Abstract
Nasopharyngeal carcinoma (NPC) is a rare malignancy in most parts of the world, but is one of the most common cancers in Southeast Asia. Both genetic and environmental factors contribute to the tumorigenesis of NPC, most notably the consumption of certain salted food items and Epstein-Barr virus infection. This review will focus on the current progress of the genetic analysis of NPC (genetic susceptibilities and somatic alterations). We will review the current advances in genomic technologies and their shaping of the future direction of NPC research.
1. INTRODUCTION TO NPC
The nasopharyngeal carcinoma (NPC) is a malignancy of the head and neck region that arises from the epithelial cells that cover the surface and line the nasopharynx. This disease was initially reported in 1901, and characterized clinically in 1922 [1]. It is a rare malignancy in the United States, accounting for 2% of all head and neck squamous cell carcinomas, with an incidence of 0.5 to 2 per 100,000. However, it is endemic in many geographical regions, including Southern China and Southeast Asia, where the observed incidence rates range from 15 and 50 per 100,000 persons. An intermediate incidence has been reported in Alaskan Eskimos and in the Mediterranean basin (North Africa, Southern Italy, Greece, and Turkey), ranging from 15 to 20 cases per 100,000 persons [2]. A male preponderance exists; with a male-to-female ratio of approximately . Overall, NPC can occur in all age groups, but has a bimodal age distribution. The incidence peaks at 50 to 60 years of age; and a small peak is observed during late childhood [3].
1.1. Anatomy
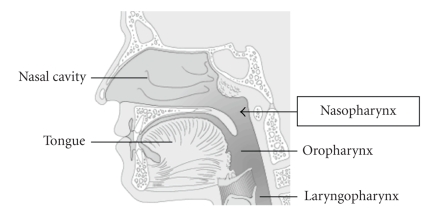
The nasopharynx (the upper part of the throat, behind the nose) is a cuboidal chamber (about 1.5 inches on each edge) located posterior to the nasal choanae (see Figure 1). It is bounded superiorly by the clivus, and inferiorly by the lower border of the soft palate. The posterior border is made up by the mucosa that overlies the superior constrictor muscles of the pharynx and the prevertebral fascia of the C1 and C2 vertebral bodies. Its lateral walls contain the Eustachian tubes' orifices. The fossa of Rosenmüller represents the most common site of origin for NPC [4].
Figure 1.
Anatomic site of NPC.
1.2. Epidemiology
In endemic regions, NPC presents as a complex disease caused by an interaction of the oncogenic gamma-herpesvirus Epstein-Barr virus (EBV) chronic infection, environmental, and genetic factors, in a multistep carcinogenic process. The EBV is spread worldwide, infecting over 95% of the adult population [5]. It is transmitted by saliva and its primary infection occurs during childhood with replication of the virus in the oropharyngeal lining cells, followed by a latent infection of B lymphocytes (primary target of the EBV). Although the infection is typically subclinical, the virus is associated with later development of several malignancies, including NPC [6]. Elevated titers of EBV associated antigens (especially of IgA class), a latent EBV infection identified in neoplastic cells of virtually all cases of NPC, and the clonal EBV genome consistently detected in invasive carcinomas and high-grade dysplastic lesions suggest a critical role of EBV in the pathogenesis of NPC in endemic areas.
Significant environmental factors contribute to NPC include the consumption of foods high in salt, exposure to nitrosamines and polycyclic hydrocarbons as important carcinogens. In nonendemic areas, the association of NPC with alcohol and tobacco use has been reported, either as weak or controversial in some series [3, 7].
Genetic studies of endemic populations revealed the association of HLA antigen haplotype with NPC: HLA-2, HLA-B17, and HLA-Bw26 double the risk of the disease, and genomic and cytogenetic studies have shown multiple aberrations in chromosomes 1, 3, 9, 11, 12, and 14. These genetic factors will be discussed in association with the current advances in genomic technologies in the following sections.
1.3. Sign and symptoms
NPC may easily escape diagnosis at early stages, and most of the cases remain undiagnosed until they present as a metastasis to the lymph nodes of the neck. The tumor is difficult to diagnose for multiple reasons including the nonspecificity of the initial symptoms and the difficulty of examining the postnasal space. Additionally, lesions can grow within the submucosa of the nasopharynx and escape endoscopic visualization [8–10]. The majority of tumors arise in the lateral walls, especially from the fossa of Rosenmuller and Eustachian tube cushions. Tumors can grow within the nasopharynx or extend to the opposite lateral wall; they can also infiltrate other structures toward the base of the skull, and invade the palate, nasal cavity, or the oropharynx. The most common presenting symptom is a painless cervical lymph node enlargement due metastasis, followed by nasal, aural, and neurological symptoms. A unilateral neck mass is reported in about 36% of cases, but other series report rates as high as 80% [3]. Only 5% of cases reported in Southern China present with distant metastases [2]. Enlargement and extension of the tumor within the nasopharynx may cause nasal obstruction-related symptoms such as congestion, nasal discharge, and bleeding. Blockage of the Eustachian tube and/or extension into the ear may result in changes in hearing or hearing loss (usually unilateral). Extension of the tumor into the base of the skull is usually associated with cranial nerve deficits. The most common distal metastatic sites are bone, lung, mediastinum, and more rarely liver [11]. Symptoms related with the distal metastatic disease include bone pain or organ dysfunction.
1.4. Pathology
With the constant advance in our understanding of this disease, the pathohistological classification of NPC has been evolving continuously. In 1978, the histological classification guideline proposed by the World Health Organization (WHO) categorized NPC into three groups: type 1 (keratinizing squamous cell carcinoma), type 2 (nonkeratinizing carcinoma), and type 3 (undifferentiated carcinoma). Types 2 and 3 have also been called lymphoepithelioma [1, 3]. The 1991 WHO classification of nasopharyngeal carcinomas divided them into two groups: squamous cell carcinoma (keratinizing squamous cell carcinoma, type 1 of the former classification), and nonkeratinizing carcinoma (types 2 and 3 of the former classification combined under a single category). The second group (nonkeratinizing carcinoma) was further subdivided into differentiated and undifferentiated carcinomas. Lymphoepithelioma-like carcinoma was considered a morphologic variant of undifferentiated carcinoma [1]. The current WHO classification keeps the 1991 terminology, and adds one additional category: basaloid squamous cell carcinoma [1, 12].
Published data indicate a probably higher proportion of keratinizing squamous cell carcinoma among all NPC in nonendemic areas compared with endemic areas. Some studies reported that squamous cell carcinoma (former WHO type 1) accounts for approximately 25% of all NPC in North America, but only 1% in endemic areas; whereas undifferentiated carcinoma (former WHO type 3) accounts for 95% of all cases in high incidence areas, but 60% of cases in North America [1, 3, 12].
1.5. Staging
The extent of the disease is the most important prognostic factor, and staging will have a great impact on the selection of treatment in patients with NPC [1]. The tumor-node-metastasis (TNM) staging system, promulgated by the American Joint Comittee on Cancer (AJCC), is the most frequent system used to classify the extent of spread of nasopharyngeal carcinomas [13]. Information about the tumor, lymph nodes, and metastasis is combined according to a process called stage grouping. Each set use Roman numerals O to IV to describe progression from earliest to most advanced stage. Therefore, according to this system, patients are designated into stages 0, I, IIA, IIB, III, IVA, IVB, and IVC [12, 13].
1.6. Diagnosis
NPC shows an extraordinarily high cure rate for early stage disease, thus early detection is critical to improve the overall prognosis and reduce morbidity and metastasis [10]. The detection of NPC is based on the clinical history and the physical examination, but a definitive diagnosis requires a biopsy of the lesion [3]. A series of radiologic tests, including a computed tomography (CT) scans with intravenous contrast and magnetic resonance imaging (MRI) of the head and neck are currently being used to assess the tumor extension and the stage of the disease [11].
EBV-related antigens in sera are also useful markers for NPC diagnosis [7, 14]. Ho et al. found an increased diagnostic sensitivity and specificity (99% and 96%, resp.) using a combination of serum protein profiles with an EBV antibody serology test [15]. A clinical history of a known metastasis but an unknown site of primary tumor with a positive serology for EBV may also help in diagnosis, redirecting the search for a primary disease at the nasopharynx. The plasma Epstein-Barr virus DNA (EBV-DNA) level has also been suggested to be a reliable indicator for staging and prognosis of NPC [16]. The EBV infection can also be detected by immunostaining of tumoral cells for latent membrane protein 1 (LMP-1), and/or in situ hybridization for EBV-encoded RNAs. Results using these techniques on paraffin-embedded tissue sections support the evidence that EBV plays a major role in the pathogenesis of the disease [17].
1.7. Treatment
External radiotherapy alone is still the primary treatment for early stage NPC. Concomitant chemoradiotherapy has been used in recent years for locally advanced disease. The management of recurrent cervical lymph node metastases in NPC after radiation and chemotherapy is a radical surgery of the lymph nodes of the neck with postoperative brachytherapy. The overall 5-year survival rate for patients with locally advanced disease is around 55–60%. The salvage surgical procedure for persistent or recurrent neck disease shows a 5-year control rate of 66% and a 5-year actuarial survival of 38% [18, 19].
2. GENETIC ANALYSES OF NPC
While nasopharyngeal carcinoma is a rare malignancy in most parts of the world, it is one of the most common cancers in Southeast Asia including areas such as southern China, Hong Kong, Singapore, Malaysia, and Taiwan. The reported incidence in these countries ranges from 10 to 53 cases per 100,000 persons. The incidence is also high among Eskimos in Alaska and Greenland and in Tunisians, ranging from 15 to 20 cases per persons [2]. A clear and specific etiology for NPC is still lacking. In general, NPC is thought to be the result of both genetic susceptibility and environmental factors, such as consumption of certain salted food items [20] and infection with EBV [21]. Familial clustering of NPC has been widely observed in both the Chinese population [22, 23], and non-Chinese patient cohort [24]. The familial risk of NPC is among the highest of any malignancy [25]. The described relative risk of NPC in first-degree relatives is about 8.0 [26, 27]. In this article, we will review the current progress on genetic analysis of nasopharyngeal carcinoma (e.g., genetic susceptibilities and somatic alterations) in relationship with recent advances in genomic technologies.
2.1. Progress on searching for genetic susceptibilities of NPC
Although perhaps not Mendelian, strong evidences suggest that genetic factors play important roles in NPC. Epidemiological studies suggest that most of the familial aggregation of NPC derives from inherited susceptibility [2]. A recent complex segregation analysis on a Chinese cohort provided additional evidence to support a multifactorial mode of inheritance for NPC [28]. However, the molecular genetic basis of NPC remains unknown. Most of the studies searching for the susceptibility genes of NPC can be loosely categorized into 2 methodologies: a positional cloning approach and a functional cloning approach. A positional cloning approach aims first to identify the genomic location (or locus) that is linked to the disease. This is followed by the identification of the disease gene (or susceptibility gene) at this particular genomic location. The functional cloning approach, also known as candidate gene-based approach, requires sufficient prior knowledge of the disease and the functional defect(s) associated with the disease. Candidate gene(s) are identified based on this knowledge. Mutations (or polymorphisms) will then be identified and investigated in the candidate gene. These approaches complement each other. Positional cloning approaches can lead to identification of a candidate gene for functional cloning studies. On the other hand, a functional cloning approach often confirms the genomic location of the susceptibility locus identified by positional cloning studies. The following sections will summarize the progress of identifying the NPC susceptibility genes based on these approaches.
2.1.1. Positional cloning approach searching for NPC susceptibility genes
Linkage analyses are the most common approaches for the identification of a disease locus (or susceptibility locus). There are several variations of linkage analysis design, based on the pedigree structure. The linkage studies usually involve genotyping of both affected individuals and healthy family members using a panel of genetic markers. Most of the linkage studies on NPC performed so far have used microsatellite markers that are essentially polymorphic tandem repeats of di- to tetranucleotide sequence motifs flanked by unique sequences. This approach is usually tedious, labor-intensive, and requires large amounts of sample DNA, allowing only a modest number of markers to be screened. However, the recent completion of the human genome project has lead to the identification of millions of single nucleotide polymorphisms (SNP), the most abundant type of polymorphism in the human genome, which will lead to another wave of intense search for the NPC susceptibility locus/gene.
Several linkage analyses studies suggested the association of susceptibility HLA haplotypes with NPC development. Most studies conducted among the Chinese population demonstrated an increased risk of NPC for individuals with HLA-A2. A recent study detected a consistent association between NPC and the prevalent Chinese HLA-A2 subtype (HLA-A*0207), but not the prevalent Caucasian subtype (HLA-A*0201) [29]. The HLA types of AW19, BW46, and B17 have also been reported to be associated with an increased risk, whereas HLA-A11 is associated with a decreased risk [30]. The involvement of HLA in NPC tumorigenesis may be through its cytotoxic T cell recognition and host immune response to EBV infection. However, it has been suspected that HLA alleles may not directly contribute to the susceptibility of NPC. Interestingly, Lu et al. (1990) reported a linkage study based on affected sib pairs which suggested that a gene closely linked to the major histocompatibility complex (MHC) region but distinct from the HLA genes confers a greatly increased risk of nasopharyngeal carcinoma [31].
A recent study provides evidence for the linkage of NPC to chromosome 3p and a fine map of NPC susceptibility locus to a 13.6-cM region on 3p21.31-21.2 [32]. These results are in agreement with several previous studies that suggest that the deletion of chromosomes 3p is a common genetic event in NPC [33, 34]. Many tumor suppressor candidate genes such as CACNA2D2, DLC1, FUS1, H37, HYAL1, RASSF1A, SEMA3B, and SEMA3F and tumor susceptibility genes such as hMLH1 have been isolated from the region [32]. These studies indicate that genes in the 3p21 may play a critical role in tumorigenesis of familial NPC. Consistent with this notion, another study detected a high frequency of loss of heterozygosity on 3p, in histologically normal nasopharyngeal epithelia and dysplastic lesions from Southern Chinese individuals, suggesting that the genetic abnormality appear to be causative for NPC [35]. Isolation and identification of susceptibility genes from 3p21 may greatly advance the understanding of the etiology and development of NPC.
A recent genome-wide scanning of 20 families with included 65 affected individuals provides evidence of a major susceptibility locus for NPC on chromosome 4p15.1-q12 [36]. The strongest linkage was observed with marker D4S405 (LOD score = 3.54) and D4S3002 (LOD score = 4.2). Interestingly, when EBV antibody titer was included as a covariate, the LOD scores reached 4.70 and 5.36 for these markers, respectively. This observation was recently confirmed by a population-based large-scale study of Han Chinese from Guangxi province using 34 microsatellites spanning an 18-megabase region of chromosome 4 (4p15.1-q12) [37].
2.1.2. Functional cloning approach searching for NPC susceptibility genes
Recent studies suggested that genetic polymorphisms in genes that metabolize carcinogens are associated with NPC susceptibility. Cytochrome P450 2E1 (CYP2E1) is one of the cytochrome P450s and is responsible for the metabolic activation of nitrosamines and the related carcinogens. The variant form of CYP2E1 has a marked difference in its activity and causes different levels of DNA damage in human cells. Nitrosamines are the effective carcinogens for NPC and are believed to be involved in the pathogenesis of NPC. Case-control studies have shown a strong association of the variant form of CYP2E1 (c2 allele) with increased risk of this disease in Chinese populations [38, 39]. Other nitrosamine metabolizing genes, such as Cytochrome P450 2A6 (CYP2A6), have also been suggested to play a role in NPC susceptibility [40].
Genetic polymorphism of glutathione S-transferase M1 (GSTM1) is a phase II enzyme known to play an important role in the detoxification of several carcinogens found in tobacco smoke, a synergistic risk factor for NPC [41]. This enzyme also modulates the induction of other enzymes and proteins that are important for cellular functions, such as DNA repair. The enzyme is therefore important to metabolize carcinogens, maintaining genomic integrity and cancer susceptibility. A recent study in the United States has reported that GSTM1 null genotype is associated with an almost twofold increase in risk for NPC [42]. The findings implied that polymorphisms of this modifier might lead to different cellular responses to environmental carcinogens among different individuals, different degrees of genetic instability or damages in the nasopharyngeal epithelial cells. Similar associations were observed in studies on Tunisian and Thai populations [43, 44].
The association of other DNA repair genes with NPC susceptibility has also been implied. Both X-ray repair cross-complementing group 1 gene (XRCC1) and 8-oxoguanine glycosylase 1 (hOGG1) are important in DNA base excision repair. While a reduced risk for NPC was observed with polymorphism of the XRCC1 gene (Arg280His), polymorphism of the hOGG1 gene (Ser326Cys) was shown to be associated with an increased risk for NPC in the Taiwan population [45]. The reduced risk of NPC associated with polymorphism in the XRCC1 gene was confirmed with a different polymorphism (Arg194Trp) recently identified in the population from Guangdong, China, particularly in males and smokers [46]. Interestingly, the higher risk of NPC was observed among those subjects with certain combined genotypes for both hOGG1 and XRCC1 polymorphisms [45], clearly suggesting that carriers of multiple putative high-risk genotypes have the highest risk of developing NPC.
The potential roles of genes that contribute to the immune response have also been studied. Signaling pathways activated by the toll-like receptor 4 (TLR4) involve the induction of anticancer immunity. Functional analyses of an SNP variant of the TLR4 gene at the -untranslated region (-UTR) suggested that it is associated with decreased mRNA stability, and leads to a reduced expression of this gene [47]. This -UTR polymorphism has been shown to be associated with a significantly increased risk for NPC. It is hypothesized that this polymorphism downregulates TLR4 expression through destabilizing the mRNA, and leads in EBV metainfective antiviral immunologic deficits and a high risk of NPC. Similarly, associations with increased risks for NPC have also been detected with polymorphism in toll-like receptor 1, 6, 10, respectively [23, 48].
The palate, lung, and nasal epithelium carcinoma-associated (PLUNC) protein gene plays a role in the innate immune response in the regions of the oral and nasal cavities. In a recent case-controlled study of Chinese population composed of 239 unrelated NPC patients and 286 healthy controls, SNPs in the promoter region of this gene (PLUNC) were significantly associated with susceptibility to NPC, [49]. These results suggest that genetic variation in PLUNC may influence susceptibility to NPC in this Chinese population.
Tremendous enthusiasm in the genetics community has been generated for the identification of millions of polymorphisms (e.g., SNPs) throughout the human genome. Recently, an increasing number of studies have been devoted to investigate the polymorphisms in a variety of cancer-related genes for their potential influence on NPC susceptibility, including matrix metalloproteinases (MMPs) [50, 51], transforming growth factor-beta1 (TGF-beta1) [52], interleukin-10 (IL-10) [53], antigen processing 1 gene (TAP1) [54], p53 [55], cyclin D1 (CCND1) [56], FAS (CD95) [57], mouse double minute 2 (MDM2) [58], and Nedd4 binding protein 2 (N4BP2) [59]. While polymorphisms in these genes have been associated with a statistically significantly increased risk of NPC, the risks are generally small and appear to be restricted to specific studies. It is apparent that the understanding of interactions of these polymorphisms and other risk factors are more important. With the continuous advances in high-throughput sequence and genotyping technologies, this list will increase rapidly.
SNPs appear to be the most abundant sequence variations between individuals. Enthusiasm for very high density SNP sets in the human genome has been largely centered on the potential use for association studies, especially in the context of measured linkage disequilibrium. Indeed, successful implementations using genome-wide association analysis have already been reported for cancer risks [60, 61]. A recent milestone publication by the Welcome Trust Case Control Consortium [62] established the “standard” for genome-wide association analysis, in term of result interpretation, quality control, population stratification, and control sample sharing. These advances in analytical approaches, together with the advent of rapid, affordable, large-scale genotyping methods that enable the cotyping of over 500,000 SNPs on each genomic sample (http://www.affymetrix.com), greatly facilitate the search for new susceptibility genes of NPC, and will lead to a better understanding of the potential interactions among susceptibility genes and between susceptibility genes and environmental factors.
2.2. Progress on profiling somatic abnormalities of the NPC genome
Tumors develop through the combined processes of genetic instability and selection, resulting in clonal expansion of cells that have accumulated the most advantageous set of genetic aberrations. Many types of instability can contribute to neoplastic development, including point mutations, chromosomal rearrangements, DNA dosage abnormalities (amplifications or deletions), alteration of microsatellite sequences, and epigenetic changes. Knowledge of genomic aberrations can have clinical implications in diagnosis, treatment, and prognostics of cancer. Four decades ago, the milestone discovery of Philadelphia chromosome (a translocation between chromosome 9 and 22, which fuses the Bcr gene and the Abl tyrosine kinase gene) [63] led to one of the first effective targeted therapies for cancer: treatment of chronic myelogenous leukemia (CML) with the tyrosine kinase inhibitor imatinib (Gleevec). Since then, many exciting clinical advances have been made based on the increasing knowledge of the tumor genome.
During the 1970s and 1980s, several genome-wide approaches were developed to measure these tumor genomic alterations including loss of heterozygosity analysis (LOH) and comparative genomic hybridization (CGH). Advances in genetics and bioengineering have refined these techniques over the past two decades, and the recent development of multicolor staining-based cytogenetic techniques such as multicolor fluorescence in situ hybridization (M-FISH) and spectral karyotyping (SKY) have further improved the ability to analyze the tumor genome [64]. The completion of the human genome project [65, 66] now makes it possible to query the cancer genome systematically in ways that were hitherto impossible. Microarrays designed to analyze targeted genomic regions relevant to chronic lymphocytic leukemia have been produced for use in clinical trials to determine the relationship between therapeutic options and genomic aberrations [67]. Association of genomic aberrations with prognosis has been found for a variety of tumor types, including prostate cancer [68], breast cancer [69], gastric cancer [70], head and neck cancer [71], lymphoma [72, 73], and NPC [74].
2.2.1. Progress on genomic profiling of NPC
Copy number analysis of NPC —
Comparative genomic hybridization (CGH) was developed to survey gene copy number abnormalities (amplifications and deletions) across a whole genome [75]. In a typical CGH analysis, fluorescently labeled disease DNA (frequently Fluorescein or FITC) and normal DNA (frequently Rhodamine or Texas Red) are cohybridized to the normal metaphase chromosomes to generate fluorescence ratios along the length of chromosomes that provide a cytogenetic representation of DNA copy number variation. CGH was the first effective approach to scanning the entire genome for variations in DNA content [76, 77]. A large number of CGH-based studies on NPC lead to the identification of consistent gain at chromosome 1q, 3q, 8q, 12 and loss at 3p, 9p, 11q, 14q [78–81]. A recent large-scale meta-analysis of CGH results revealed several genomic “hotspots” that show consistent copy number alterations in NPC [82]. These findings provided foundation for further identifications of the corresponding oncogenes and tumor suppressor genes in NPC.
While chromosome-based CGH provided critical hints for identifying candidate genes for NPC, it has a limited mapping resolution (20 Mb). Array-based CGH is a second-generation approach in which fluorescence ratios on microarrayed DNA elements provide a locus-by-locus measure of gene copy number variation [83, 84]. Using this approach, frequent amplifications were detected for several oncogene loci, including MYCL1 at 1p34.3 (66.7%), TERC at 3q26.3 (46.7%), ESR at 6q25.1 (46.7%), and PIK3CA at 3q26.3 (40%) [85].
Although the array-based CGH can potentially increase mapping resolution, most of the early arrays used for the CGH studies have utilized large genomic clones, for example, bacterial artificial chromosomes (BACs), which have a limited spatial sensitivity. In addition, large genomic clones also suffer from reduced specificity due to their inclusion of common repeats (e.g., Alu and long interspersed nuclear elements (LINEs)), redundant sequences (e.g., low copy repeats (LCRs), also known as segmental duplications), and segments of extensive sequence similarity (pseudogenes or paralogous genes) [86]. Recently, several additional higher-density tools for CGH analysis have become available with the completion of the human genome sequence. These include cDNA array-based CGH [87, 88], oligonucleotide array-based CGH [89, 90], tiling array-based CGH [84], and copy number analysis using high-density SNP microarrays [91–94]. Tiling and SNP array-based approaches have drawn most attention due to their high resolution. Tiling arrays have the potential to resolve small (gene level) gains and losses (resolution 40 kb) that might be missed by marker-based genomic arrays which contain large number of gaps due to the distance between the targeted probes [84, 95]. We can envision that in the near future, we will have the ability to survey copy number changes at close to bp resolution using tiling arrays that contain billions of overlapping probes covering the entire genome. The SNP array-based approach provides the unique advantage of concurrent CGH and LOH analysis, which we discuss in further detail below [92, 93].
Loss of heterozygosity (LOH) analysis of NPC —
Chromosomal aberrations include segments of allelic imbalance identifiable by loss of heterozygosity (LOH) at polymorphic loci, which can be used to identify regions harboring tumor suppressor genes. Allelic losses, which are caused by mitotic recombination, gene conversion, or nondisjunction cannot be detected by CGH and thus require LOH analysis for their identification. This approach is “favored” by the Knudson two-hit hypothesis [96, 97] for hunting the tumor-suppressor genes. Traditionally, polymorphic markers, such as restriction fragment length polymorphisms (RFLPs) and microsatellite markers, have been used to detect LOH through allelotypic comparisons of DNA from a cancer sample and a matched normal sample [98]. However, this approach is time consuming, and labor intensive, and requires a large amount of sample DNA, allowing only a modest number of markers to be screened. Most of the early LOH studies were focused on individual chromosomes, and only a few genome-wide LOH studies have been performed on NPC [34, 99–101]. The most frequent LOH were observed at chromosome 3p, 9p, and 14q, which is in agreement with the CGH based findings.
The mapping of the human genome has allowed for the identification of millions of SNP loci (http://www.ncbi.nlm.nih.gov/SNP), which makes them ideal markers for various genetic analyses, including LOH. Because of their abundance, even spacing, and stability across the genome, SNPs have significant advantages over RFLPs and microsatellite markers as a basis for high-resolution whole genome allelotyping with accurate copy number measurements. High-density oligonucleotide arrays have recently been generated to support large-scale high throughput SNP analysis [102]. It is now possible to genotype over 500,000 SNP markers using the Affymetrix Mapping 500K SNP oligonucleotide array. LOH patterns generated by SNP array analysis have a high degree of concordance with previous microsatellite analyses on the same cancer samples [103]. Additionally, shared regions of LOH from SNP arrays can cluster lung cancer samples into subtypes [104], and distinct patterns of LOH are found to associate with specific clinical features in primary breast, bladder, head and neck, and prostate tumors [93, 105–108]. While SNP array has not been utilized in NPC studies, it is expected that large scale SNP array-based LOH profiles will be generated on NPC in the near future. It is worth noting that a high-density SNP array is also a very powerful tool for identifying susceptibility gene(s) using either linkage or association study designs. One might envision that with a single high-throughput genomic platform, large-scale population-based study, searching for genetic susceptibility of NPC (inherited risk factors) can be performed concurrently with genomic profiling of NPC (somatic mutations).
Cytogenetic analysis of NPC —
Cytogenetics has be widely used since the introduction of chromosome-banding techniques (keryotyping) in 1969 [109, 110]. One major drawback of these approaches is the requirement of in vitro culture and metaphase preparation of the cells of interest. Due to the poor tumor growth in vitro, only a limited number of karyotyping-based studies have been performed on primary NPC, which suggested genomic aberrations of 3q and 5q [111, 112]. Nevertheless, cytogenetic approaches will always have their place in the genomic profiling due to the ability to directly visualize chromosomal abnormalities. To obtain the cytogenetic information, cell lines and xenografts have been used frequently for the karyotyping studies on NPC, where many structural and numerical alterations found on 1p, 3p, 3q, 5q, 9p, 12, 11q, 13q, 14q, 16q, and X [113–118]. Among these alterations, deletion of 3p and gain of 3q are the most frequent events [119, 120]. More importantly, these cytogenetic techniques complement CGH and LOH by providing information on chromosomal structural rearrangements that are not resolved by DNA copy number analyses. For example, balanced translocations are one of the more common genomic abnormalities in cancer [121], but they cannot be detected by CGH or LOH. An experienced cytogeneticist, however, can readily detect many forms of chromosomal rearrangements of NPC using classical cytogenetic techniques, such as karyotyping [122].
The advances in the labeling techniques lead to the development of fluorescence in situ hybridization (FISH) method, which has proven to be an excellent choice for independent validation of other genomic methods. Fan et al. [123] reported FISH-based studies to validate the frequent amplification of c-myc and Int-2 that was initially discovered by CGH analysis. Recently, with the introduction of several new labeling techniques, such as spectral karyotyping (SKY), multicolor FISH (M-FISH), cross-species color banding (Rx-FISH), and multicolor chromosome banding, it is possible to carry out discovery studies using the cytogenetic methods. These techniques permit the simultaneous visualization of all chromosomes in different colors, and thus considerably improve the detection of translocations or deletions. For example, both SKY and M-FISH use a combinatorial labeling scheme with spectrally distinguishable fluorochromes. The chromosome-specific probe pools (chromosome painting probes) are generated from flow-sorted chromosomes and then amplified and fluorescently labeled by degenerate oligonucleotide-primed polymerase chain reaction. With the introduction of these techniques in 1996 [124–126], the comprehensive analysis of complex chromosomal rearrangements present in tumor karyotypes was greatly improved. A recent SKY analysis on NPC cell lines confirmed most of the abnormalities identified previously by CGH and LOH and illustrated additional breakpoints on a number of apparently balanced chromosomes, including 3p21, 3q26, 5q31, 6p21-p25, 7p14-p22 and 8q22 [127].
2.2.2. Genome-wide expressional microarray analysis of NPC
The use of microarray and other global profiling technologies has led to a significant number of exciting new biological discoveries, and important correlations between gene-expression patterns and disease states. Never before could a small sample of RNA from two different conditions reveal so much information at the transcriptional level. Microarray-based expression profiling on tumor tissues have been used to identify molecular signatures that can promote the precise classification and prognostication of various types of cancers. Historically, only a few expression profiling analysis studies have been performed on NPC [128–130]. The limited amount of available clinical materials and heavy infiltration of non-cancer cells present major difficulties for these studies. With advances in preamplification technologies and microdissection tools, comprehensive expression profiling of NPC is possible. In the past couple of years, several genome-wide expression profiling studies have been devoted to identify candidate genes (e.g., genes involved in regulations of Ras activity, cell cycle, and WNT pathway) [131, 132], investigate the disease etiology (e.g., EBV infection, host responses, and hypoxia) [133–135], and evaluate the therapeutic effectiveness on NPC [136]. With the continuation of advances in genome-wide expressional microarray technology, comprehensive expression profiling of NPC is now starting to take the center stage. This should lead to substantial translational outcomes that will advance the management of this disease.
2.2.3. Comprehensive genomic approaches
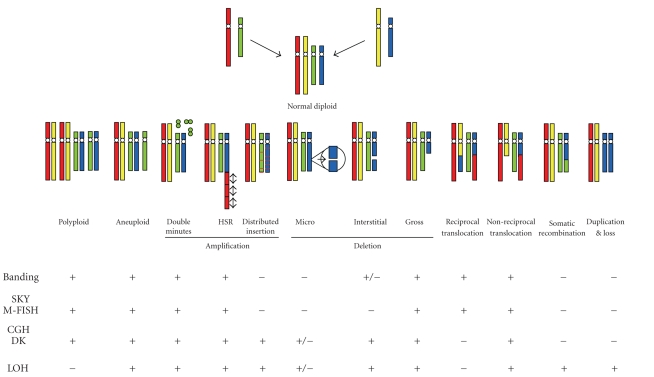
A major challenge confronting the identification of the molecular genetic factors that contribute to the NPC tumorigenesis is the diversity of the genetic alterations. Among these are germline variations (such as the susceptibility genes described in previous section) that lead to hereditary cancer predispositions, the acquisition of transforming DNA or RNA sequences from cancer viruses (e.g., EBV for NPC), somatic mutations in the cancer genome (e.g., copy number change, translocation, LOH), and epigenetic mechanisms (such as DNA methylation or histone modification) that promote oncogenesis by modifying cancer-related genes. Somatic genomic alterations such as point mutations, genomic amplifications or deletions, loss of allelic heterozygosity, and chromosomal translocations are believed to play a central role in the development of most solid tumors, including NPC. All of these mechanisms result in dysregulated expression of oncogenes and tumor suppressor genes, but none of the existing genomic techniques can capture all of these genetic changes in a single analysis (see Figure 2). This represents a major obstacle to the comprehensive analysis of tumor genomes and their relationship to clinical phenotypes or disease progression.
Figure 2.
Identification of chromosomal abnormalities using various genomic and cytogenetic approaches. “+” and “−” denote effectiveness and ineffectiveness of the methods for the detection of a specific chromosomal abnormality. Banding: chromosome banding or karyotyping analysis; SKY: spectral karyotyping analysis; M-FISH: multicolor fluorescence in situ hybridization; CGH: comparative genomic hybridization; DK: digital karyotyping analysis; LOH: loss of heterozygosity. Adapted from [137] with kind permission of Future Drugs Ltd.
A more practical approach to overcome this problem is to combine a selective set of molecular genetic technologies such as CGH, LOH, and various molecular cytogenetic analyses for comprehensive screening of genomic alterations with high resolution. Each of these techniques has their own unique advantages, but they also have their own limitations which have motivated efforts to combine these approaches as shown in Figure 2. In this instance, the SNP array-based LOH and CGH analyses provide a high-resolution mapping of copy number abnormalities, but offer little information on chromosomal structure/spatial changes (e.g., translocations, the most common class of somatic mutation registered in the cancer gene census [121]). On the other hand, modern cytogenetic techniques provide a clear picture of the gross chromosomal structure/spatial alterations, but have limited resolution. Therefore, strategically combining a complementary set of genetic tests is a logical approach for characterizing a complex cancer genome. This has been successfully attempted to investigate the immortalization of nasopharyngeal epithelial cells [138], where karyotyping, spectral karyotyping (SKY) and array CGH were utilized concurrently to reveal a gain of 17q21–q25 fragment on 11p15 chromosome, with the specific derivative chromosome 11: der(11)t(11;17)(p15.1;q21.1).
This multimodal approach can be extended to combine DNA structural analyses with additional genome functional activity at the RNA and/or protein levels. Recent technical advances in microarray-based gene expression analysis have offered substantial improvement in diagnosis, treatment, and prognosis of cancer patients. This continuous progress in microarray-based expression analysis and the large public depositories of microarray data have motivated new efforts to extract additional biological information from these data in addition to the static RNA transcript levels. One such attempt involves inferring the chromosomal structural changes from spatially-linked changes in microarray expression data [139–142]. Several array CGH studies have shown a genome-wide correlation of gene expression with copy number alterations and have proved useful in individual amplicon refinement [143, 144]. For example, through tissue microarray FISH and RT-PCR, a minimally amplified region around ERBB2 was identified in a large number of breast tumors. In addition, gene amplification was found to be correlated with increased gene expression in a subset of those samples [145]. Recently, several groups have observed that chromosomal alterations can lead to regional gene expression biases in human tumors and tumor-derived cell lines [139–141, 146, 147]. A recent study also demonstrated the correlation between SNP array-based LOH profiles and expression profiles [105]. These studies suggest that a fraction of gene expression values (15–25%) are regulated in concordance with chromosomal DNA content [139–141, 146, 147]. Several statistical methods have been developed and have shown promising results for detecting DNA copy number abnormalities based on differential gene expression [139–142]. With the recent growth in transcriptomic profiling studies of NPC, these techniques for “reverse inference” of DNA alterations from RNA expression data will become a valuable approach for genomic profiling that can provide cross-validation of functional genomic alterations at multiple biological levels when combined with DNA-based approaches such as CGH and LOH. These attempts for strategic integration of genomic information at multiple levels provide an exciting paradigm to introduce the system biology (or more specifically system genomics) concept into NPC research. Further strategies for implementing a comprehensive database that contains additional levels of genomic information such as alternative splicing and methylation status have also been suggested.
3. FUTURE DIRECTIONS
The high susceptibility of individuals in Southern Asia to NPC is still puzzling. The recent advances in the single nucleotide polymorphism and haplotype analyses, genome-wide screening, and association studies may help to decipher the inheritable genetic components for this enigmatic cancer. The cellular genes involved in DNA damage and its association with EBV entry or latency should be focused upon and further explored. Recently deployed technologies, such as high-density SNP array, will play a critical role in the search of these susceptibility genes. This same platform has also been successfully adapted to perform LOH and CGH profiling of the cancer genome, which place it in a unique position in the area of NPC research.
Previous molecular studies on NPC have focused on DNA and chromosomal levels, but few on transcriptomic and proteomic profiles. Small biopsy material and heavy infiltration of non-cancer cells present major difficulties for transcriptomic and proteomic studies. With advances in microdissection and preamplification technology, comprehensive expression profiling of NPC is now starting to take center stage. This should lead to substantial translational outcomes that will advance the management of this disease.
While substantial amount of information on the genomic alteration of NPC have been accumulated, the recent advances in genomic technologies (e.g., high-density SNP array) and the vast resources created by Human Genome Project will lead to more comprehensive results. Strategic integration of the data streams from multiple experimental applications (e.g., CGH, LOH, and expression microarray) at different biological levels (e.g., DNA, RNA and protein levels) will greatly enhance our ability to capture the precise portrait of the NPC genome.
Table 1.
The most frequent genomic abnormalities of NPC.
| Frequent abnormalities | |
|---|---|
| CGH | Gain: 1q, 3q, 8q, 12p, 12q and loss: 3p, 9p, 11q, 14q, 16q |
| LOH | 3p, 9p, and 14q, |
| Karyotyping | Gain: 3q and loss: 3q |
ACKNOWLEDGMENTS
This work was supported in part by NIH PHS Grant no DE014847, DE016569, CA114688, and a research grant from Prevent Cancer Foundation. We thank Ms. Katherine Long and Ms. Minghua Chai for excellent editorial assistance.
References
- 1.Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365(9476):2041–2054. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 2.Chan AT, Teo PM, Johnson PJ. Nasopharyngeal carcinoma. Annals of Oncology. 2002;13(7):1007–1015. doi: 10.1093/annonc/mdf179. [DOI] [PubMed] [Google Scholar]
- 3.Jeyakumar A, Brickman TM, Jeyakumar A, Doerr T. Review of nasopharyngeal carcinoma. Ear, Nose, & Throat Journal. 2006;85(3):168–170, 172-173, 184. [PubMed] [Google Scholar]
- 4.Diaz EM, Kies MS, Sturgis EM, et al. Neoplasms of the head and neck. In: Kufe DW, Frei E III, Holland JF, et al., editors. Holland Frei - Cancer Medicine 7. Vol. 1149. Columbia, BC, Canada: Decker; 2006. pp. 1211–1275. [Google Scholar]
- 5.Rickinson AB, Kieff E. Epstein-Barr virus. In: Knipe DM, Howley PM, editors. Field’s Virology. Philadelphia, Pa, USA: Lippincott, Williams & Wilkins; 2001. pp. 2575–2627. [Google Scholar]
- 6.Chang ET, Adami H-O. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiology Biomarkers and Prevention. 2006;15(10):1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 7.Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5(5):423–428. doi: 10.1016/s1535-6108(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 8.Wei WI, Sham JST, Zong Y-S, Choy D, Ng MH. The efficacy of fiberoptic endoscopic examination and biopsy in the detection of early nasopharyngeal carcinoma. Cancer. 1991;67(12):3127–3130. doi: 10.1002/1097-0142(19910615)67:12<3127::aid-cncr2820671231>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 9.Leong J-L, Fong K-W, Low W-K. Factors contributing to delayed diagnosis in nasopharyngeal carcinoma. Journal of Laryngology and Otology. 1999;113(7):633–636. doi: 10.1017/s0022215100144718. [DOI] [PubMed] [Google Scholar]
- 10.Krishna SM, James S, Kattoor J, Balaram P. Serum EBV DNA as a biomarker in primary nasopharyngeal carcinoma of Indian origin. Japanese Journal of Clinical Oncology. 2004;34(6):307–311. doi: 10.1093/jjco/hyh055. [DOI] [PubMed] [Google Scholar]
- 11.Brennan B. Nasopharyngeal carcinoma. Orphanet Journal of Rare Diseases. 2006;1:23. doi: 10.1186/1750-1172-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JKC, Pilch BZ, Kuo TT, Wenig BM, Lee AWM. Tumors of the nasopharynx: introduction. In: Barnes L, Eveson JW, Reichart P, Sidransky D, editors. Pathology and Genetics of Head and Neck Tumours (World Health Organization Classification of Tumours) Lyon, France: IARC press; 2005. pp. 82–84. [Google Scholar]
- 13.Greene FL, Page DL, Fleming ID, et al. American Joint Committee on Cancer: Cancer Staging Manual. New York, NY, USA: Springer; 2002. [Google Scholar]
- 14.Cho WC. Nasopharyngeal carcinoma: molecular biomarker discovery and progress. Molecular Cancer. 2007;6:1. doi: 10.1186/1476-4598-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho DW, Yang ZF, Wong BY, et al. Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry serum protein profiling to identify nasopharyngeal carcinoma. Cancer. 2006;107(1):99–107. doi: 10.1002/cncr.21970. [DOI] [PubMed] [Google Scholar]
- 16.Shao JY, Zhang Y, Li YH, et al. Comparison of Epstein-Barr virus DNA level in plasma, peripheral blood cell and tumor tissue in nasopharyngeal carcinoma. Anticancer Research. 2004;24(6):4059–4066. [PubMed] [Google Scholar]
- 17.Pathmanathan R, Prasad U, Sadler R, Flynn K, Raab-Traub N. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. New England Journal of Medicine. 1995;333(11):693–698. doi: 10.1056/NEJM199509143331103. [DOI] [PubMed] [Google Scholar]
- 18.Wei WI. Nasopharyngeal cancer: current status of management: a New York head and neck society lecture. Archives of Otolaryngology - Head and Neck Surgery. 2001;127(7):766–769. [PubMed] [Google Scholar]
- 19.Wei WI, Mok VWK. The management of neck metastases in nasopharyngeal cancer. Current Opinion in Otolaryngology and Head and Neck Surgery. 2007;15(2):99–102. doi: 10.1097/MOO.0b013e3280148a06. [DOI] [PubMed] [Google Scholar]
- 20.Ho JHC. An epidemiologic and clinical study of nasopharyngeal carcinoma. International Journal of Radiation Oncology Biology Physics. 1978;4(3-4):183–198. [PubMed] [Google Scholar]
- 21.Chien YC, Chen JY, Liu MY, et al. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. New England Journal of Medicine. 2001;345(26):1877–1882. doi: 10.1056/NEJMoa011610. [DOI] [PubMed] [Google Scholar]
- 22.Jia WH, Feng BJ, Xu ZL, et al. Familial risk and clustering of nasopharyngeal carcinoma in Guangdong, China. Cancer. 2004;101(2):363–369. doi: 10.1002/cncr.20372. [DOI] [PubMed] [Google Scholar]
- 23.Zeng YX, Jia WH. Familial nasopharyngeal carcinoma. Seminars in Cancer Biology. 2002;12(6):443–450. doi: 10.1016/s1044579x02000871. [DOI] [PubMed] [Google Scholar]
- 24.Levine PH, Pocinki AG, Madigan P, Bale S. Familial nasopharyngeal carcinoma in patients who are not Chinese. Cancer. 1992;70(5):1024–1029. doi: 10.1002/1097-0142(19920901)70:5<1024::aid-cncr2820700503>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Suárez C, Rodrigo JP, Ferlito A, Cabanillas R, Shaha AR, Rinaldo A. Tumours of familial origin in the head and neck. Oral Oncology. 2006;42(10):965–978. doi: 10.1016/j.oraloncology.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Friborg J, Wohlfahrt J, Koch A, Storm H, Olsen OR, Melbye M. Cancer susceptibility in nasopharyngeal carcinoma families—a population-based cohort study. Cancer Research. 2005;65(18):8567–8572. doi: 10.1158/0008-5472.CAN-04-4208. [DOI] [PubMed] [Google Scholar]
- 27.Friborg J, Wohlfahrt J, Melbye M. Familial risk and clustering of nasopharyngeal carcinoma in Guangdong, China. Cancer. 2005;103(1):211–212. doi: 10.1002/cncr.20759. [DOI] [PubMed] [Google Scholar]
- 28.Jia WH, Collins A, Zeng YX, et al. Complex segregation analysis of nasopharyngeal carcinoma in Guangdong, China: evidence for a multifactorial mode of inheritance (complex segregation analysis of NPC in China) European Journal of Human Genetics. 2005;13(2):248–252. doi: 10.1038/sj.ejhg.5201305. [DOI] [PubMed] [Google Scholar]
- 29.Hildesheim A, Apple RJ, Chen CJ, et al. Association of HLA class I and II alleles and extended haplotypes with nasopharyngeal carcinoma in Taiwan. Journal of the National Cancer Institute. 2002;94(23):1780–1789. doi: 10.1093/jnci/94.23.1780. [DOI] [PubMed] [Google Scholar]
- 30.Liebowitz D. Nasopharyngeal carcinoma: the Epstein-Barr virus association. Seminars in Oncology. 1994;21(3):376–381. [PubMed] [Google Scholar]
- 31.Lu SJ, Day NE, Degos L, et al. Linkage of a nasopharyngeal carcinoma susceptibility locus to the HLA region. Nature. 1990;346(6283):470–471. doi: 10.1038/346470a0. [DOI] [PubMed] [Google Scholar]
- 32.Xiong W, Zeng ZY, Xia JH, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Research. 2004;64(6):1972–1974. doi: 10.1158/0008-5472.can-03-3253. [DOI] [PubMed] [Google Scholar]
- 33.Deng L, Jing N, Tan G, et al. A common region of allelic loss on chromosome region 3p25.3–26.3 in nasopharyngeal carcinoma. Genes Chromosomes and Cancer. 1998;23(1):21–25. doi: 10.1002/(sici)1098-2264(199809)23:1<21::aid-gcc4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 34.Lo KW, Teo PM, Hui AB, et al. High resolution allelotype of microdissected primary nasopharyngeal carcinoma. Cancer Research. 2000;60(13):3348–3353. [PubMed] [Google Scholar]
- 35.Chan AS, To KF, Lo KW, et al. High frequency of chromosome 3p deletion in histologically normal nasopharyngeal epithelia from Southern Chinese. Cancer Research. 2000;60(19):5365–5370. [PubMed] [Google Scholar]
- 36.Feng BJ, Huang W, Shugart YY, et al. Genome-wide scan for familial nasopharyngeal carcinoma reveals evidence of linkage to chromosome 4. Nature Genetics. 2002;31(4):395–399. doi: 10.1038/ng932. [DOI] [PubMed] [Google Scholar]
- 37.Guo XC, Scott K, Liu Y, et al. Genetic factors leading to chronic Epstein-Barr virus infection and nasopharyngeal carcinoma in South East China: study design, methods and feasibility. Human Genomics. 2006;2(6):365–375. doi: 10.1186/1479-7364-2-6-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hildesheim A, Anderson LM, Chen CJ, et al. CYP2E1 genetic polymorphisms and risk of nasopharyngeal carcinoma in Taiwan. Journal of the National Cancer Institute. 1997;89(16):1207–1212. doi: 10.1093/jnci/89.16.1207. [DOI] [PubMed] [Google Scholar]
- 39.Hildesheim A, Chen CJ, Caporaso NE, et al. Cytochrome P4502E1 genetic polymorphisms and risk of nasopharyngeal carcinoma: results from a case-control study conducted in Taiwan. Cancer Epidemiology Biomarkers and Prevention. 1995;4(6):607–610. [PubMed] [Google Scholar]
- 40.Tiwawech D, Srivatanakul P, Karalak A, Ishida T. Cytochrome P450 2A6 polymorphism in nasopharyngeal carcinoma. Cancer Letters. 2006;241(1):135–141. doi: 10.1016/j.canlet.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 41.Friborg JT, Yuan JM, Wang R, Koh WP, Lee HP, Yu MC. A prospective study of tobacco and alcohol use as risk factors for pharyngeal carcinomas in Singapore Chinese. Cancer. 2007;109(6):1183–1191. doi: 10.1002/cncr.22501. [DOI] [PubMed] [Google Scholar]
- 42.Nazar-Stewart V, Vaughan TL, Burt RD, Chen C, Berwick M, Swanson GM. Glutathione S-transferase M1 and susceptibility to nasopharyngeal carcinoma. Cancer Epidemiology Biomarkers and Prevention. 1999;8(6):547–551. [PubMed] [Google Scholar]
- 43.Bendjemana K, Abdennebi M, Gara S, et al. Genetic polymorphism of gluthation-S transferases and N-acetyl transferases 2 and nasopharyngeal carcinoma: the Tunisia experience. Bulletin du Cancer. 2006;93(3):297–302. [PubMed] [Google Scholar]
- 44.Tiwawech D, Srivatanakul P, Karalak A, Ishida T. Glutathione S-transferase M1 gene polymorphism in Thai nasopharyngeal carcinoma. Asian Pacific Journal of Cancer Prevention. 2005;6(3):270–275. [PubMed] [Google Scholar]
- 45.Cho EY, Hildesheim A, Chen CJ, et al. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer Epidemiology Biomarkers and Prevention. 2003;12(10):1100–1104. [PubMed] [Google Scholar]
- 46.Cao Y, Miao XP, Huang MY, et al. Polymorphisms of XRCC1 genes and risk of nasopharyngeal carcinoma in the Cantonese population. BMC Cancer. 2006;6:167. doi: 10.1186/1471-2407-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song C, Chen LZ, Zhang RH, Yu XJ, Zeng YX. Functional variant in the -untranslated region of toll-like receptor 4 is associated with nasopharyngeal carcinoma risk. Cancer Biology and Therapy. 2006;5(10):1285–1291. doi: 10.4161/cbt.5.10.3304. [DOI] [PubMed] [Google Scholar]
- 48.Zhou X-X, Jia W-H, Shen G-P, et al. Sequence variants in toll-like receptor 10 are associated with nasopharyngeal carcinoma risk. Cancer Epidemiology Biomarkers and Prevention. 2006;15(5):862–866. doi: 10.1158/1055-9965.EPI-05-0874. [DOI] [PubMed] [Google Scholar]
- 49.He Y, Zhou G, Zhai Y, et al. Association of PLUNC gene polymorphisms with susceptibility to nasopharyngeal carcinoma in a Chinese population. Journal of Medical Genetics. 2005;42(2):172–176. doi: 10.1136/jmg.2004.022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou G, Zhai Y, Cui Y, et al. Functional polymorphisms and haplotypes in the promoter of the MMP2 gene are associated with risk of nasopharyngeal carcinoma. Human Mutation. 2007;28(11):1091–1097. doi: 10.1002/humu.20570. [DOI] [PubMed] [Google Scholar]
- 51.Nasr HB, Mestiri S, Chahed K, et al. Matrix metalloproteinase-1 (-1607) 1G/2G and -9 (-1562) C/T promoter polymorphisms: susceptibility and prognostic implications in nasopharyngeal carcinomas. Clinica Chimica Acta. 2007;384(1-2):57–63. doi: 10.1016/j.cca.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 52.Wei Y-S, Zhu Y-H, Du B, et al. Association of transforming growth factor-1 gene polymorphisms with genetic susceptibility to nasopharyngeal carcinoma. Clinica Chimica Acta. 2007;380(1-2):165–169. doi: 10.1016/j.cca.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 53.Wei Y-S, Kuang X-H, Zhu Y-H, et al. Interleukin-10 gene promoter polymorphisms and the risk of nasopharyngeal carcinoma. Tissue Antigens. 2007;70(1):12–17. doi: 10.1111/j.1399-0039.2007.00806.x. [DOI] [PubMed] [Google Scholar]
- 54.Hassen E, Farhat K, Gabbouj S, Jalbout M, Bouaouina N, Chouchane L. TAP1 gene polymorphisms and nasopharyngeal carcinoma risk in a Tunisian population. Cancer Genetics and Cytogenetics. 2007;175(1):41–46. doi: 10.1016/j.cancergencyto.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 55.Sousa H, Santos AM, Catarino R, et al. Linkage of TP53 codon 72 pro/pro genotype as predictive factor for nasopharyngeal carcinoma development. European Journal of Cancer Prevention. 2006;15(4):362–366. doi: 10.1097/00008469-200608000-00010. [DOI] [PubMed] [Google Scholar]
- 56.Catarino RJ, Breda E, Coelho V, et al. Association of the A870G cyclin D1 gene polymorphism with genetic susceptibility to nasopharyngeal carcinoma. Head and Neck. 2006;28(7):603–608. doi: 10.1002/hed.20377. [DOI] [PubMed] [Google Scholar]
- 57.Bel Hadj Jrad B, Mahfouth W, Bouaouina N, et al. A polymorphism in FAS gene promoter associated with increased risk of nasopharyngeal carcinoma and correlated with anti-nuclear autoantibodies induction. Cancer Letters. 2006;233(1):21–27. doi: 10.1016/j.canlet.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 58.Zhou G, Zhai Y, Cui Y, et al. MDM2 promoter SNP309 is associated with risk of occurrence and advanced lymph node metastasis of nasopharyngeal carcinoma in Chinese population. Clinical Cancer Research. 2007;13(9):2627–2633. doi: 10.1158/1078-0432.CCR-06-2281. [DOI] [PubMed] [Google Scholar]
- 59.Zheng M-Z, Qin H-D, Yu X-J, et al. Haplotype of gene Nedd4 binding protein 2 associated with sporadic nasopharyngeal carcinoma in the Southern Chinese population. Journal of Translational Medicine. 2007;5:36. doi: 10.1186/1479-5876-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu N, Wang C, Hu Y, et al. Genome-wide association study in esophageal cancer using GeneChip mapping 10K array. Cancer Research. 2005;65(7):2542–2546. doi: 10.1158/0008-5472.CAN-04-3247. [DOI] [PubMed] [Google Scholar]
- 61.Ellis NA, Kirchhoff T, Mitra N, et al. Localization of breast cancer susceptibility loci by genome-wide SNP linkage disequilibrium mapping. Genetic Epidemiology. 2006;30(1):48–61. doi: 10.1002/gepi.20101. [DOI] [PubMed] [Google Scholar]
- 62.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. Journal of the National Cancer Institute. 1960;25:85–109. [PubMed] [Google Scholar]
- 64.Liehr T, Heller A, Starke H, Claussen U. FISH banding methods: applications in research and diagnostics. Expert Review of Molecular Diagnostics. 2002;2(3):217–225. doi: 10.1586/14737159.2.3.217. [DOI] [PubMed] [Google Scholar]
- 65.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 66.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 67.Schwaenen C, Nessling M, Wessendorf S, et al. Automated array-based genomic profiling in chronic lymphocytic leukemia: development of a clinical tool and discovery of recurrent genomic alterations. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(4):1039–1044. doi: 10.1073/pnas.0304717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paris PL, Andaya A, Fridlyand J, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Human Molecular Genetics. 2004;13(13):1303–1313. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- 69.Callagy G, Pharoah P, Chin S-F, et al. Identification and validation of prognostic markers in breast cancer with the complementary use of array-CGH and tissue microarrays. Journal of Pathology. 2005;205(3):388–396. doi: 10.1002/path.1694. [DOI] [PubMed] [Google Scholar]
- 70.Weiss MM, Kuipers EJ, Postma C, et al. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cellular Oncology. 2004;26(5-6):307–317. doi: 10.1155/2004/454238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosin MP, Cheng X, Poh C, et al. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clinical Cancer Research. 2000;6(2):357–362. [PubMed] [Google Scholar]
- 72.Martinez-Climent JA, Alizadeh AA, Segraves R, et al. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood. 2003;101(8):3109–3117. doi: 10.1182/blood-2002-07-2119. [DOI] [PubMed] [Google Scholar]
- 73.Rubio-Moscardo F, Climent J, Siebert R, et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005;105(11):4445–4454. doi: 10.1182/blood-2004-10-3907. [DOI] [PubMed] [Google Scholar]
- 74.Yan W, Song L, Wei W, Li A, Liu J, Fang Y. Chromosomal abnormalities associated with neck nodal metastasis in nasopharyngeal carcinoma. Tumor Biology. 2005;26(6):306–312. doi: 10.1159/000089289. [DOI] [PubMed] [Google Scholar]
- 75.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258(5083):818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 76.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nature Genetics. 2005;37(6):S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 77.Pinkel D, Albertson DG. Comparative genomic hybridization. Annual Review of Genomics and Human Genetics. 2005;6:331–354. doi: 10.1146/annurev.genom.6.080604.162140. [DOI] [PubMed] [Google Scholar]
- 78.Chen Y-J, Ko J-Y, Chen P-J, et al. Chromosomal aberrations in nasopharyngeal carcinoma analyzed by comparative genomic hybridization. Genes Chromosomes and Cancer. 1999;25(2):169–175. [PubMed] [Google Scholar]
- 79.Hui AB-Y, Lo K-W, Leung S-F, et al. Detection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridisation. International Journal of Cancer. 1999;82(4):498–503. doi: 10.1002/(sici)1097-0215(19990812)82:4<498::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 80.Fang Y, Guan X-Y, Guo Y, et al. Analysis of genetic alterations in primary nasopharyngeal carcinoma by comparative genomic hybridization. Genes Chromosomes and Cancer. 2001;30(3):254–260. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1086>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 81.Chien G, Yuen PW, Kwong D, Kwong YL. Comparative genomic hybridization analysis of nasopharygeal carcinoma: consistent patterns of genetic aberrations and clinicopathological correlations. Cancer Genetics and Cytogenetics. 2001;126(1):63–67. doi: 10.1016/s0165-4608(00)00392-7. [DOI] [PubMed] [Google Scholar]
- 82.Li X, Wang E, Zhao Y-D, et al. Chromosomal imbalances in nasopharyngeal carcinoma: a meta-analysis of comparative genomic hybridization results. Journal of Translational Medicine. 2006;4:4. doi: 10.1186/1479-5876-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genetics. 1998;20(2):207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 84.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nature Genetics. 2004;36(3):299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 85.Hui AB, Lo KW, Teo PM, To KF, Huang DP. Genome wide detection of oncogene amplifications in nasopharyngeal carcinoma by array based comparative genomic hybridization. International Journal of Oncology. 2002;20(3):467–473. [PubMed] [Google Scholar]
- 86.Mantripragada KK, Buckley PG, Diaz De Ståhl T, Dumanski JP. Genomic microarrays in the spotlight. Trends in Genetics. 2004;20(2):87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 87.Pollack JR, Perou CM, Alizadeh AA, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genetics. 1999;23(1):41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 88.Zhou X, Jordan RCK, Mok S, Birrer MJ, Wong DTW. DNA copy number abnormality of oral squamous cell carcinoma detected with cDNA array-based comparative genomic hybridization. Cancer Genetics and Cytogenetics. 2004;151(1):90–92. doi: 10.1016/j.cancergencyto.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 89.Brennan C, Zhang Y, Leo C, et al. High-resolution global profiling of genomic alterations with long oligonucleotide microarray. Cancer Research. 2004;64(14):4744–4748. doi: 10.1158/0008-5472.CAN-04-1241. [DOI] [PubMed] [Google Scholar]
- 90.Lucito R, Healy J, Alexander J, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Research. 2003;13(10):2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bignell GR, Huang J, Greshock J, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Research. 2004;14(2):287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao X, Li C, Paez JG, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Research. 2004;64(9):3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 93.Zhou X, Mok SC, Chen Z, Li Y, Wong DTW. Concurrent analysis of loss of heterozygosity (LOH) and copy number abnormality (CNA) for oral premalignancy progression using the Affymetrix 10 K SNP mapping array. Human Genetics. 2004;115(4):327–330. doi: 10.1007/s00439-004-1163-1. [DOI] [PubMed] [Google Scholar]
- 94.Zhao X, Weir BA, LaFramboise T, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Research. 2005;65(13):5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 95.Davies JJ, Wilson IM, Lam WL. Array CGH technologies and their applications to cancer genomes. Chromosome Research. 2005;13(3):237–248. doi: 10.1007/s10577-005-2168-x. [DOI] [PubMed] [Google Scholar]
- 96.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proceedings of the National Academy of Sciences of the United States of America. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knudson AG. Hereditary cancer: two hits revisited. Journal of Cancer Research and Clinical Oncology. 1996;122(3):135–140. doi: 10.1007/BF01366952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vogelstein B, Fearon ER, Kern SE, et al. Allelotype of colorectal carcinomas. Science. 1989;244(4901):207–211. doi: 10.1126/science.2565047. [DOI] [PubMed] [Google Scholar]
- 99.Mutirangura A, Tanunyutthawongese C, Pornthanakasem W, et al. Genomic alterations in nasopharyngeal carcinoma: loss of heterozygosity and Epstein-Barr virus infection. British Journal of Cancer. 1997;76(6):770–776. doi: 10.1038/bjc.1997.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shao JY, Wang HY, Huang XM, et al. Genome-wide allelotype analysis of sporadic primary nasopharyngeal carcinoma from Southern China. International Journal of Oncology. 2000;17(6):1267–1275. doi: 10.3892/ijo.17.6.1267. [DOI] [PubMed] [Google Scholar]
- 101.Shao J-Y, Huang X-M, Yu X-J, et al. Loss of heterozygosity and its correlation with clinical outcome and Epstein-Barr virus infection in nasopharyngeal carcinoma. Anticancer Research. 2001;21(4 B):3021–3029. [PubMed] [Google Scholar]
- 102.Wang DG, Fan J-B, Siao C-J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280(5366):1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 103.Lindblad-Toh K, Tanenbaum DM, Daly MJ, et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nature Biotechnology. 2000;18(9):1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- 104.Jänne PA, Li C, Zhao X, et al. High-resolution single-nucleotide polymorphism array and clustering analysis of loss of heterozygosity in human lung cancer cell lines. Oncogene. 2004;23(15):2716–2726. doi: 10.1038/sj.onc.1207329. [DOI] [PubMed] [Google Scholar]
- 105.Wang ZC, Lin M, Wei L-J, et al. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Research. 2004;64(1):64–71. doi: 10.1158/0008-5472.can-03-2570. [DOI] [PubMed] [Google Scholar]
- 106.Hoque MO, Lee C-CR, Cairns P, Schoenberg M, Sidransky D. Genome-wide genetic characterization of bladder cancer: a comparison of high-density single-nucleotide polymorphism arrays and PCR-based microsatellite analysis. Cancer Research. 2003;63(9):2216–2222. [PubMed] [Google Scholar]
- 107.Lieberfarb ME, Lin M, Lechpammer M, et al. Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Research. 2003;63(16):4781–4785. [PubMed] [Google Scholar]
- 108.Zhou X, Li C, Mok SC, Chen Z, Wong DTW. Whole genome loss of heterozygosity profiling on oral squamous cell carcinoma by high-density single nucleotide polymorphic allele (SNP) array. Cancer Genetics and Cytogenetics. 2004;151(1):82–84. doi: 10.1016/j.cancergencyto.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 109.Caspersson T, Zech L, Modest EJ, Foley GE, Wagh U, Simonsson E. Chemical differentiation with fluorescent alkylating agents in Vicia faba metaphase chromosomes. Experimental Cell Research. 1969;58(1):128–140. doi: 10.1016/0014-4827(69)90123-2. [DOI] [PubMed] [Google Scholar]
- 110.Caspersson T, Zech L, Modest EJ, Foley GE, Wagh U, Simonsson E. DNA-binding fluorochromes for the study of the organization of the metaphase nucleus. Experimental Cell Research. 1969;58(1):141–152. doi: 10.1016/0014-4827(69)90124-4. [DOI] [PubMed] [Google Scholar]
- 111.Kristensen M, Quek HH, Chew CT, Chan SH. A cytogenetic study of 74 nasopharyngeal carcinoma biopsies. Annals of the Academy of Medicine Singapore. 1991;20(5):597–600. [PubMed] [Google Scholar]
- 112.Mitelman F, Mark Vendel E, Mineur A, Giovanella B, Klein G. A 3q+ marker chromosome in EBV-carrying nasopharyngeal carcinomas. International Journal of Cancer. 1983;32(6):651–655. doi: 10.1002/ijc.2910320602. [DOI] [PubMed] [Google Scholar]
- 113.Zhang S, Wu Y, Zeng Y, Zech L, Klein G. Cytogenetic studies on an epithelioid cell line derived from nasopharyngeal carcinoma. Hereditas. 1982;97(1):23–28. doi: 10.1111/j.1601-5223.1982.tb00706.x. [DOI] [PubMed] [Google Scholar]
- 114.Chang Y-S, Lin S-Y, Lee P-F, Durff T, Chung H-C, Tsai M-S. Establishment and characterization of a tumor cell line from human nasopharyngeal carcinoma tissue. Cancer Research. 1989;49(23):6752–6757. [PubMed] [Google Scholar]
- 115.Huang DP, Ho JHC, Chan WK, Lau WH, Lui M. Cytogenetics of undifferentiated nasopharyngeal carcinoma xenografts from Southern Chinese. International Journal of Cancer. 1989;43(5):936–939. doi: 10.1002/ijc.2910430535. [DOI] [PubMed] [Google Scholar]
- 116.Bernheim A, Rousselet G, Massaad L, Busson P, Tursz T. Cytogenetic studies in three xenografted nasopharyngeal carcinomas. Cancer Genetics and Cytogenetics. 1993;66(1):11–15. doi: 10.1016/0165-4608(93)90141-8. [DOI] [PubMed] [Google Scholar]
- 117.Lin C-T, Chan W-Y, Chen W, et al. Characterization of seven newly established nasopharyngeal carcinoma cell lines. Laboratory Investigation. 1993;68(6):716–727. [PubMed] [Google Scholar]
- 118.Hui ABY, Cheung S-T, Fong Y, Lo K-W, Huang DP. Characterization of a new EBV-associated nasopharyngeal carcinoma cell line. Cancer Genetics and Cytogenetics. 1998;101(2):83–88. doi: 10.1016/s0165-4608(97)00231-8. [DOI] [PubMed] [Google Scholar]
- 119.Lo K-W, Huang PWSD, Lee CKJ. Genetic changes in nasopharyngeal carcinoma. Chinese Medical Journal. 1997;110(7):548–549. [PubMed] [Google Scholar]
- 120.Lo K-W, Huang DP. Genetic and epigenetic changes in nasopharyngeal carcinoma. Seminars in Cancer Biology. 2002;12(6):451–462. doi: 10.1016/s1044579x02000883. [DOI] [PubMed] [Google Scholar]
- 121.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nature Reviews Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Waghray M, Parhar RS, Taibah K, Al-Sedairy S. Rearrangements of chromosome arm 3q in poorly differentiated nasopharyngeal carcinoma. Genes Chromosomes and Cancer. 1992;4(4):326–330. doi: 10.1002/gcc.2870040409. [DOI] [PubMed] [Google Scholar]
- 123.Fan C-S, Wong N, Leung S-F, et al. Frequent c-myc and Int-2 overrepresentations in nasopharyngeal carcinoma. Human Pathology. 2000;31(2):169–178. doi: 10.1016/s0046-8177(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 124.Schröck E, Du Manoir S, Veldman T, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273(5274):494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 125.Liyanage M, Coleman A, du Manoir S, et al. Multicolour spectral karyotyping of mouse chromosomes. Nature Genetics. 1996;14(3):312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- 126.Speicher MR, Ballard SG, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nature Genetics. 1996;12(4):368–375. doi: 10.1038/ng0496-368. [DOI] [PubMed] [Google Scholar]
- 127.Wong N, Hui ABY, Fan B, et al. Molecular cytogenetic characterization of nasopharyngeal carcinoma cell lines and xenografts by comparative genomic hybridization and spectral karyotyping. Cancer Genetics and Cytogenetics. 2003;140(2):124–132. doi: 10.1016/s0165-4608(02)00657-x. [DOI] [PubMed] [Google Scholar]
- 128.Ma YH, Zhang LJ, Wang XQ, Zeng MS, Li B, Zeng YX. Rapid idenfication of differentially expressed genes in subtractive library of nasopharyngeal carcinoma using cDNA chip. Acta Scientiarum Naturalium Universitatis Sunyatseni. 2000;39(6A):192–196. [Google Scholar]
- 129.Xie L, Xu L, He Z, et al. Identification of differentially expressed genes in nasopharyngeal carcinoma by means of the atlas human cancer cDNA expression array. Journal of Cancer Research and Clinical Oncology. 2000;126(7):400–406. doi: 10.1007/PL00008488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang LJ, Fang Y, Ma YH, et al. Gene expression profiling in nasopharyngeal carcinoma determined by high density cDNA microarray. Ai Zheng. 2002;21(6):588–592. [PubMed] [Google Scholar]
- 131.Zeng Z, Zhou Y, Xiong W, et al. Analysis of gene expression identifies candidate molecular markers in nasopharyngeal carcinoma using microdissection and cDNA microarray. Journal of Cancer Research and Clinical Oncology. 2007;133(2):71–81. doi: 10.1007/s00432-006-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zeng Z-Y, Zhou Y-H, Zhang W-L, et al. Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Human Pathology. 2007;38(1):120–133. doi: 10.1016/j.humpath.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 133.Sung FL, Hui EP, Tao Q, et al. Genome-wide expression analysis using microarray identified complex signaling pathways modulated by hypoxia in nasopharyngeal carcinoma. Cancer Letters. 2007;253(1):74–88. doi: 10.1016/j.canlet.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 134.Lee Y-CG, Hwang Y-C, Chen K-C, et al. Effect of Epstein-Barr virus infection on global gene expression in nasopharyngeal carcinoma. Functional and Integrative Genomics. 2007;7(1):79–93. doi: 10.1007/s10142-006-0035-2. [DOI] [PubMed] [Google Scholar]
- 135.Sengupta S, Den Boon JA, Chen I-H, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Research. 2006;66(16):7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 136.Soo RA, Wu J, Aggarwal A, et al. Celecoxib reduces microvessel density in patients treated with nasopharyngeal carcinoma and induces changes in gene expression. Annals of Oncology. 2006;17(11):1625–1630. doi: 10.1093/annonc/mdl283. [DOI] [PubMed] [Google Scholar]
- 137.Zhou X, Yu T, Cole SW, Wong DTW. Advancement in characterization of genomic alterations for improved diagnosis, treatment and prognostics in cancer. Expert Review of Molecular Diagnostics. 2006;6(1):39–50. doi: 10.1586/14737159.6.1.39. [DOI] [PubMed] [Google Scholar]
- 138.Li HM, Man C, Jin Y, et al. Molecular and cytogenetic changes involved in the immortalization of nasopharyngeal epithelial cells by telomerase. International Journal of Cancer. 2006;119(7):1567–1576. doi: 10.1002/ijc.22032. [DOI] [PubMed] [Google Scholar]
- 139.Crawley JJ, Furge KA. Identification of frequent cytogenetic aberrations in hepatocellular carcinoma using gene-expression microarray data. Genome Biology. 2002;3(12):RESEARCH0075. doi: 10.1186/gb-2002-3-12-research0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou X, Cole SW, Hu S, Wong DTW. Detection of DNA copy number abnormality by microarray expression analysis. Human Genetics. 2004;114(5):464–467. doi: 10.1007/s00439-004-1087-9. [DOI] [PubMed] [Google Scholar]
- 141.Zhou X, Cole SW, Rao N, et al. Identification of discrete chromosomal deletion by binary recursive partitioning of microarray differential expression data. Journal of Medical Genetics. 2005;42(5):416–419. doi: 10.1136/jmg.2004.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Myers CL, Dunham MJ, Kung SY, Troyanskaya OG. Accurate detection of aneuploidies in array CGH and gene expression microarray data. Bioinformatics. 2004;20(18):3533–3543. doi: 10.1093/bioinformatics/bth440. [DOI] [PubMed] [Google Scholar]
- 143.Pollack JR, Sørlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(20):12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wolf M, Mousses S, Hautaniemi S, et al. High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia. 2004;6(3):240–247. doi: 10.1593/neo.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kauraniemi P, Kuukasjärvi T, Sauter G, Kallioniemi A. Amplification of a 280-kilobase core region at the ERBB2 locus leads to activation of two hypothetical proteins in breast cancer. American Journal of Pathology. 2003;163(5):1979–1984. doi: 10.1016/S0002-9440(10)63556-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Phillips JL, Hayward SW, Wang Y, et al. The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis. Cancer Research. 2001;61(22):8143–8149. [PubMed] [Google Scholar]
- 147.Virtaneva K, Wright FA, Tanner SM, et al. Expression profiling reveals fundamental biological differences in acute myeloid leukemia with isolated trisomy 8 and normal cytogenetics. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(3):1124–1129. doi: 10.1073/pnas.98.3.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]