Abstract
Trypanosoma brucei
thwarts the host immune response by replacing its variant surface glycoprotein (VSG). The actively transcribed VSG is located in one of ~20 telomeric expression sites (ES). Antigenic variation can occur by transcriptional switching, reciprocal translocations, or duplicative gene conversion events among ES or with the large repertoire of telomeric and non-telomeric VSG. In recently isolated strains, duplicative gene conversion occurs at a high frequency and predominates, but the switching frequency decreases dramatically upon laboratory-adaptation. Uniquely, T. brucei telomeres grow—apparently indefinitely—at a steady rate of 6–12 base pairs (bp) per population doubling (PD), but the telomere adjacent to an active ES undergoes frequent truncations. Using two-dimensional gel electrophoresis, we demonstrate that all of the chromosome classes of fast-switching and minimally propagated T. brucei have shorter telomeres than extensively propagated Lister 427 clones, suggesting a link between laboratory adaptation, telomere growth, and VSG switching rates.
Index Descriptors: Trypanosoma brucei, Antigenic Variation, Isolation History, Propagation History, Telomere Length, Strain Phylogeny, Variant Surface Glycoprotein
1. Introduction
Antigenic variation allows several pathogens to evade immune responses and prolong an infection. Borrelia burgdorferi, Pneumocystis carinii and Trypanosoma brucei, all evade the host immune response by sequential expression of different surface antigens that, in T. brucei, form a homogeneous, dense, coat, overlying the cytoplasmic membrane (Cross, 1975; Kutty, et al., 2001; Singh and Girschick, 2004). The T. brucei genome encodes hundreds of VSG (Berriman, et al., 2002), and the single transcribed VSG is located in a telomere-proximal expression site (ES) (de Lange and Borst, 1982). T. brucei has ~20 telomeric ES, whose transcription is strictly monoallelic (Vanhamme, et al., 2000). Antigenic switching can occur by a coupled activation and inactivation of transcription between two ES (in-situ switch), by reciprocal DNA translocation between two ES, or by a duplicative gene conversion event that places a new VSG into the transcribed ES (Horn and Cross, 1997; McCulloch, et al., 1997; Myler, et al., 1984; Rudenko, et al., 1996; van der Ploeg, et al., 1984a). Gene-conversion-mediated switching predominates in pleomorphic strains (Robinson, et al., 1999). Although we know the general molecular mechanisms underlying antigenic switching, little is known about its regulation.
The rate of VSG switching varies substantially among strains of T. brucei. Recent isolates have a high switching frequency of ~10−2–10−4/population doubling (PD) (Barry, 1997; Robinson, et al., 1999; Turner, 1997; Turner and Barry, 1989), and grow to lower densities in vitro, which complicates laboratory studies. Thus, isolates were often passaged sequentially in mice in the absence of immune selection (Turner, 1997). After several months, such parasites become monomorphic and more virulent, with as much as a 10,000-fold reduction of switching frequency (Horn and Cross, 1997; Lamont, et al., 1986; McCulloch, et al., 1997; Turner, 1997; Turner and Barry, 1989; van Deursen, et al., 2001). Unfortunately, nothing is known about the molecular changes that occur during laboratory adaptation — especially what mediates the reduction in switching frequency. Identifying the mediators of these changes would improve our understanding of antigenic variation.
The intriguing location of VSG ES led us to investigate, from several directions, whether telomere structure might play a role in regulating antigenic variation. T. brucei telomeres consist of hexameric TTAGGG-repeats that, together with their interacting proteins, protect chromosome ends from being recognized as double stranded breaks (Blackburn and Challoner, 1984; de Lange, 2002; van Steensel, et al., 1998). Uniquely, T. brucei telomeres grow at a steady rate of 6–12 bp/PD, but the ES telomere adjacent to the single transcribed VSG undergoes frequent truncations (Bernards, et al., 1983; Dreesen and Cross, 2006a; Horn and Cross, 1997; Myler, et al., 1988; Pays, et al., 1983; van der Ploeg, et al., 1984b). Although the mechanism and frequency of these truncations remains unknown, if they fall within the telomeric TTAGGG repeats they are elongated by telomerase at a rate that is inversely proportional to telomere length (Dreesen and Cross, 2006b; Glover and Horn, 2006; Horn, et al., 2000).
When the reverse-transcriptase component of telomerase is deleted, telomeres shorten at a rate of 3–6 bp/PD (Dreesen, et al., 2005). Upon reaching a discreet length, short silent ES telomeres stabilize within a distinct size range, maintained by a telomerase-independent mechanism (Dreesen and Cross, 2006b). Although a short telomere at an active ES is also stabilized, at least temporarily, subsequent breakage appears to catalyze its replacement by a different VSG and its associated telomere, through duplicative gene conversion (Dreesen and Cross, 2006a). To explain the apparent difference between active and silent ES, we recently proposed (Dreesen and Cross, 2006a; Dreesen, et al., 2007) that a double-stranded break at the transcribed ES could fall into the subtelomeric region and be repaired by break-induced replication, using another ES as template, with consequent duplication and activation of the resident VSG. We have speculated that telomere breakage and repair could increase the rate of antigenic switching at a short active ES telomere (Dreesen and Cross, 2006a; Dreesen, et al., 2007).
In this study, we measured telomere lengths in clones of some recent isolates whose histories were known, in the recently laboratory-adapted strain used for the genome project, and in several clones of the extensively propagated and widely used Lister 427 strain. We found that minimally propagated and recently laboratory-adapted strains have shorter telomeres than the extensively propagated Lister 427.
2. Materials and methods
2.1. History of T. brucei strains
The detailed lineages of T. brucei cell lines used in this study and by other laboratories are available at http://tryps.rockefeller.edu/trypanosome_pedigrees.html. The strain abbreviations used in this paper are indicated in parentheses and are summarized in Table 1. T. brucei S42 (RUMP102), STIB348 (RUMP106) and T. b. rhodesiense s427 (RUMP151) are isolates that have not been extensively propagated in the laboratory. S42 was isolated from a female warthog near Kirawira, Tanzania, in March 1966 (Baker, et al., 1967). It was minimally passaged before being tsetse-transmitted in November 1974. STIB366D was derived from a single metacyclic cell and was minimally propagated before freezing (by GAMC) as RUMP102. STIB246 was isolated from a Hartebeest in Serengeti in 1971 (Geigy and Kauffmann, 1973). After growth in mice and rats for a total of ~6 weeks, and cloning as STIB348, it was transmitted through tsetse in July 1975 (Leo Jenni, Swiss Tropical Institute, Basel). STIB348U was derived from a single metacyclic cell and frozen (by GAMC) as RUMP106 after passage through 2 irradiated mice. T. b. rhodesiense RUMP151 is derived from the true s427, which was isolated in 1960 from a sheep in south-east Uganda (Cunningham and Vickerman, 1962) and frozen as EATRO217. After its initial isolation, s427/EATRO217 was passaged through ~20 mice before being frozen (by GAMC) as RUMP151, which is highly resistant to human serum (GAMC, unpublished data), consistent with its isolation from an area of epidemic human sleeping sickness ((Cunningham and Vickerman, 1962) and M.P. Cunningham, personal communication). TREU927, was isolated in Kiboko, Kenya, in 1969 or 1970, and preserved after 3–11 passages (Goedbloed, et al., 1973). A clone of TREU927 (GUTat 10.1), after 27 passages in mice to render it more virulent (van Deursen, et al., 2001), was the source of DNA used for the first T. brucei complete genome sequence (Berriman, et al., 2005). The procyclic 927 cells from which we isolated DNA were derived from GUTat 10.1 that had been differentiated to procyclic forms in the laboratory of Scott Landfear and further propagated for 2–4 weeks. The origin of Lister 427 is unknown. It is not related to s427, as was generally assumed (Cross, 1975), and was maintained by frequent syringe passage in mice during indeterminate intervals from 1961–1967. Lister 427 ‘single-marker’ (SM) bloodstream and 29-13 procyclic forms (Wirtz, et al., 1999) are closely related to other extensively propagated clones of Lister 427 that have a low switching frequency of ~10−6–10−7/PD (Horn and Cross, 1997; Lamont, et al., 1986). The relative positions of clones RUMP150 and RUMP501 (these were tsetse-transmitted in 1983 and 2004, respectively) in the Lister 427 lineage are clear (http://tryps.rockefeller.edu/trypanosome_pedigrees.html), and their genotypic identity has been verified on several occasions (C.M.R. Turner and G.A.M. Cross, unpublished data). RUMP150 is genetically indistinguishable from the SM and 29-13 clones, but the degree of propagation of Lister 427 prior to its earlier incarnation as RUMP501 cannot be verified. RUMP100 and RUMP502 are descendants of EATRO795, isolated from a Zebu cow in Kenya (September, 1964), which has a high switching frequency (Onyango, et al., 1966; Robinson, et al., 1999; Turner, 1997; Turner and Barry, 1989). The ancestors of RUMP100 were recloned several times, with minimal passaging, prior to infecting a mouse with a single metacyclic cell, yielding STIB367H, from which the RUMP100 population was frozen (by GAMC) after one additional mouse passage. RUMP502 is one passage removed from GUP2900 (GUG359). Although we lack documentation of the lineage prior to GUP2900, RUMP502 was recently verified to be genetically indistinguishable from a verified sample of EATRO795 (C. M. R. Turner, personal communication).
Table 1. T. brucei.
strains and clone designations
| Strain and clone | Origin | Year | References |
|---|---|---|---|
| Minimally propagated clones | |||
| EATRO795-RUMP100/502 | Kenya | 1964 | Onyango et al, 1966; Turner, 1997; Robinson et al., 1999. |
| S42-RUMP102 | Tanzania | 1966 | Baker et al., 1967. |
| STIB246/348-RUMP106 | Tanzania | 1971 | Geigy and Kauffmann, 1973. |
| s427/EATRO217-RUMP151 | Uganda | 1960 | Cunningham and Vickerman, 1962. |
|
| |||
| Intermediate laboratory-adapted clone | |||
| TREU927 | Kenya | 1969/70 | Goedbloed et al., 1973; van Deursen et al., 2001; Berriman et al., 2005. |
|
| |||
| Extensively propagated (origin unknown) Lister 427 clones | |||
| SM (bloodstream forms) | Wirtz et al., 1999. | ||
| 29-13 (procyclic forms) | |||
| RUMP150 | |||
| RUMP501 | |||
2.2. Cultivation of T. brucei
Lister 427 clones were cultured in HMI-9 at 37°C (Doyle, et al., 1980; Hirumi and Hirumi, 1989) or in SDM-79 at 27°C (Brun and Schonenberger, 1979). TREU927 procyclic forms were grown in Cunningham’s medium (Cunningham, 1977). All other strains were grown for three days in female CD-1 AKA ICR (Charles River) mice. Where larger quantities were required, trypanosomes were grown for a further 3 days in male Sprague-Dawley rats (Charles River), then purified by centrifugation and passage through a DEAE cellulose column (Taylor, et al., 1974).
2.3. DNA isolation and terminal restriction fragment analysis
DNA was isolated from freshly harvested cells, as previously described (Munoz-Jordan, et al., 2001), except for clones 100 and 502, where it was isolated from frozen cell pellets that had been stored at −70°C, which did not affect DNA quality. Briefly, ~2 × 108 cells were washed twice in TDB (5 mM KCl, 80 mM NaCl, 1 mM MgSO4, 20 mM Na2HPO4, 2 mM NaH2PO4, 20 mM glucose, pH 7.7), resuspended in 1ml TNE (10 mM Tris pH 7.4, 100 mM NaCl, 10 mM EDTA), lysed by adding 1 ml TNES (TNE with 1% SDS) supplemented with 100 μg/ml proteinase K (Roche), and incubated overnight at 55°C. DNA was extracted with phenol/chloroform/isoamyl-alcohol (PCI), precipitated and resuspended in TNE with 100 μg/ml RNase A (Sigma). After incubation for 3 h at 37°C, TNES with 100 μg/ml proteinase K was added, and samples were incubated overnight at 55°C. Lastly, DNA was PCI-extracted, precipitated and resuspended in T10E1 (10 mM Tris pH 7.4, 1 mM EDTA). Restriction enzyme digestion took place overnight according to the manufacture’s conditions (New England Biolabs). DNA restriction fragments were separated on a 0.8% agarose gel. Equal loading was ensured by measuring DNA concentration after digestion and by staining of agarose gels with Ethidium Bromide. Bulk telomere length was analyzed by denaturing in-gel hybridization: gels were dried at room temperature, denatured, neutralized, and hybridized at 50°C using a radiolabeled (TTAGGG)4-probe in Church Mix [0.5 M NaPO4 (pH 7.2), 1 mM EDTA (pH 8.0), 7% SDS and 1% BSA]. Gels were subsequently washed three times for 30 min with 4x SSC (1x SSC is 15 mM tri-sodium citrate and 150 mM NaCl) and once for 30 min with 4x SSC + 0.1% SDS at 20°C, then exposed to a phosphorimager screen for various times (Dreesen, et al., 2005; Hemann and Greider, 1999). Signals were quantified using ImageQuant software (Molecular Dynamics).
2.4. 2-dimensional DNA electrophoresis
DNA agarose plugs were prepared and chromosome-sized DNA was separated by Rotating Agarose Gel Electrophoresis (Stratagene, Inc), as described previously (Navarro and Cross, 1996; Navarro and Cross, 1998). Gels were stained with 2 μg/ml Ethidium bromide in 0.5 × TAE. Entire lanes, containing MBC, IC and MC were cut from the gel, placed in 15 ml Falcon tubes, pre-incubated in digestion buffer, and digested overnight with RsaI, MboI and AluI, using gentle rotation. Fresh aliquots of enzymes were added the next morning and incubation extended for 20 h. Digested plugs were equilibrated in 0.5 × TAE for 30 min and embedded in 0.8% Agarose in 0.5 × TAE. Digested fragments were separated in the second dimension for 1 h at 90 v and 15 h at 40 v. In-gel hybridization was performed as described above (Dreesen, et al., 2005; Hemann and Greider, 1999).
2.5 Telomere size distribution
Using Image Quant software, each gel lane was graphically partitioned into 30 equally sized rectangle. The hybridization signal in each compartment was quantified and subsequently represented as the percentage of the total signal in the entire lane.
3. Results
3.1 Telomere length in laboratory-adapted less-propagated strains
Based on our experimental results with telomerase-deficient cells, we had the idea that telomere length at the active ES might affect the frequency of antigenic variation. This notion led us to measure telomere length in clones derived from some well-characterized pleomorphic and monomorphic strains, whose history was fairly clear.
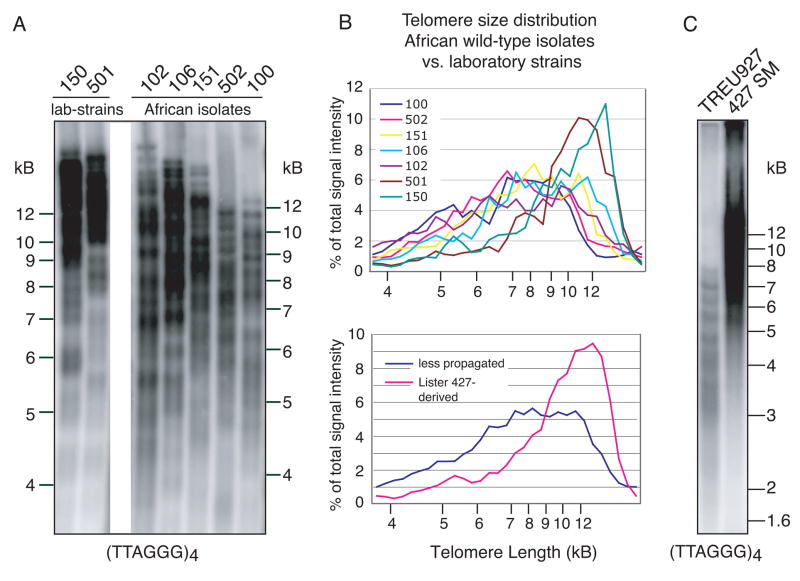
The telomeres of the four Lister 427-derived clones (only 150 and 501 are shown in Fig. 1A) are strikingly similar in length, ranging from 3–20 kB, with an average of ~15 kB, as previously shown (Dreesen, et al., 2005; Munoz-Jordan, et al., 2001). Telomeres of less propagated isolates ranged in size from 3–12 kB, with an average of ~8–10 kB (Fig. 1A), and quantification of these data confirmed that the average telomere length was shorter in less propagated strains than in two clones of Lister 427 (Fig. 1B). TREU927 is of particular interest, since it was recently laboratory-adapted and used for the T. brucei genome project (Berriman, et al., 2005). As described elsewhere (van Deursen, et al., 2001), TREU927 clone 4 was passaged 27 times in mice to increase its virulence, and predominately expresses VSG GUTat 10.1. Even after 4 more months in vitro culture and ~ 75 additional doublings as procyclic forms, our sample of TREU927 has much shorter telomeres than the extensively in vitro cultured Lister 427 SM (Fig. 1C).
Fig. 1.
Analysis of telomere length in recently isolated and laboratory-adapted T. brucei. Genomic DNA from laboratory-adapted strains and wild-type isolates was digested with AluI plus MboI and separated on an 0.8% agarose gel. Telomere restriction fragments were visualized by in-gel hybridization using a radiolabeled (TTAGGG)4 probe. (A) Lister 427 clones RUMP150 and 501 have long telomeres (average ~15 kB). Less propagated isolates (clones RUMP102, 106, 151, 502 and 100) have shorter telomeres (average ~ 8–10 kB). (B) Upper panel: telomere size distribution as a percentage of total telomere signal. Lower panel: average telomere size distribution of two laboratory-adapted clones (RUMP150 and 501) and five recent isolates (RUMP102, 106, 151, 502 and 100). (C) Comparison of telomere length between recently laboratory-adapted TREU927 and extensively propagated Lister 427 SM clone. Despite equal loading, signal intensity for TREU927 is lower, due to its shorter telomere tracts. Please note that the resolving power of agarose gels at sizes larger than ~15 kB is limited, which is the average telomere length in Lister 427 SM (Munoz-Jordan, et al., 2001). Size markers are as indicated.
Laboratory-adapted TREU927 and Lister 427 SM stably express one particular VSG. Constitutive elongation of all non-transcribed telomeres could account for the telomere length difference between the extensively propagated Lister 427 and the recently adapted strain TREU927. We predict that continuous in vitro growth of TREU927, with stable expression of GUTat 10.1, would eventually result in similarly elongated telomeres.
3.2 Telomere length analysis by 2-dimensional gel electrophoresis
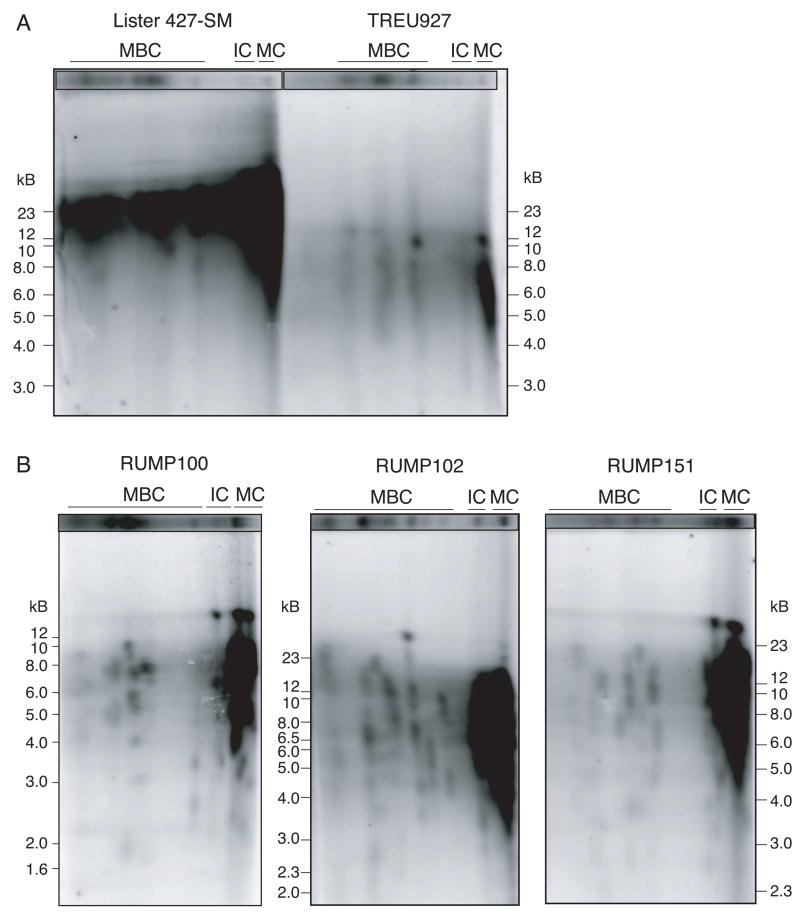
The T. brucei genome consists of 11 diploid megabase chromosomes (MBC), which contain most VSG ES and all essential housekeeping genes, a few intermediate chromosomes (IC), whose function remains unclear but which often contain an ES, and ~ 100 minichromosomes (MC) that contain only 177-bp repeats, VSGs and telomeric repeats. To address whether telomere length varied among the different chromosome types, we used 2-dimensional gel electrophoresis to separate chromosomes and then their telomeres. In the first dimension, MBC, IC and MC were separated by Rotating Agarose Gel Electrophoresis. Under the conditions used, MBC were well separated and, as previously described, we observed extensive size polymorphisms among different strains (Fig. 2A) (Melville, et al., 1998; Melville, et al., 2000). MC are not well separated under these conditions and migrate at the bottom of the gel (Fig. 2A). To analyze telomere lengths on individual chromosome types of Lister 427 clones, entire lanes (shown in Fig. 2A) were cut out of the gel, digested with restriction enzymes, and perpendicularly embedded in a conventional 0.8% agarose gel. The Ethidium-bromide-stained chromosomal separations are graphically inserted on top of each gel and serve as a loading indicator for the telomere blots (Fig. 2B and C). Telomere restriction fragments were separated in the second dimension and visualized by in-gel hybridization (Dreesen and Cross, 2006b; Dreesen, et al., 2005). Within the well-separated MBC telomeres, individual spots representing clusters of telomeres of particular lengths, can be distinguished (Fig. 2B, arrows). Consistent with previous observations using individual telomeric VSG as hybridization probes, all MBC telomeres migrate well above the 12 kB molecular weight marker (Dreesen and Cross, 2006a; Dreesen and Cross, 2006b; Dreesen, et al., 2005; Horn and Cross, 1997; Munoz-Jordan, et al., 2001). The abundant MC telomeres (~200) migrate within the same size range (12–23 kB) (Fig. 2B, circle). As a control, we visualized telomere length in telomerase-deficient T. brucei that had been continuously cultured for over 2.5 years (Fig. 2C). We previously reported that long-term cultured telomerase-deficient cells lose MC (Dreesen and Cross, 2006b), so the telomere signal was very low (Fig. 2C). Furthermore, this control confirmed that the telomere signal observed in Lister 427 SM was derived from terminal TTAGGG repeats.
Fig. 2.
Telomere size distribution among chromosome classes analyzed by 2-dimensional gel electrophoresis. (A) Chromosome size polymorphism among T. brucei clones RUMP151, 102, 100, TREU927 and Lister 427 SM. Chromosomes were separated by Rotating Agarose Gel Electrophoresis and stained with Ethidium bromide. (B) Entire lanes from (A) were digested with a mixture of MboI, AluI and RsaI, and perpendicularly embedded in 0.8% agarose to separate telomere terminal restriction fragments (Ethidium-bromide-stained chromosomal DNA profiles were graphically inserted on top of the gel). Telomere restriction fragments were visualized using a (TTAGGG)4-probe. MBC are well separated (arrows) whereas IC and MC (circled) are not. Exposure time was 6 h. (C) Direct comparison of Lister 427-SM parental and extensively propagated telomerase-deficient clone. Exposure time was 15 hours. Size markers are as indicated. It is unclear whether the genomic rearrangements (*) in the TERT null clone are derived from MC or IC.
Next, we compared telomere lengths between the Lister 427 SM clone, the recently laboratory-adapted genome strain TREU927, and minimally propagated strains represented by RUMP clones 100, 102 and 151 (Fig. 3). Two-dimensional gel electrophoresis confirmed that the telomeres of TREU927, especially those in the MC range, are very short, ranging from 3–10 kB (Fig. 1C and 3A). Similarly, in RUMP100, a clone of the well-characterized rapid-switching EATRO795 strain, the telomeres run from 5–10 kB (Fig. 3B, left panel). Most 102 and 151 MBC telomeres also run at significantly lower sizes (Fig. 3B, middle and right panels). No systematic size differences were observed between MBC, IC and MC telomeres.
Fig. 3.
Telomere size distribution among chromosome classes of Lister 427-SM and several less propagated clones, analysed by two-dimensional gel electrophoresis. (A) SM and TREU927. Ethidium-bromide-stained chromosomal separations are shown on the top and serve as a loading indicator. Exposure time was ~48 hours. Size markers are indicated on the right. (B) RUMP100 (left), 102 (middle) and 151 (right). Exposure times were optimized to most effectively visualize individual MBC telomeres and telomere clusters.
4. Discussion
In this study, we analyzed telomere length in clones representing five ‘recent’ T. brucei isolates, the recently laboratory-adapted TREU927, and the extensively propagated Lister 427. We found that the recent isolates have much shorter telomeres than extensively propagated clones, and this is true for MBC, IC and MC. This is the most extensive analysis of telomere length in different T. brucei isolates. Telomeres in different T. cruzi isolates vary in size from 1–10 kB (Freitas-Junior, et al., 1999). More recently, telomere length was monitored during growth of Leishmania major, L. tarentolae and Crithidia fasciculata (Genest and Borst, 2007). Telomeres of L. major and L. tarentolae, but not C. fasciculata, grew by ~1 bp/PD. In contrast, T. brucei telomeres grow by 6–12 bp/PD. Furthermore, a short transcribed ES telomere is exceedingly rapidly elongated by telomerase, but elongation is counteracted by frequent telomere breakage (Bernards, et al., 1983; Dreesen and Cross, 2006b; Glover and Horn, 2006; Horn and Cross, 1997; Horn, et al., 2000; Pays, et al., 1983).
Regular and apparently unlimited growth of transcriptionally silent telomeres, which is so far unique to T. brucei, can explain why clones that have been extensively propagated in vitro have long telomeres. We would predict that longer propagation of more recently adapted strains, in the absence of immune selection, would also lead to an increase in telomere length, although we cannot exclude the possibility that the observed differences are due to inherent differences in telomere biology among various strains. We cannot yet explain why the non-transcribed telomeres of more recent isolates would be inherently shorter. However, if trypanosomes in the wild are switching VSG at far higher rates than after laboratory adaptation, which is a highly favored idea, supported by experimental data (Horn and Cross, 1997; Lamont, et al., 1986; Robinson, et al., 1999; Turner, 1997; Turner and Barry, 1989), the frequent breakage of transcriptionally active ES telomeres, coupled with frequent inter-telomeric VSG recombination events among all chromosome classes, would counter the intrinsic lengthening of silent telomeres that occurs in the absence of selection for such recombination events, under the usual in-vitro propagation conditions. Furthermore, the generally elevated rate of recombination at chromosome ends, as observed in other organisms, could lead to a general involvement non-expression site telomeres, resulting in their overall shortening.
We therefore speculate that telomere growth and VSG switching are inversely related.
Acknowledgments
We thank Marco Sanchez and Scott Landfear for supplying T. brucei 927 procyclic forms, Leo Jenni for various tsetse-transmitted clones, Jenny Li for technical assistance, Michael Turner for checking strain genotypes for us, on several occasions, and Piet Borst for communicating unpublished results. We are grateful to Titia de Lange, Ed Louis, Heinrich zu Dohna, Pradeep Patnaik and the members of the Cross laboratory for inspiring discussions, and to Veena Mandava and Luisa Figueiredo for suggestions on the manuscript. This work was supported by NIH grants AI21729 and AI50614.
Abbreviations
- EATRO
East African Trypanosomiasis Research Organization
- ES
expression site
- IC
intermediate-size chromosomes
- GU
Glasgow University
- MBC
megabase chromosomes
- MC
minichromosomes
- PD
population doubling
- RUMP
Rockefeller University Molecular Parasitology
- STIB
Swiss Tropical Institute Basel
- TREU
Trypanosomiasis Research Edinburgh University
- VSG
variant surface glycoprotein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker JR, Sachs R, Laufer I. Trypanosomes of wild mammals in an area northwest of the Sergenti National Park, Tanzania. Zeitschrift fur Tropenmedizin und Parasitologie. 1967;18:280–284. [PubMed] [Google Scholar]
- Barry JD. The relative significance of mechanisms of antigenic variation in African trypanosomes. Parasitology Today. 1997;13:212–218. doi: 10.1016/s0169-4758(97)01039-9. [DOI] [PubMed] [Google Scholar]
- Bernards A, Michels PA, Lincke CR, Borst P. Growth of chromosome ends in multiplying trypanosomes. Nature. 1983;303:592–597. doi: 10.1038/303592a0. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Berriman M, Hall N, Sheader K, Bringaud F, Tiwari B, Isobe T, Bowman S, Corton C, Clark L, Cross GA, Hoek M, Zanders T, Berberof M, Borst P, Rudenko G. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol Biochem Parasitol. 2002;122:131–140. doi: 10.1016/s0166-6851(02)00092-0. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Challoner PB. Identification of a telomeric DNA sequence in Trypanosoma brucei. Cell. 1984;36:447–457. doi: 10.1016/0092-8674(84)90238-1. [DOI] [PubMed] [Google Scholar]
- Brun R, Schonenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Tropica. 1979;36:289–292. [PubMed] [Google Scholar]
- Cross GAM. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- Cunningham I. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. The Journal of Protozoology. 1977;24:325–329. doi: 10.1111/j.1550-7408.1977.tb00987.x. [DOI] [PubMed] [Google Scholar]
- Cunningham MP, Vickerman K. Antigenic analysis in the Trypanosoma brucei group, using the agglutination reaction. Trans R Soc Trop Med Hyg. 1962;56:48–59. doi: 10.1016/0035-9203(62)90088-3. [DOI] [PubMed] [Google Scholar]
- de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- de Lange T, Borst P. Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature. 1982;299:451–453. doi: 10.1038/299451a0. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Hirumi H, Hirumi K, Lupton EN, Cross GAM. Antigenic variation in clones of animal-infective Trypanosoma brucei derived and maintained in vitro. Parasitology. 1980;80:359–369. doi: 10.1017/s0031182000000810. [DOI] [PubMed] [Google Scholar]
- Dreesen O, Cross GAM. Consequences of telomere shortening at an active VSG expression site in telomerase-deficient Trypanosoma brucei. Eukaryotic Cell. 2006a;5:2114–2119. doi: 10.1128/EC.00059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesen O, Cross GAM. Telomerase-Independent Stabilization of Short Telomeres in Trypanosoma brucei. Molecular and Cellular Biology. 2006b;26:4911–4919. doi: 10.1128/MCB.00212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesen O, Li B, Cross GAM. Telomere structure and shortening in telomerase-deficient Trypanosoma brucei. Nucleic Acids Research. 2005;33:4536–4543. doi: 10.1093/nar/gki769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesen O, Li B, Cross GAM. Telomere structure and function in trypanosomes: a proposal. Nature Reviews Microbiology. 2007;5:70–75. doi: 10.1038/nrmicro1577. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior LH, Porto RM, Pirrit LA, Schenkman S, Scherf A. Identification of the telomere in Trypanosoma cruzi reveals highly heterogeneous telomere lengths in different parasite strains. Nucleic Acids Research. 1999;27:2451–2456. doi: 10.1093/nar/27.12.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigy R, Kauffmann M. Sleeping sickness survey in the Serengeti area (Tanzania) 1971. I. Examination of large mammals for trypanosomes. Acta Tropica. 1973;30:12–23. [PubMed] [Google Scholar]
- Genest PA, Borst P. Analysis of telomere length variation in Leishmania over time. Molecular and Biochemical Parasitology. 2007;151:213–215. doi: 10.1016/j.molbiopara.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Glover L, Horn D. Repression of polymerase I-mediated gene expression at Trypanosoma brucei telomeres. EMBO Reports. 2006;7:93–99. doi: 10.1038/sj.embor.7400575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedbloed E, Ligthart GS, Minter DM, Wilson AJ, Dar FK, Paris J. Serological studies of trypanosomiasis in East Africa. II. Comparisons of antigenic types of Trypanosoma brucei subgroup organisms isolated from wild tsetse flies. Annals of Tropical Medicine and Parasitology. 1973;67:31–43. [PubMed] [Google Scholar]
- Hemann MT, Greider CW. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Research. 1999;27:3964–3969. doi: 10.1093/nar/27.20.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. Journal of Parasitology. 1989;75:985–989. [PubMed] [Google Scholar]
- Horn D, Cross GAM. Analysis of Trypanosoma brucei VSG expression site switching in vitro. Molecular and Biochemical Parasitology. 1997;84:189–201. doi: 10.1016/s0166-6851(96)02794-6. [DOI] [PubMed] [Google Scholar]
- Horn D, Spence C, Ingram AK. Telomere maintenance and length regulation in Trypanosoma brucei. EMBO Journal. 2000;19:2332–2339. doi: 10.1093/emboj/19.10.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutty G, Ma L, Kovacs JA. Characterization of the expression site of the major surface glycoprotein of human-derived Pneumocystis carinii. Molecular Microbiology. 2001;42:183–193. doi: 10.1046/j.1365-2958.2001.02620.x. [DOI] [PubMed] [Google Scholar]
- Lamont GS, Tucker RS, Cross GAM. Analysis of antigen switching rates in Trypanosoma brucei. Parasitology. 1986;92:355–367. doi: 10.1017/s003118200006412x. [DOI] [PubMed] [Google Scholar]
- McCulloch R, Rudenko G, Borst P. Gene conversions mediating antigenic variation in Trypanosoma brucei can occur in variant surface glycoprotein expression sites lacking 70-base-pair repeat sequences. Molecular and Cellular Biology. 1997;17:833–843. doi: 10.1128/mcb.17.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville SE, Leech V, Gerrard CS, Tait A, Blackwell JM. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Molecular and Biochemical Parasitology. 1998;94:155–173. doi: 10.1016/s0166-6851(98)00054-1. [DOI] [PubMed] [Google Scholar]
- Melville SE, Leech V, Navarro M, Cross GAM. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Molecular Biochemical Parasitology. 2000;111:261–273. doi: 10.1016/s0166-6851(00)00316-9. [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Cross GAM, de Lange T, Griffith JD. t-loops at trypanosome telomeres. EMBO Journal. 2001;20:579–588. doi: 10.1093/emboj/20.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myler PJ, Aline RF, Jr, Scholler JK, Stuart KD. Changes in telomere length associated with antigenic variation in Trypanosoma brucei. Molecular and Biochemical Parasitology. 1988;29:243–250. doi: 10.1016/0166-6851(88)90079-5. [DOI] [PubMed] [Google Scholar]
- Myler PJ, Allison J, Agabian N, Stuart K. Antigenic variation in African trypanosomes by gene replacement or activation of alternate telomeres. Cell. 1984;39:203–211. doi: 10.1016/0092-8674(84)90206-x. [DOI] [PubMed] [Google Scholar]
- Navarro M, Cross GAM. DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Molecular and Cellular Biology. 1996;16:3615–3625. doi: 10.1128/mcb.16.7.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M, Cross GAM. In situ analysis of a variant surface glycoprotein expression-site promoter region in Trypanosoma brucei. Molecular and Biochemical Parasitology. 1998;94:53–66. doi: 10.1016/s0166-6851(98)00049-8. [DOI] [PubMed] [Google Scholar]
- Onyango RJ, Van Hoeve K, De Raadt P. The epidemiology of Trypanosoma rhodesiense sleeping sickness in Alego location, Central Nyanza, Kenya. I. Evidence that cattle may act as reservoir hosts of trypanosomes infective to man. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1966;60:175–182. doi: 10.1016/0035-9203(66)90024-1. [DOI] [PubMed] [Google Scholar]
- Pays E, Laurent M, Delinte K, Van Meirvenne N, Steinert M. Differential size variations between transcriptionally active and inactive telomeres of Trypanosoma brucei. Nucleic Acids Research. 1983;11:8137–8147. doi: 10.1093/nar/11.23.8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Burman N, Melville SE, Barry JD. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Molecular and Cellular Biology. 1999;19:5839–5846. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G, McCulloch R, Dirks-Mulder A, Borst P. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Molecular and Biochemical Parasitology. 1996;80:65–75. doi: 10.1016/0166-6851(96)02669-2. [DOI] [PubMed] [Google Scholar]
- Singh SK, Girschick HJ. Molecular survival strategies of the Lyme disease spirochete Borrelia burgdorferi. The Lancet Infectious Disease. 2004;4:575–583. doi: 10.1016/S1473-3099(04)01132-6. [DOI] [PubMed] [Google Scholar]
- Taylor AE, Lanham SM, Williams JE. Influence of methods of preparation on the infectivity, agglutination, activity, and ultrastructure of bloodstream trypanosomes. Experimental Parasitology. 1974;35:196–208. doi: 10.1016/0014-4894(74)90023-x. [DOI] [PubMed] [Google Scholar]
- Turner CM. The rate of antigenic variation in fly-transmitted and syringe-passaged infections of Trypanosoma brucei. FEMS Microbiology Letters. 1997;153:227–231. doi: 10.1111/j.1574-6968.1997.tb10486.x. [DOI] [PubMed] [Google Scholar]
- Turner CM, Barry JD. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology. 1989;99:67–75. doi: 10.1017/s0031182000061035. [DOI] [PubMed] [Google Scholar]
- van der Ploeg LH, Cornelissen AW, Michels PA, Borst P. Chromosome rearrangements in Trypanosoma brucei. Cell. 1984a;39:213–221. doi: 10.1016/0092-8674(84)90207-1. [DOI] [PubMed] [Google Scholar]
- van der Ploeg LH, Liu AY, Borst P. Structure of the growing telomeres of Trypanosomes. Cell. 1984b;36:459–468. doi: 10.1016/0092-8674(84)90239-3. [DOI] [PubMed] [Google Scholar]
- van Deursen FJ, Shahi SK, Turner CM, Hartmann C, Guerra-Giraldez C, Matthews KR, Clayton CE. Characterisation of the growth and differentiation in vivo and in vitro-of bloodstream-form Trypanosoma brucei strain TREU 927. Molecular and Biochemical Parasitology. 2001;112:163–171. doi: 10.1016/s0166-6851(00)00359-5. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Vanhamme L, Poelvoorde P, Pays A, Tebabi P, Van Xong H, Pays E. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Molecular Microbiology. 2000;36:328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Molecular and Biochemical Parasitology. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]