIX.flag.75.13R4.MYC.HIS protein could not restore heat-stability of the pIX-gene deleted virus (Figure 2e).
To study the functionality of the scFv in the capsid we assayed the ligand (β-galactosidase)-binding capability of the 13R4 on the surface of the adenovirus. To this end, CsCl purified pIX.flag.75.13R4.MYC.HIS-loaded HAdV-5ΔpIX.CMV.GFP/LUC and wt HAdV-5 were separately mixed with 100 μg β-galactosidase. The viruses were purified using continuous CsCl density gradients to separate free β-galactosidase from the β-galactosidase bound to the 13R4 on the surface of the adenoviral particles. To examine if the 13R4 scFv was able to bind β-galactosidase both viruses were analyzed by Western analysis (Figure 3a) and immunoelectron microscopy (Figure 3b). Both assays showed that β-galactosidase was bound specifically to viruses loaded with the 13R4 scFv.
Figure 3.
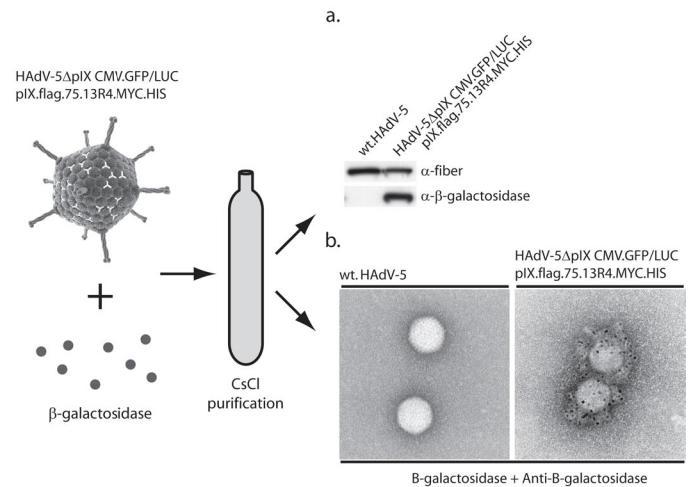
To test if the HAdV-5ΔpIX.CMV.GFP/LUC particles loaded with pIX.flag.75.13R4.MYC.HIS were able to bind specifically β-galactosidase on the outside of the virion, viruses were mixed with β-galactosidase and purified via the standard CsCl purification protocol. A. Western analysis. The captured β-galactosidase was detected using anti-β-galactosidase. Wt HAdV-5 was used as negative control. B. Immunoelectron microscopic analysis of β-galactosidase bound to HAdV-5ΔpIX.CMV.GFP/LUC loaded with pIX.flag.75.13R4.MYC.HIS. The β-galactosidase was only detected on the viruses that carried the 13R4 scFv fused to pIX.
For another approach to study the ligand (β-galactosidase)-binding capability of the 13R4 on the surface of the adenovirus the pIX.flag.75.13R4.MYC.HIS-loaded HAdV-5ΔpIX.CMV.GFP/LUC particles were trapped in DAKO IDEIA microwells and incubated with β-galactosidase. The DAKO kit was developed for demonstration of HAdV in clinical specimens. It contains microwell strips pre-coated with an anti HAdV antibody. After blocking and washing, the particles were exposed to the substrate (ONPG). Compared to wt HAdV-5, the particles that contain the pIX.flag.75.13R4.MYC.HIS molecules showed significant β-galactosidase activity (Figure 4), demonstrating that the 13R4 scFv can bind β-galactosidase on the surface of the virions. From the absorbance value of 0.5, a path length of 0.6 cm, and a molar absorption of ~ 4500 M−1cm−1,25 we can calculate that the concentration of o-nitrophenol formed is 1.85 × 10−4M, with corresponds to 37 nmoles in the sample volume. Since the reaction was developed for one h, this is the equivalent of 37/60 = 0.61 Miller units.25 Since the E.coli β-galactosidase has a specific activity of 58.000 units/nmole of monomer, there is in the well 0.61/58.000 = 1.05 × 10−5 nmoles of monomer, i.e. 2.6 × 10−6 nmoles of β-galactosidase tetramer, or 1.6 × 109 β-galactosidase tetramers. The DAKO wells used to capture the adenoviruses seem to be saturated between 1 × 106 and 5 × 106 viruses (data obtained from Dako Diagnostics; http://www.dako.co.uk/products). This means that about 1.6 ×109/5 × 106= 320 to 1.6 × 109/106 = 1600 β-galactosidase tetramers would be bound to each particle. Since each particle contains 240 pIX molecules in the capsid, this number suggests that the majority of the inserted scFv is active and accessible in the capsid.
Figure 4.
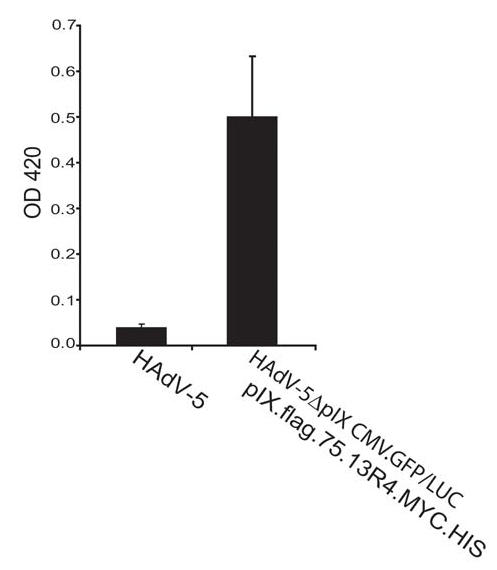
DAKO IDEIA β-galactosidase binding assay. To measure the binding capability of 13R4 on the surface of the virion to its native ligand β-galactosidase, the HAdV-5ΔpIX.CMV.GFP/LUC loaded with pIX.flag.75.13R4.MYC.HIS were incubated with β-galactosidase and trapped on a DAKO IDEIA strips. The bound β-galactosidase was detected by measuring OD420 (left y-asis) after adding ONPG together with the Z-buffer (n=3). As negative control an identical number of wt HAdV-5 particles were used.
Discussion
Here we demonstrate the functional incorporation of a hyper-stable scFv (13R4), directed against β-galactosidase, fused with pIX into the adenovirus capsid. To test whether the pIX.flag.75.13R4.MYC.HIS (51.8 kDa) is able to incorporate into the viral capsid a 911 cell line was created that produced the pIX.flag.75.13R4.MYC.HIS proteins. To this end 911 cells were exposed to the lentiviral vector LV-CMV-pIX.flag.75.13R4.MYC.HIS-IRES-NPTII. The production of pIX.flag.75.13R4.MYC.HIS proteins in the 911 cells was homogeneous as was seen earlier with pIX and pIX.flag.75.MYC proteins.23 Propagation of HAdV-5ΔpIX.CMV.GFP/LUC on the 911- pIX.flag.75.13R4.MYC.HIS cells did result in virus production levels that are similar to the levels obtained after propagation on the standard 911 helper cell line. Western analysis showed that pIX.flag.75.13R4.MYC.HIS was incorporated into the HAdV-5ΔpIX.CMV.GFP/LUC as efficient as wt pIX (14.3 kDa) in wt HAdV-5 particles. Infection experiments using HAdV-5ΔpIX.CMV.GFP/LUC with or without pIX.flag.75.13R4.MYC.HIS on 911 helper cells resulted in similar luciferase levels 24 hours post infection (data not shown). Despite the complete loading, the particles that have the pIX.flag.75.13R4.MYC.HIS incorporated in their capsid are not heat stable. This may be attributed to sterical hindrance, since we have seen similar effects with large pIX-fusion proteins (data not shown).
These data demonstrate that insertions of ligands up to at least 2.5 times the molecular weight of wt pIX can be incorporated into the capsid without decreasing the incorporation efficiency and without impairing the fiber and penton-base mediated internalization. Retaining the capacity of the RGD motif in the penton-base to bind αv integrins may be important for retargeted viruses, and may depend on the ligand used for retargeting.
The 13R4.MYC.HIS fusion protein was located on the surface of the adenovirus capsid as shown by immunoelectronmicroscopy. The number of gold particles on the adenovirus particles appeared to be less then in previous studies using pIX.flag.75.MYC.19, 23 This might be due to the differences between the antibodies used for detection of the epitopes. Furthermore, the structure of the 13R4 scFv could have impaired the accessibility of the C-terminal HIS epitope. Alternatively, the positively charged HIS tag may have associated with the acidic-loop regions of the hexon molecules, making them less accessible to antibodies. Both by Western analysis and by immunoelectron microscopy we demonstrated that HAdV-5ΔpIX.CMV.GFP/LUC particles loaded with pIX.flag.75.13R4.MYC.HIS bind β-galactosidase, whereas wt HAdV-5 does not. The detection of β-galactosidase in both assays was done using the GAL-13 antibody. This antibody does not recognize denatured or reduced β-galactosidase (data from Sigma–Aldrich; http://www.sigmaaldrich.com/sigma/datasheet/g8021dat.pdf). Nevertheless, in our Western analysis we were able to detect β-galactosidase using this antibody. The results of the DAKO IDEIA β-galactosidase binding assay using the HAdV-5ΔpIX.CMV.GFP/LUC particles loaded with pIX.flag.75.13R4.MYC.HIS show binding of native β-galactosidase tetramers. The capacity to bind β-galactosidase was specific for the particles harboring the pIX.flag.75.13R4.MYC.HIS molecules. We estimated that approximately 320 to 1600 β-galactosidase tetramers are bound per virus particle. Since the particle contains 240 pIX molecules in the capsid, this number suggests that the majority of the inserted scFv is active and accessible in the capsid. This is the first time that it has been shown that pIX can be used as anchor to incorporate large targeting ligands such as the model scFv 13R4 into the HAdV-5 capsid.
Unfortunately, the β-galactosidase cannot be used as receptor on cell surfaces. This makes it difficult to formally show that the scFv can mediate adenovirus retargeting. Future studies using for retargeting biological relevant scFv, such as the scFv-αHER226, will show the applicability of these retargeting moieties. Alternatively 13R4 can be used as scaffold for loop grafting.27
Hyper-stable scFv can be produced without the need of stabilizing disulfide bounds. The development of techniques that facilitate the creation of large libraries of hyper-stable scFv will be of great importance for use in therapeutic agents such as gene transfer vectors described in this study. Traditional techniques involve isolation of VH and VL domains to construct scFv from an original hybridoma or in vitro display systems to screen and select for specific scFv that consequently should be tested for their ability to fold into functional scFv in the cytoplasm. Although these techniques have been shown effective for standard scFv, they rarely yield of the hyper-stable scFv variants. Other techniques such as intracellular antigen capturing (IAC) and CDR grafting are promising approaches to create effective scFv that can be used for retargeting of adenoviruses to specific cells or tissues.28–30
The efficient incorporation of the relatively large 13R4.MYC.HIS fusion protein is promising for future retargeting strategies. The fact that the model scFv used in this study is biological active on the surface of the adenovirus is supporting the feasibility of retargeting adenovirus vectors by inserting of scFv that are directed against specific cellular receptors. However, it should be noticed that formal proof of effective retargeting via these pIX modifications still needs to be provided. In this light it is important to be aware that the efficiency of retargeting can depend on the capsid protein to which the scFv is added.18 Therefore it is necessary to compare side by side the retargeting efficiency with a single scFv fused with different capsid proteins (i.e. pIX and fiber).
Methods
Cells
The HAdV-5 El-transformed cell line 91131 was maintained at 37°C in a humidified atmosphere of 5% CO2 in DMEM medium (Gibco-BRL, Breda, The Netherlands) supplemented with 8% fetal bovine serum (Gibco-BRL) and 0.3 % glucose (J.T. Baker, Deventer, The Netherlands). The 911 cells were used to propagate and titer adenovirus vectors. Infections of the cells with HAdVs were carried out in infection medium containing 2% horse serum.
Production of recombinant lentiviruses
The lentiviral vectors used in this study were described in a earlier studie.23 The lentivirus vectors were derived from the plasmid pLV-CMV-eGFP. Plasmid pLV-CMV-pIX.flag.75.13R4.MYC.HIS-IRES-NPTII, has been constructed by standard cloning procedures.23 The gene for pIX.flag.75 was obtained from the pCDNA3.1-based construct pAd5pIX.MYC.flag.75.MYC.19 The gene encoding the scFv 13R4 was subcloned from the plasmid pPM163R4.9 The lentiviral vectors were produced and quantified as described previously.23, 32 For titer estimations is was assumed that 1 ng p24 equals to 2 × 103 transducing units in an infection assay.33
Lentiviral transduction
For transduction, the lentiviral supernatant was added to fresh medium together with 8 μg/ml Polybrene (Sigma Aldrich, Zwijndrecht, The Netherlands). After overnight incubation, the medium was replaced with fresh medium. Cells transduced with lentiviral vectors containing the neomycine selection gene were cultured in medium supplemented with 200 mg.l−1 G418 (Invitrogen, Breda, The Netherlands).
Adenovirus vectors
The HAdV-CMV.GFP/LUC and HAdV-5ΔpIX.CMV.GFP/LUC were made as described previously19. The vectors carry a green fluorescent protein (GFP) and a firefly luciferase (LUC) transgene, each under the control of the human cytomegalovirus (CMV) immediate-early promoter. HAdV-5 was obtained from the virus collection of the Department of Molecular Cell Biology of the Leiden University Medical Center. The concentration of the adenovirus particles was measured by a standard OD260 protocol.34 Heat-inactivation studies of adenoviruses were performed as described previously.
Western analysis
Cell lysates were made in RIPA lysis buffer (50 mM Tris.Cl pH=7.5, 150 mM NaCl, 0.1% SDS, 0.5% DOC and 1% NP40). Protein concentrations were measured via the standard method with the BCA protein assay (Pierce, Perbio Science B.V., Etten-Leur, The Netherlands). Virus lysates were prepared by adding 5×109 virus particles directly to Western sample buffer. The Western blotting and detection procedures have been previously described.19, 23 For detection of β-galactosidase the monoclonal anti-β-galactosidase clone GAL-13 was used (1:2000, Sigma Aldrich, Zwijndrecht, The Netherlands).
Immunohistochemistry assays
For immunohistochemistry assays, the 911-pIX.flag.75.13R4 cells were grown on glass cover slips in 6-wells plates, fixed in methanol, washed with PBS containing 0.05% Tween-20, and incubated with primary antibody, anti-flag (ANTI-flag® M2 Affinity Gel Freezer-Safe; Sigma Aldrich Chemie, Zwijndrecht, The Netherlands) (1:1000 diluted in PBS, 3%BSA) for 60 minutes at room temperature. The cells were washed and incubated with secondary FITC-conjugated goat anti-mouse serum (1:100 diluted in PBS, 3%BSA) for 30 minutes at room temperature. Nuclei were visualized using propidium iodide. Subsequently, the cells were washed and mounted on object glasses using Dabco/Glycerol (Glycerol, 0.02M Tris.Cl pH=8.0 and 1 μg/ml 2.4-diamidino-2-phenylindole). Cells were visualized with a Leica DM-IRBE microscope.
β-galactosidase binding
CsCl purified viruses (1 × 1011 particles per well) were incubated with 100 μg β-galactosidase (Sigma Aldrich, Zwijndrecht, The Netherlands) for 60 minutes on room temperature. Subsequently, the viruses were subjected to a continuous CsCl density gradient similar to the standard CsCl density gradient protocol during adenovirus purification. The purified viruses were further analyzed via Western analysis and immunoelectron microscopy. For the DAKO IDEIA β-galactosidase binding assay purified viruses (5 × 109 particles per well) were trapped together with β-galactosidase (Sigma Aldrich, Zwijndrecht, The Netherlands) on DAKO IDEIA microwell strips (Dako Ltd, Ely, United Kingdom). After 60 minutes incubation on room temperature, the microwell strips were washed 8 times according to the IDEIA kit instructions. After adding 60 μl ONPG (stock was 4 mg/ml in sodium-phosphate buffer, pH7.5) (Sigma Aldrich, Zwijndrecht, The Netherlands) and 60 μl Z-buffer (100 mM NaH2PO4/Na2HPO4pH 7.5; 10 mM KCl; 1 mM MgSO4 and 50 mM 2-mercaptoethanol) the reaction was stopped after 60 minutes by adding 50 μl Na2CO3 (stock was 1 M Na2CO3). The ONPG incubations were performed at 37°C. The conversion of ONPG as a measurement for β-galactosidase activity was determined by assaying the OD420.
Immunoelectron microscopy
The presence of the modified pIX molecules in the viral capsids and the presence of β-galactosidase on the surface of the viral particles was visualized with antisera and gold-labeled protein-A as described.19 For the detection of the HIS epitope the penta-HIS antibody was used (QIAGEN Benelux B.V., Venlo, The Netherlands) (1:200 diluted in PBS-2% BSA). For detection of β-galactosidase the same antibody was used as for Western analysis (1:200 diluted in PBS-2% BSA). Subsequently, these samples were fixed in 1.5% glutaraldehyde in cacodylate buffer and negatively stained with 1% uranyl acetate for 15 min. The viruses were examined with a Philips CM-10 transmission electron microscope at 100 kV.
Acknowledgments
We thank Ronald W.A.L. Limpens (Leiden University Medical Center) for help with immune-affinity electron microscopy, and participants in the GIANT program for stimulating discussions. This work was supported by the Technology Foundation STW (program LGN66.3977), and the European Union through the 6th Framework Program GIANT (Contract no.: 512087).
References
- 1.St George JA. Gene therapy progress and prospects: adenoviral vectors. Gene Ther. 2003;10:1135–1141. doi: 10.1038/sj.gt.3302071. [DOI] [PubMed] [Google Scholar]
- 2.Magnusson MK, Hong SS, Henning P, Boulanger P, Lindholm L. Genetic retargeting of adenovirus vectors: functionality of targeting ligands and their influence on virus viability. J Gene Med. 2002;4:356–370. doi: 10.1002/jgm.285. [DOI] [PubMed] [Google Scholar]
- 3.Biocca S, Ruberti F, Tafani M, Pierandrei-Amaldi P, Cattaneo A. Redox state of single chain Fv fragments targeted to the endoplasmic reticulum, cytosol and mitochondria. Biotechnology (N Y) 1995;13:1110–1115. doi: 10.1038/nbt1095-1110. [DOI] [PubMed] [Google Scholar]
- 4.Hedley SJ, Auf der MA, Hohn S, Escher D, Barberis A, Glasgow JN, et al. An adenovirus vector with a chimeric fiber incorporating stabilized single chain antibody achieves targeted gene delivery. Gene Ther. 2006;13:88–94. doi: 10.1038/sj.gt.3302603. [DOI] [PubMed] [Google Scholar]
- 5.Cattaneo A, Biocca S. The selection of intracellular antibodies. Trends Biotechnol. 1999;17:115–121. doi: 10.1016/s0167-7799(98)01268-2. [DOI] [PubMed] [Google Scholar]
- 6.Wickham TJ. Genetic targeting of adenoviral vectors. In: Curiel DT, Douglas JT, editors. Vector targeting for therapeutic gene delivery. Wiley-Liss; Hoboken: 2002. pp. 143–170. [Google Scholar]
- 7.Curiel DT. Strategies to alter the tropism of adenoviral vectors via genetic capsid modification. In: Curiel DT, Douglas JT, editors. Vector Targeting for Therapeutic Gene Delivery. Wiley-Liss: Hoboken; New Jersey, U.S.A.: 2002. pp. 171–200. [Google Scholar]
- 8.Tavladoraki P, Girotti A, Donini M, Arias FJ, Mancini C, Morea V, et al. A single-chain antibody fragment is functionally expressed in the cytoplasm of both Escherichia coli and transgenic plants. Eur J Biochem. 1999;262:617–624. doi: 10.1046/j.1432-1327.1999.00443.x. [DOI] [PubMed] [Google Scholar]
- 9.Martineau P, Jones P, Winter G. Expression of an antibody fragment at high levels in the bacterial cytoplasm. J Mol Biol. 1998;280:117–127. doi: 10.1006/jmbi.1998.1840. [DOI] [PubMed] [Google Scholar]
- 10.Ohage EC, Wirtz P, Barnikow J, Steipe B. Intrabody construction and expression. II. A synthetic catalytic Fv fragment. J Mol Biol. 1999;291:1129–1134. doi: 10.1006/jmbi.1999.3020. [DOI] [PubMed] [Google Scholar]
- 11.Proba K, Worn A, Honegger A, Pluckthun A. Antibody scFv fragments without disulfide bonds made by molecular evolution. J Mol Biol. 1998;275:245–253. doi: 10.1006/jmbi.1997.1457. [DOI] [PubMed] [Google Scholar]
- 12.Jung S, Honegger A, Pluckthun A. Selection for improved protein stability by phage display. J Mol Biol. 1999;294:163–180. doi: 10.1006/jmbi.1999.3196. [DOI] [PubMed] [Google Scholar]
- 13.Saban SD, Nepomuceno RR, Gritton LD, Nemerow GR, Stewart PL. CryoEM structure at 9A resolution of an adenovirus vector targeted to hematopoietic cells. J Mol Biol. 2005;349:526–537. doi: 10.1016/j.jmb.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 14.Marsh MP, Campos SK, Baker ML, Chen CY, Chiu W, Barry MA. CryoEM of protein IX-modified adenoviruses suggests a new position for the C-terminus of protein IX. J Virol. 2006;80:11881–11886. doi: 10.1128/JVI.01471-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabry CM, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RW, et al. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 2005;24:1645–1654. doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saban SD, Silvestry M, Nemerow GR, Stewart PL. Visualization of α-helices in a 6 Angstrom resolution cryoEM structure of adenovirus allows refinement of capsid protein assignments. J Virol. 2006;80:12049–12059. doi: 10.1128/JVI.01652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vellinga J, van der Heijdt S, Hoeben RC. The adenovirus capsid: major progress in minor proteins. J Gen Virol. 2005;86:1581–1588. doi: 10.1099/vir.0.80877-0. [DOI] [PubMed] [Google Scholar]
- 18.Campos SK, Barry MA. Comparison of adenovirus fiber, protein IX, and hexon capsomeres as scaffolds for vector purification and cell targeting. Virology. 2006;349:453–462. doi: 10.1016/j.virol.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 19.Vellinga J, Rabelink MJ, Cramer SJ, van den Wollenberg DJ, Van der MH, Leppard KN, et al. Spacers increase the accessibility of peptide ligands linked to the carboxyl terminus of adenovirus minor capsid protein IX. J Virol. 2004;78:3470–3479. doi: 10.1128/JVI.78.7.3470-3479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campos SK, Parrott MB, Marsh M, Chiu W, Barry MA. Metabolically biotinylated viruses for vector targetting, virus purification, and capsid imaging. Mol Ther. 2004;9:S390. doi: 10.1016/j.ymthe.2004.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le LP, Everts M, Dmitriev IP, Davydova JG, Yamamoto M, Curiel DT. Fluorescently labeled adenovirus with pIX-EGFP for vector detection. Mol Imaging. 2004;3:105–116. doi: 10.1162/15353500200404100. [DOI] [PubMed] [Google Scholar]
- 22.Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 23.Vellinga J, Uil TG, de Vrij J, Rabelink MJ, Lindholm L, Hoeben RC. A system for efficient generation of adenovirus protein IX-producing helper cell lines. J Gene Med. 2006;8:147–154. doi: 10.1002/jgm.844. [DOI] [PubMed] [Google Scholar]
- 24.Vellinga J, van den Wollenberg DJ, van der Heijdt S, Rabelink MJ, Hoeben RC. The coiled-coil domain of the adenovirus type 5 protein IX is dispensable for capsid incorporation and thermostability. J Virol. 2005;79:3206–3210. doi: 10.1128/JVI.79.5.3206-3210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. [Google Scholar]
- 26.Lombardi A, Sperandei M, Cantale C, Giacomini P, Galeffi P. Functional expression of a single-chain antibody specific for the HER2 human oncogene in a bacterial reducing environment. Protein Expr Purif. 2005;44:10–15. doi: 10.1016/j.pep.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Jung S, Pluckthun A. Improving in vivo folding and stability of a single-chain Fv antibody fragment by loop grafting. Protein Eng. 1997;10:959–966. doi: 10.1093/protein/10.8.959. [DOI] [PubMed] [Google Scholar]
- 28.der Maur AA, Zahnd C, Fischer F, Spinelli S, Honegger A, Cambillau C, et al. Direct in vivo screening of intrabody libraries constructed on a highly stable single-chain framework. J Biol Chem. 2002;277:45075–45085. doi: 10.1074/jbc.M205264200. [DOI] [PubMed] [Google Scholar]
- 29.Ewert S, Honegger A, Pluckthun A. Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods. 2004;34:184–199. doi: 10.1016/j.ymeth.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Worn A, Pluckthun A. Stability engineering of antibody single-chain Fv fragments. J Mol Biol. 2001;305:989–1010. doi: 10.1006/jmbi.2000.4265. [DOI] [PubMed] [Google Scholar]
- 31.Fallaux FJ, Kranenburg O, Cramer SJ, Houweling A, van Ormondt H, Hoeben RC, et al. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum Gene Ther. 1996;7:215–222. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- 32.Carlotti F, Bazuine M, Kekarainen T, Seppen J, Pognonec P, Maassen JA, et al. Lentiviral vectors efficiently transduce quiescent mature 3T3-L1 adipocytes. Mol Ther. 2004;9:209–217. doi: 10.1016/j.ymthe.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 33.Barry SC, Harder B, Brzezinski M, Flint LY, Seppen J, Osborne WR. Lentivirus vectors encoding both central polypurine tract and posttranscriptional regulatory element provide enhanced transduction and transgene expression. Hum Gene Ther. 2001;12:1103–1108. doi: 10.1089/104303401750214311. [DOI] [PubMed] [Google Scholar]
- 34.Mittereder N, March KL, Trapnell BC. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mautino MR, Ramsey WJ, Reiser J, Morgan RA. Modified human immunodeficiency virus-based lentiviral vectors display decreased sensitivity to trans-dominant Rev. Hum Gene Ther. 2000;11:895–908. doi: 10.1089/10430340050015509. [DOI] [PubMed] [Google Scholar]
- 37.Sirven A, Pflumio F, Zennou V, Titeux M, Vainchenker W, Coulombel L, et al. The human immunodeficiency virus type-1 central DNA flap is a crucial determinant for lentiviral vector nuclear import and gene transduction of human hematopoietic stem cells. Blood. 2000;96:4103–4110. [PubMed] [Google Scholar]
- 38.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 39.Swick AG, Janicot M, Cheneval-Kastelic T, McLenithan JC, Lane MD. Promoter-cDNA-directed heterologous protein expression in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 1992;89:1812–1816. doi: 10.1073/pnas.89.5.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]


