Abstract
We have been able to convert a small α/β protein, acylphosphatase, from its soluble and native form into insoluble amyloid fibrils of the type observed in a range of pathological conditions. This was achieved by allowing slow growth in a solution containing moderate concentrations of trifluoroethanol. When analyzed with electron microscopy, the protein aggregate present in the sample after long incubation times consisted of extended, unbranched filaments of 30–50 Å in width that assemble subsequently into higher order structures. This fibrillar material possesses extensive β-sheet structure as revealed by far-UV CD and IR spectroscopy. Furthermore, the fibrils exhibit Congo red birefringence, increased fluorescence with thioflavine T and cause a red-shift of the Congo red absorption spectrum. All of these characteristics are typical of amyloid fibrils. The results indicate that formation of amyloid occurs when the native fold of a protein is destabilized under conditions in which noncovalent interactions, and in particular hydrogen bonding, within the polypeptide chain remain favorable. We suggest that amyloid formation is not restricted to a small number of protein sequences but is a property common to many, if not all, natural polypeptide chains under appropriate conditions.
A range of human disorders is associated with the extracellular deposition of insoluble protein aggregates known as amyloid fibrils (1). These include Alzheimer’s disease, type II diabetes, primary and secondary systemic amyloidosis, and the spongiform encephalopathies such as Creutzfeldt-Jakob disease. Although it is still debatable as to whether amyloid fibril deposition is itself the primary origin of the pathological condition with which it is associated, a variety of experimental observations suggests that there is a strong causal link between fibril formation and the onset of pathological symptoms (2, 3). Electron microscopy shows that the amyloid fibrils associated with the various diseases all appear straight and unbranched, and are 40–120 Å in diameter (4). X-ray fiber diffraction studies indicate a characteristic cross-β structure in which the polypeptide chains form β-strands oriented perpendicular to the long axis of the fibril, resulting in β-sheets propagating in the direction of the fibril (4). This overall structure is common to all amyloid fibrils despite the lack of sequence homologies among the amyloidogenic proteins and the absence of structural similarities in the folds that these proteins adopt under normal nonpathological conditions.
This observation of a common structure for amyloid indicates that it is stabilized by interactions associated with the common covalent structure of proteins, such as backbone hydrogen bonding or hydrophobic interactions, rather than through specific interactions of the different side chains (1, 4). Recently, the SH3 domain of the p85α subunit of phosphatidylinositol 3-kinase, a protein that is not associated with any of the known amyloid diseases, was found readily to form amyloid fibrils in vitro under acidic conditions (5). This prompted us to explore whether other natural proteins might assemble into such fibrils if appropriate conditions could be found. In this paper we describe experiments to design rationally such conditions using as a model system human muscle acylphosphatase, an α/β protein consisting of 98 residues (6) and whose stability and folding properties have been studied extensively (7–9). In this manuscript we show that under appropriate conditions, this small protein forms a partially denatured state from which a protein aggregate that contains amyloid protofilaments and fibrils develops slowly.
MATERIALS AND METHODS
Protein Preparation.
Muscle acylphosphatase was purified as described (10). Protein purity was checked by specific activity measurements, SDS/PAGE, and electrospray mass spectrometry. Protein concentration was determined spectrophotometrically by using a ɛ280 value of 1.42 ml mg−1 cm−1.
IR Spectroscopy.
IR spectra were acquired by using a Bio-Rad 1750 IR spectrometer equipped with a narrow band detector. CaF2 windows and 12.5-μm myelin spacers were used, and the whole cell was thermostated by using an F8 Haake circulating water bath. Protein samples were dissolved at a concentration of 10 mg/ml in 25% (vol/vol) d3-trifluoroethanol (TFE), 50 mM d3-acetate/D2O buffer, pD 5.5. The spectra were recorded at 25°C with a resolution of 2 cm−1. Water vapor was expelled from the sample compartment by using dry nitrogen gas. Baseline correction and self-deconvolution of the spectra were performed by using the win-ir software.
Circular Dichroism.
Muscle acylphosphatase was incubated at a concentration of 0.375 mg/ml (34 μM) in 25% (vol/vol) TFE, 50 mM acetate buffer, pH 5.5, 25°C with constant stirring. Aliquots were withdrawn at regular time intervals for the far-UV CD analysis. CD spectra were acquired by means of a Jasco (Easton, MD) J-720 spectropolarimeter and cuvettes of 1-mm path length. The same aliquots withdrawn for the CD analysis were also used for electron microscopy and the other spectroscopic analysis described below.
Electron Microscopy.
Electron micrographs were acquired by using a JEM 1010 transmission electron microscope at 80-kV excitation voltage. A 3-μl sample of protein solution was placed and dried for 5 min on a Formvar and carbon-coated grid. The sample was then negatively stained with 3 μl of 1% phosphotungstic acid solution and observed at magnifications of ×25,000–100,000.
Congo Red and Thioflavine T Staining.
For Congo red birefringence experiments, aliquots of protein were air-dried onto glass slides. The resulting films were stained with a saturated solution of Congo red and sodium chloride and corrected to pH 10.0 with 1% sodium hydroxide. The stained slides were examined with an optical microscope between crossed polarizers. Absorption spectra of Congo red dye in the presence of muscle acylphosphatase aggregate were recorded as described (11). Fluorescence spectra of the thioflavine T dye in the presence of muscle acylphosphatase aggregates were acquired as described (12). In both cases, the spectra of the protein aggregates alone and of the dye alone were acquired as controls.
RESULTS
There is increasing evidence that amyloid fibrils do not develop directly from the native conformations of proteins but from precursors which are only partially folded (5, 13–16). We decided, therefore, to explore conditions for fibril formation in which acylphosphatase is denatured but retains considerable amounts of residual structure. We chose for this purpose to use TFE, a solvent known to stabilize partially folded proteins (17, 18). An extensive series of experiments was carried out in which solutions of proteins at concentrations between 0.15 and 10 mg/ml (14 and 900 μM) were incubated in aqueous TFE solutions of different molar ratios. Aggregation phenomena, indicated by light-scattering effects, were observed to occur readily between 18% and 35% TFE (vol/vol) at high protein concentrations. We chose to investigate in particular detail solutions of acylphosphatase in 25% (vol/vol) TFE; at this proportion of alcohol, the protein is just above its midpoint of unfolding at pH 5.5 and 25°C. Under these conditions, protein concentrations above 4 mg/ml resulted in rapidly formed gelatinous precipitates, which analysis by electron microscopy showed to be amorphous. At lower protein concentrations, however, conversion of the solutions to gels or precipitates took place only over periods of hours or days. This delay enabled the sequence of events taking place after diluting a solution of the protein into these conditions to be readily followed by a variety of techniques. These included far-UV CD and Fourier-transform IR spectroscopy to probe the protein conformation as well as techniques to provide evidence for the formation of amyloid fibrils, such as Congo red and thioflavine T binding tests, Congo red birefringence, and electron microscopy.
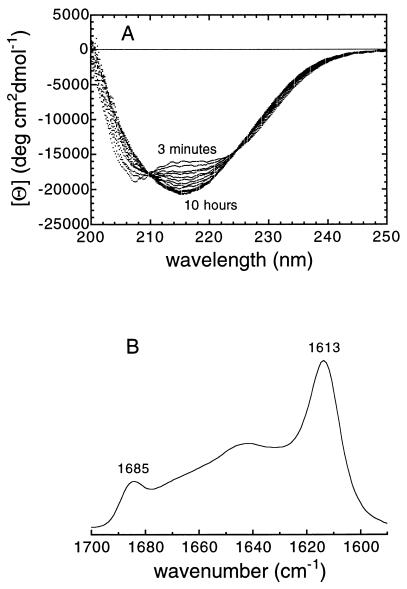
Acylphosphatase, incubated at a concentration of 34 μM in 25% (vol/vol) TFE, pH 5.5, 25°C, was found to unfold from its native state on a time scale of seconds to give a denatured state with a far-UV CD spectrum indicative of extensive α-helical content. Such transitions are common on introduction of proteins into TFE solutions (18). Far-UV CD analysis revealed, however, the presence of a subsequent slow transition, completed within 2–3 hours, from this α-helical conformation to one containing a considerable content of β-sheet structure (Fig. 1A). The presence of two isodichroic points at 210 and 225 nm in the CD spectra acquired over this period indicated that this transition at least approximates to a two-state process. The concomitant appearance of two bands at 1,685 and 1,613 cm−1 in the amide I region of the IR spectrum (Fig. 1B) suggests that such β-structure results from hydrogen bonding within protein aggregates (19). Moreover, electron micrographs reveal clearly the presence of aggregates in samples recovered at this stage of the reaction. These are, however, granular in appearance, with no evidence for extended fibrils characteristic of amyloid formation (Fig. 2A).
Figure 1.
(A) Far-UV CD spectra of muscle acylphosphatase acquired during the aggregation process. The first and last spectra reported in the figure were acquired after 3 min and 10 hr after the initiation of the reaction, respectively. The spectra show a slow two-state transition between two conformations containing significant amounts of α-helical and β-sheet structure, respectively. After 10 hr of incubation, the spectra do not change their appearance, but undergo a progressive reduction of signal and a shift of the negative peak toward the higher wavelengths, as a result of the accumulation of protein aggregates. (B) Amide I region of the IR spectrum of muscle acylphosphatase denatured in TFE. The spectrum was recorded after a period of 2 hr from the start of the reaction. The two peaks at 1,613 and 1,685 cm−1 are indicative of intermolecular β structure (19).
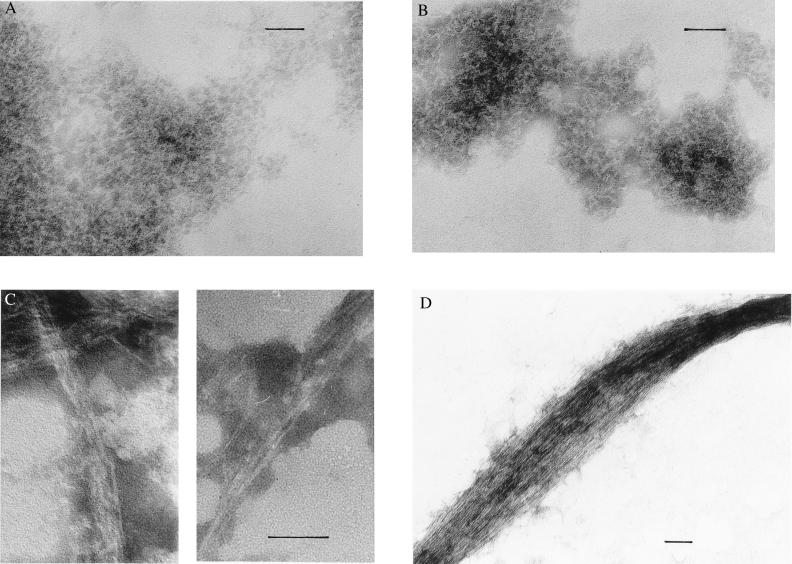
Figure 2.
Electron micrographs showing the morphological development of acylphosphatase aggregates. (A) Aggregate of granular aspect formed 72 min after initiation of the reaction. (B) Short fibrils formed after 32 hr. (C) Protofilaments formed after 14 days. (D) Clusters of protofilaments formed after 45 days. In C, the two micrographs show fibrils from two different preparations. (Bar = 0.05 μm.)
After 10 hours, the intensity of the CD signal started to decrease, probably as a consequence of formation of more extended aggregates. Electron micrographs from samples recovered after a period of ≈32 hours reveal the presence of short filaments, indicating that a fibrillar protein aggregate was beginning to form (Fig. 2B). In similar samples recovered after a period of 14 days, the fibrillar material is clearly evident (Fig. 2C). The fibrils revealed by the electron micrographs are long and unbranched with diameters of ≈30–50 Å. This fibrillar material appears similar to the protofilaments observed in the early stages of other amyloidogenic systems, given the small diameter and the absence of higher order structures in which two or more filaments twist around each other (4). After incubation for 1–2 months, the electron micrographs demonstrate structures in which large bundles of protofibrils are formed, giving rope-like structures of various diameters, some of them as large as 800–1,500 Å in width (Fig. 2D). At this stage, more than 50% of acylphosphatase appears to be present in fibrillar form rather than in a soluble form or as amorphous aggregates. The persistence of protofilaments also is apparent for other protein sequences incubated under similar conditions (C. E. MacPhee and C.M.D., unpublished observations). The fact that individual protofilaments, rather than assemblies of ≈2–5 such structures, are present in these experiments suggests that the small clusters of protofilaments observed in many amyloid fibrils are held together by hydrophobic interactions that are disrupted in TFE solutions.
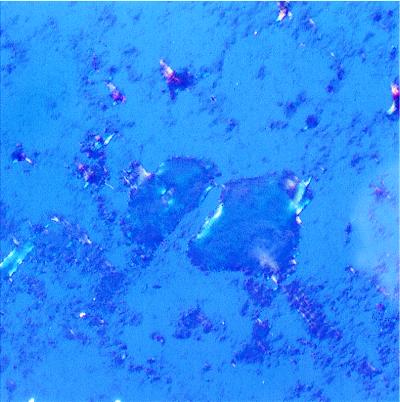
A series of optical tests was carried out to investigate further that this fibrillar material meets all criteria to be classified as amyloid. A >20-fold increase of the 482 nm fluorescence (excitation 440 nm) of the dye thioflavine T was observed in the presence of aliquots of protein aggregate. In addition, these samples produced a red shift of the maximum light absorption of the dye Congo red. A spectrum with a maximum intensity at 540 nm was exhibited by a mixture of protein aggregates and Congo red after subtraction of the absorption spectra of the two components taken separately. Both of these observations are indicative of the presence of amyloid-based structure (11, 12). Finally, the addition of Congo red to a sample with acylphosphatase fibrils produced the characteristic green birefringence under cross-polarized light (Fig. 3). The development of green birefringence is highly diagnostic for the presence of amyloid (20). In summary, the acylphosphatase aggregates formed after long incubation times respond positively to all diagnostic tests for the presence of amyloid fibrils. Moreover, the various structural changes observed during their formation appear similar to those that have been observed when fibrils are formed in vitro from proteins associated with amyloidotic diseases (21).
Figure 3.
Optical microscope image obtained under cross-polarized light and containing acylphosphatase aggregates stained with Congo red. The protein aggregate was obtained after 14 days of incubation in 25% (vol/vol) TFE. The photograph shows the blots of green birefringence coming from regions rich in amyloid fibrils.
DISCUSSION
The findings reported here, along with the previous observations of fibrils formed by the SH3 domain at low pH (5), shows that amyloid fibril formation is not simply a feature of the few proteins found in the amyloid plaques that accumulate in vivo. We have succeeded in designing conditions to produce fibrils from a protein of choice that is not associated with any of the amyloid diseases identified so far. Unlike the SH3 domain, acylphosphatase does not possess an exceptionally high propensity to form β-sheet structure. Moreover, the partially denatured state of acylphosphatase that is initially present under the conditions studied is a conformation containing a significant amount of α-helical structure but little, if any, β-sheet structure. It appears, therefore, that the conformation of the partially folded state is not by itself a critical feature of fibril formation. Rather, we suggest that the basis for amyloidogenesis is the presence of partially denaturing conditions that destabilize the native fold of the protein but do not preclude noncovalent interactions between the various groups within the protein. Such conditions are likely to result in partially folded states of proteins, such as those observed here and for several of the naturally occurring amyloidogenic proteins under conditions where fibrils form readily (1). Moreover, such species are likely to have a high propensity to aggregate as both hydrophobic residues, and main-chain hydrogen bond donors and acceptors are at least partially exposed.
If aggregation occurs between proteins in their native conformations, the intramolecular interactions within the native fold, which render the majority of hydrophobic groups and the main-chain hydrogen bond acceptors and donors inaccessible, reduce very substantially the potential for intermolecular bonding. Such bonding is therefore likely to be weak and the corresponding aggregation processes reversible. By contrast, aggressively denaturing conditions, such as high concentrations of urea or guanidinium chloride, destabilize all of the noncovalent interactions that the polypeptide chains can potentially form, and a soluble highly unfolded state is usually favored under these conditions (22). Conditions in which partially folded states of proteins are formed are therefore likely to be favorable for aggregation compared with those in which native or highly denatured states are present. In the case of TFE, very high concentrations of the alcohol [>35% (vol/vol)] do not favor aggregation of the protein, probably because of increased intramolecular hydrogen bonding and a reduced hydrophobic effect. These findings are consistent with the results of studies in which N-acylation of peptides was used to vary their α-helical propensity and amyloidogenic potential (23, 24). Interestingly, only peptides with acyl chains of medium length and having medium propensity to form α-helical structure were found to generate β-sheet structure. Peptides with very short or very long acyl chains were not found to form such structure, an observation attributed to the initial α-helical conformation being too unstable or too stable, respectively, for the ordered aggregation process to take place (23).
To obtain ordered structures rather than amorphous precipitates, it appears that the solution conditions for some proteins must be such that the nucleation and growth of the fibrils is slow. In this sense, the process of fibril formation is likely to be similar to that of crystal growth, which is well established to take place under near-equilibrium conditions (25). In the present experiments, the slow development of ordered fibrils through structural reorganization under carefully chosen solution conditions illustrates this feature clearly. It is important to make it clear that the particular conditions we have used are not suggested to be universally appropriate for fibril formation by proteins. As with the formation of crystals, the conditions for optimum growth are likely to be highly dependent on the properties of the particular system under study. We believe, however, that conditions that will generally give rise to efficient formation of amyloid will be partially denaturing and will be such as to allow the slow and controlled growth of aggregated species.
These findings have implications for understanding the origin of amyloid fibril formation in disease states. In a normally functioning organism, the propensities of polypeptide chains to aggregate will be reduced by the formation and intrinsic stability of the native state under physiological conditions. Moreover, the proteolytic machinery for the degradation of unfolded and misfolded polypeptide chains, and the presence of molecular chaperones that restrict the intermolecular interactions of unfolded polypeptides, constitute a further element for the prevention of aggregation (26). Aggregation can, however, occur if proteins are exposed to destabilizing conditions where such controls are not effective. It has been proposed that local environments in which the pH is low, for example in lysosomes, may be particularly favorable for formation of amyloid (13, 27). Other factors, such as the presence of amphipathic compounds such as phospholipids, have also been suggested to be involved, particularly in facilitating elongation of fibrils (28).
The present results suggest that, provided appropriate conditions are maintained over prolonged periods of time, the formation of ordered amyloid protofilaments and fibrils could be an intrinsic property of many polypeptide chains, rather than being a phenomenon limited to very few aberrant sequences. Understanding the origin of a particular amyloidosis, therefore, is likely to require an understanding of the specific kinetic and thermodynamic factors that influence the balance of the equilibria between the various possible states that are accessible to a given amyloidogenic protein. The ability to design conditions under which fibrillation can be observed with a wider range of proteins than those so far identified in specific diseases gives us the opportunity to investigate in detail the mechanism of the underlying process and hence to explore the factors that predispose individual sequences to form ordered aggregates under particular conditions. This could be an important factor in the development of strategies to combat the formation of amyloid deposition in those diseases with which they are associated.
Acknowledgments
This is a contribution from the Oxford Centre for Molecular Sciences, which is funded by the Engineering and Physical Sciences Research Council, the Biotechnology and Biological Sciences Research Council, and the Medical Research Council. The work has also been supported by funds from the Consiglio Nazionale delle Ricerche (Target Project Biotechnology), from Ministero dell’Universitá e della Ricerca Scientifica e Technologica (Project Structural Biology), from the European Community (Contract ERB BIO4-CT96-0517), and from the Cassa di Risparmio di Firenze. F.C. was supported by a grant from the European Community (EC contract no. B104-CT96-5113). A.C. is funded by the Wellcome Trust, which also purchased the electron microscope. The research of C.M.D. is supported in part by the Wellcome Trust and through an International Research Scholars award from the Howard Hughes Medical Institute.
ABBREVIATION
- TFE
trifluoroethanol
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
A Commentary on this article begins on page 3342.
References
- 1.Kelly J W. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 2.Coelho T. Curr Opin Neurol. 1996;9:355–359. [PubMed] [Google Scholar]
- 3.Lansbury P T., Jr Acc Chem Res. 1996;29:317–321. [Google Scholar]
- 4.Sunde M, Blake C C F. Adv Protein Chem. 1997;50:123–159. doi: 10.1016/s0065-3233(08)60320-4. [DOI] [PubMed] [Google Scholar]
- 5.Guijarro J I, Sunde M, Jones J A, Campbell I D, Dobson C M. Proc Natl Acad Sci USA. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefani M, Taddei N, Ramponi G. Cell Mol Life Sci. 1997;53:141–151. doi: 10.1007/PL00000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiti F, van Nuland N A J, Taddei N, Magherini F, Stefani M, Ramponi G, Dobson C M. Biochemistry. 1998;37:1447–1455. doi: 10.1021/bi971692f. [DOI] [PubMed] [Google Scholar]
- 8.van Nuland N A J, Chiti F, Taddei N, Raugei G, Ramponi G, Dobson C M. J Mol Biol. 1998;283:883–891. doi: 10.1006/jmbi.1998.2009. [DOI] [PubMed] [Google Scholar]
- 9.Chiti F, Taddei N, van Nuland N A J, Magherini F, Stefani M, Ramponi G, Dobson C M. J Mol Biol. 1998;283:893–903. doi: 10.1006/jmbi.1998.2010. [DOI] [PubMed] [Google Scholar]
- 10.Modesti A, Taddei N, Bucciantini M, Stefani M, Colombini B, Raugei G, Ramponi G. Protein Expression Purif. 1995;6:799–805. doi: 10.1006/prep.1995.0011. [DOI] [PubMed] [Google Scholar]
- 11.Klunk W E, Pettegrew J W, Abraham D J. J Histochem Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 12.LeVine H., III Amyloid. 1995;2:1–6. [Google Scholar]
- 13.Lai Z, Lashuel H A, Kelly J W. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 14.Booth D R, Sunde M, Bellotti V, Robinson C V, Hutchinson W L, Fraser P E, Hawkins P N, Dobson C M, Radford S E, Blake C C F, Pepys M B. Nature (London) 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 15.Prusiner S B. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 16.Wood S J, Mackenzie L, Maleef B, Hurle M R, Wetzel R. J Biol Chem. 1996;271:4086–4092. doi: 10.1074/jbc.271.8.4086. [DOI] [PubMed] [Google Scholar]
- 17.Thomas P D, Dill K A. Protein Sci. 1993;2:2050–2065. doi: 10.1002/pro.5560021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiraki K, Nishikawa K, Goto Y. J Mol Biol. 1995;245:180–194. doi: 10.1006/jmbi.1994.0015. [DOI] [PubMed] [Google Scholar]
- 19.Muga A, Arrondo J L R, Bellon T, Sancho J, Bernabeu C. Arch Biochem Biophys. 1993;300:451–457. doi: 10.1006/abbi.1993.1061. [DOI] [PubMed] [Google Scholar]
- 20.Missmahal H P. In: Amyloidosis. Mandema E, Ruinen L, Scholten J H, Cohen A S, editors. Amsterdam: Excerpta Medica; 1968. pp. 22–29. [Google Scholar]
- 21.Lomakin A, Chung D S, Benedek G B, Kirschner D A, Teplow D B. Proc Natl Acad Sci. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanford C. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 23.Mihara H, Takahashi Y. Curr Opin Struct Biol. 1997;7:501–508. doi: 10.1016/s0959-440x(97)80113-3. [DOI] [PubMed] [Google Scholar]
- 24.Mihara H, Takahashi Y, Ueno A. Biopolymers. 1998;47:83–92. doi: 10.1002/(SICI)1097-0282(1998)47:1<83::AID-BIP9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 25.Mullin J W. Crystallization. London: Butterworths; 1972. pp. 174–232. [Google Scholar]
- 26.Hendrick J P, Hartl F U. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 27.Barrow C J, Zagorski M G. Science. 1991;253:179–182. doi: 10.1126/science.1853202. [DOI] [PubMed] [Google Scholar]
- 28.McLaurin J, Franklin T, Chakrabartty A, Fraser P E. J Mol Biol. 1998;278:183–194. doi: 10.1006/jmbi.1998.1677. [DOI] [PubMed] [Google Scholar]