Abstract
Indoor mold concentrations were measured in the dust of moldy homes (MH) and reference homes (RH) by quantitative PCR (QPCR) assays for 82 species or related groups of species (assay groups). About 70% of the species and groups were never or only rarely detected. The ratios (MH geometric mean : RH geometric mean) for 6 commonly detected species (Aspergillus ochraceus, A. penicillioides, A. unguis, A. versicolor, Eurotium group, and Cladosporium sphaerospermum) were > 1 (Group I). Logistic regression analysis of the sum of the logs of the concentrations of Group I species resulted in a 95% probability for separating MH from RH. These results suggest that it may be possible to evaluate whether a home has an abnormal mold condition by quantifying a limited number of mold species in a dust sample. Also, four common species of Aspergillus were quantified by standard culturing procedures and their concentrations compared to QPCR results. Culturing underestimated the concentrations of these four species by 2 to 3 orders of magnitude compared to QPCR.
Introduction
Mold growth in homes, schools and other buildings has become a major issue of public concern. The use of a combination of methods, including visual inspections, moisture testing and culture- and/or microscopy-based microbial analyses of air and surface samples, is considered to be the best means of identifying building mold problems.1,2 Performing all of these procedures can require considerable time, labor and expense, however, and their results are only as valid as the thoroughness and competence of the inspectors and analysts performing them. The development of more rapid, inexpensive and standardized methods for mold analysis and interpretation of the results could greatly contribute to reducing these costs and uncertainties.
Analyses of house dust samples have been suggested to provide a better indication of cumulative exposures to molds than short-duration air samples.1,3 Findings of gradual increases in mold concentrations in floor dust, despite regular vacuuming,4 suggest that this matrix may serve as a reservoir of fungal contamination. A DNA-based method for quantitative measurement of different species or closely related groups of indoor molds has been developed at the United States Environmental Protection Agency5 and has been used for the analysis of selected target organisms in environmental samples, including building dust.6–10 In this study, 82 QPCR assays were applied to quantify indoor molds in dust samples from MH and RH and the data were used to develop a prototype logistic regression analysis-based approach for the presumptive differentiation of these two categories of homes. Four species of Aspergillus were also quantified by widely used culture-based analysis and the results compared to QPCR analysis.
Experimental
Selection of homes and dust sampling
The study homes were selected in a larger, ongoing study in Cincinnati, OH on the interactions between diesel exhausts, aeroallergens, genetics and atopy on children’s health. Information on housing characteristics and conditions was collected during walk-through investigations of the study homes and from questionnaires provided to the occupants. In addition, any visible moisture and mold damage in the homes was recorded.
Eighteen homes, having a total area of at least 0.2 m2 of visible mold growth, were selected as moldy-homes (MH) and 19 reference homes (RH) homes were selected randomly from those with no visible moisture or mold damage or mold damage history. During a walk-through, dust samples were collected from the floor of the primary activity room of the child participating in the larger study. Samples were collected by vacuuming with a Filter Queen Majestic® vacuum cleaner (Health-Mor, HMI Industries Inc., Seven Hills, OH) for 2 min m−2 of floor area sampled. Total sampling area was 2 m2 for carpets. For hard floors, the entire open floor area in the room was sampled. Information on the area and the material vacuumed was recorded. Dust samples were sifted (355 μm sieve) and the fine dust was stored at −20 °C.
Fungal cultures and rDNA sequences
Species names, culture collection sources and relevant GenBank sequence accession numbers of standard cultures used for the QPCR assays are listed in Table 1. The species assignments were verified, or in some cases revised, from macro- and microscopic examinations of the cultures and by comparisons of their rDNA sequences with those of previously published sequences in GenBank.
Table 1.
Fungal cultures, rDNA sequences, QPCR assays and calibration standard curve parameters used for target organism quantification
| Standard cultures and rDNA sequences
|
QPCR assays and standard curve parameter values
|
||
|---|---|---|---|
| Species, strain, GenBank accession # | Assay name | Slope (b) | y-intercept (a) |
| Acremonium strictum, ATCC 34717, cf. AY625058 | Astrc | −3.73 | 23.83 |
| Alternaria alternata, EGS 35-193, cf. AY625056 | Aaltr | −4.00 | 23.76 |
| Aspergillus caespitosus, SRRC 308, AY373841 | Acaes | −3.53 | 19.85 |
| Aspergillus candidus, NRRL 303, cf. AY373842 | Acand3 | −3.39 | 20.23 |
| Aspergillus carbonarius, SRRC 15, AY373844 | Acarb | −3.58 | 20.40 |
| Aspergillus cervinus, SRRC 371, AY373845 | Acerv | −4.23 | 32.73 |
| Aspergillus clavatus, SRRC 17, cf. AY373845 | Aclav | −3.92 | 26.33 |
| Aspergillus flavipes, NRRL 302, cf. AY373849 | Aflvp2 | −3.85 | 26.28 |
| Aspergillus flavus, ATCC 16883, cf. AY373848 | Aflav | −3.61 | 22.74 |
| Aspergillus fumigatus, NRRL 163, cf. AY373851 | Afumi | −3.56 | 20.96 |
| Aspergillus niger, ATCC 16888, AY373852 | Anigr | −3.38 | 21.01 |
| Aspergillus niveus, SRRC 333, AY373853 | Anive | −3.90 | 28.37 |
| Aspergillus ochraceus, NRRL 398, AY373856 | Aochr1 | −3.60 | 23.44 |
| Aspergillus paradoxus, SRRC 336, AY373860 | Apard | −3.47 | 19.74 |
| Aspergillus parasiticus, NRRL 502, AY373859 | Apara | −3.50 | 24.07 |
| Aspergillus penicillioides, ATCC 16910, AY373862 | Apeni2 | −3.55 | 23.13 |
| Aspergillus puniceus, SRRC 2155, AY373862 | Apuni | −3.53 | 22.04 |
| Aspergillus restrictus, ATCC 16912, AY373864 | Arest | −3.83 | 25.35 |
| Aspergillus sclerotiorum, ATCC 16892, AY373866 | Asclr | −3.56 | 22.30 |
| Aspergillus sydowii, NRRL 250, AY373866 | Asydo3 | −3.39 | 25.29 |
| Aspergillus tamarii, NRRL 427, cf. AY373870 | Atama2 | −3.67 | 24.12 |
| Aspergillus terreus, ATCC 1012, AY373871 | Aterr2 | −3.46 | 23.06 |
| Aspergillus unguis, SRRC 344, AY373872 | Aungu | −3.29 | 21.16 |
| Aspergillus ustus, NRRL 275, AY373877 | Austs2 | −3.45 | 21.09 |
| Aspergillus versicolor, NRRL 238, cf. AY373882 | Avers2 | −3.71 | 27.24 |
| Aspergillus wentii, NRRL 377, cf. AY373884 | Awent | −4.63 | 30.76 |
| Aureobasidium pullulans, EPA 701, AY625057 | Apull | −3.38 | 20.93 |
| Chaetomium globosum, ATCC 32404, cf. AY625061 | Cglob | −3.52 | 20.28 |
| Cladosporium cladosporioides 1, ATCC 6721, AY625059 | Cclad1 | −3.42 | 20.30 |
| Cladosporium cladosporioides 2, ATCC 16022, AY625060 | Cclad2 | −3.42 | 19.75 |
| Cladosporium herbarum, ATCC 28987, AY625062 | Cherb | −3.53 | 19.00 |
| Cladosporium sphaerospermum, UAMH 7686, AY625063 | Cspha | −3.57 | 20.30 |
| Emericella nidulans, NRRL 2395, AY373888 | Anidu2 | −3.69 | 24.32 |
| Emericella variecolor, SRRC 268, AY373893 | Avari | −3.92 | 24.65 |
| Eurotium amstelodami, NRRL 90, AY373885 | Eamst | −3.46 | 20.56 |
| Epicoccum nigrum, UAMH 3247, AY625063 | Enigr | −3.49 | 21.99 |
| Memnoniella echinata, UAMH 6594, AF081470 | Mem | −3.69 | 20.77 |
| Mucor racemosus, NRRL 1428, AY625074 | Muc1 | −3.32 | 18.82 |
| Paecilomyces variotii, ATCC 22319, AY373941 | Pvari2 | −3.45 | 20.97 |
| Penicillium atramentosum, NRRL 795, AF033492 | Patra | −3.77 | 25.82 |
| Penicillium aurantiogriseum, FRR 971, AY380455 | PenGrp1 | −4.04 | 25.33 |
| Penicillium brevicompactum, FRR 862, AY373898 | Pbrev | −3.97 | 25.91 |
| Penicillium canescens, FRR 910, AY373901 | Pcane2 | −3.81 | 27.02 |
| Penicillium chrysogenum, EPA 467, AY373903 | Pchry | −3.50 | 22.43 |
| Penicillium citreonigrum, FRR 2046, AY373908 | Pcteo | −3.51 | 29.15 |
| Penicillium citrinum, FRR 1841, AY373904 | Pcitr | −4.22 | 27.49 |
| Penicillium coprophilum, NRRL 13627, AF033469 | Pcopr | −3.85 | 29.03 |
| Penicillium corylophilum, FRR 802, AY373906 | Pcory | −3.72 | 24.36 |
| Penicillium crustosum, FRR 1669, AY373907 | PenGrp2 | −4.27 | 27.46 |
| Penicillium decumbens, FRR 741, AY373909 | Pdecu2 | −4.28 | 28.26 |
| Penicillium digitatum, FRR 1313, AY373910 | Pdigi | −3.63 | 22.82 |
| Penicillium expansum, ATCC 7861, AY373912 | Pexpa | −3.47 | 25.32 |
| Penicillium fellutanum, FRR 746, AY373913 | Pfell | −3.99 | 29.49 |
| Penicillium glandicola, FRR 2036, AY373916 | Pglan | −3.59 | 24.98 |
| Penicillium griseofulvum, FRR 3571, AY373917 | Pgris | −3.45 | 23.61 |
| Penicillium implicatum, FRR 2061, AY380455 | Pimpl | −3.62 | 22.17 |
| Penicillium islandicum, NRRL 10127, cf. AY373919 | Pisla | −3.49 | 22.22 |
| Penicillium italicum, ATCC 48114, AY373920 | Pital | −3.42 | 26.66 |
| Penicillium melinii, FRR 2041, AY373923 | Pmeli | −3.48 | 19.10 |
| Penicillium miczynskii, FRR 1077, AY373924 | Pmicz | −3.59 | 24.23 |
| Penicillium olsonii, NRRL 28496, cf. AY373925 | Polsn | −3.78 | 24.63 |
| Penicillium oxalicum, NRRL 787, AF033438 | Poxal | −3.96 | 25.74 |
| Penicillium purpurogenum, FRR 1061, AY373926 | Ppurp | −3.68 | 22.85 |
| Penicillium raistrickii, FRR 1044, AY373927 | Prais3 | −3.58 | 23.42 |
| Penicillium restrictum, NRRL 1748, AF033457 | Prest2 | −3.56 | 22.93 |
| Penicillium roquefortii, FRR 849, AY373929 | Proqu | −4.24 | 26.34 |
| Penicillium sclerotiorum, FRR 2074, AY373930 | Psclr | −3.32 | 19.48 |
| Penicillium simplicissimum, NRRL 1075, AF033440 | Psimp2 | −3.42 | 23.74 |
| Penicillium spinulosum, FRR 1750, AY373933 | Pspin2 | −3.32 | 24.81 |
| Penicillium variabile, FRR 1290, AY373936 | Pvarb2 | −3.34 | 20.52 |
| Rhizopus stolonifer, ATCC 14037, AY625075 | Rstol | −3.62 | 18.51 |
| Scopulariopsis brevicaulis, UAMH 7771, AY625065 | SCbrv | −3.39 | 19.24 |
| Scopulariopsis chartarum, ATCC 16279, AY625066 | SCchr | −3.39 | 20.02 |
| Stachybotrys chartarum, UAMH 6417, AF206273 | Stac | −3.55 | 18.96 |
| Trichoderma asperellum, ATCC 38501, cf. AJ230669 | Taspr1 | −3.32 | 21.51 |
| Trichoderma atroviride, EPA 405, AY625067 | Tviri | −3.75 | 22.92 |
| Trichoderma harzianum, NRRL 13019, AY625068 | Tharz | −3.89 | 25.70 |
| Trichoderma longibrachiatum, UAMH 9515, AY625069 | Tlong | −3.42 | 22.68 |
| Ulocladium atrum, EGS 30-188, cf. AY625072 | Uatrm | −3.61 | 17.01 |
| Ulocladium chartarum, EGS 36-055, cf. AY625071 | Uchar | −3.63 | 22.46 |
| Ulocladium botrytis, EGS 13-030, cf. AY625070 | Ubotr | −3.58 | 18.75 |
| Wallemia sebi, UAMH 7897, AY625073 | Wsebi | −3.43 | 21.34 |
QPCR assays and standard curves
Methods have been reported for preparing conidia or spore suspensions from fungal cultures, extracting DNA, performing QPCR analyses and preparing standard calibration curves for target conidia or spore equivalents versus delta cycle threshold values (ΔCT = CT,target −CT,reference), using co-extracted DNA from Geotrichum candidum as an exogenous reference.6,9 Methods for estimating the amplification factors and extrapolating spore or conidia sensitivities of the assays from the standard curves have also been described.6,9 All primer and probe sequences used in the assays as well as known species comprising the assay groups are at the website: www.epa.gov/nerlcwww/moldtech.htm. Primers and probes were synthesized commercially (Applied Biosystems, Foster City, CA; Integrated DNA Technologies, Coralville, IA; Sigma Genosys, Woodlands, TX).
DNA extractions and QPCR enumeration of molds
Mixed, positive control suspensions, containing approximately 104 or 105 spores or conidia ml−1 of each of the standard cultures listed in Table 1 were prepared as previously described.9 Dust samples and positive control suspensions were extracted by a rapid bead-milling method.8 Briefly, 90 μl of the suspensions or 5 mg of dust and 90 μl of AE buffer (Qiagen, Valencia, CA) were added together with 10 μl of a 2 × 108 conidia ml−1 reference suspension of G. candidum to sterile 2 ml conical bottom, screw cap tubes (506–636; PGC Scientifics, Gaithersburg, MD), containing 0.3 g of glass beads (G-1277; Sigma, St. Louis, MO) and 100 and 300 μl of lysis and binding buffer, respectively from an Elu-Quik DNA Purification Kit (Schleicher and Schuell, Keene, NH). The tubes were shaken in a Mini Bead-Beater (Biospec Products, Bartlesville, OH) for 1 min at a maximum speed and then centrifuged for 1 min at 8 000g to pellet the glass beads and debris. The supernatants were further purified using a DNeasy kit (Qiagen, Valencia, CA).
QPCR assays for target organism and G. candidum reference DNA in the extracts were prepared using a ‘‘Universal Master Mix’’ of PCR reagents (Applied Biosystems, Foster City, CA) and performed in an Applied Biosystems Prism model 7700 sequence detection instrument, as previously described.6 Numbers of spores or conidia detected in dust samples (N) were calculated using the equation: log10(N) = (ΔCT − a)/b, where ΔCT was the difference in observed CT values between the target and reference organisms (CT,target − CT,reference) for the respective dust sample and a and b (Table 1) were the mean y-intercept and slope parameter values from the standard calibration curves for each target assay group. Parallel analyses of method negative control samples, containing AE buffer only, were performed at a frequency of approximately one per each six test samples analyzed. Mixed, positive control conidia suspensions were analyzed at a frequency of nearly one per each test sample.
Culture based analyses of molds
Fungal colony-forming units (CFU) were isolated from dust samples by standard culturing protocols.11 Dust samples of 50 mg each were added to 15 ml sterile tubes (Corning Inc., Corning, NY) along with 4.5 ml of buffer (0.0425 g l−1 KH2PO4, 0.25 g l−1 MgSO4 · 7H2O, 0.008 g l−1 NaOH 0.02% (v/v) Tween 80). The samples were shaken at 450 rpm (gyrotory® shaker, model G76, New Brunswick Scientific, Edison, NJ) for 1 h at room temperature. A series of dilutions were prepared for each sample and plated on 2% malt extract agar (MEA) (Difco, Becton Dickinson and Company, Sparks, MD) and dichloran-18-glycerol agar (DG18) (Oxoid LTD., Basingstoke, Hampshire, England) with chloramphenicol at a concentration of 100 mg l−1. The samples were incubated at 25 °C for 7 days. Blank media and blank buffer cultivations were used for quality assurance. Colonies of Aspergillus fumigatus, A. niger, A. ochraceus and A. versicolor were identified based on colony appearance and species identifications were confirmed based on conidial structures and appearance using high resolution light microscopy (Labophot 2, Nikon Corp., Tokyo, Japan). The confirmed colonies were enumerated and CFU concentrations calculated per gram of dust.
Statistical analyses
Species (or assay groups) measured by QPCR at average concentrations of less than 2 spores per 5 mg of dust in either MH or RH were eliminated from further analysis. The remaining species or assay groups were each compared by calculating the ratio of the GM in MH to GM in RH. Assay results were categorized into those that had GM ratios > 1.0 (Group I) and those < 1.0 (Group II).
The differences in the concentrations of measured spores of the species or assay groups between MH and RH were analyzed for statistical significance using the Mann-Whitney U test (SPSS statistical package, version 10, SPSS inc. Chicago, IL). Logistic regression analysis was performed using SAS Proc Probit (SAS Institute Inc., Cary, NC) to estimate the probability of whether a home could be predicted to be a MH based on the sum of the logs of the concentration of species or assay groups.
Results
QPCR assay standard curves and control sample analyses
Standard curve parameter values for the QPCR assays used in this study are listed in Table 1. Calculated estimates of the amplification factors and extrapolated mean sensitivities of the Stachybotrys, Aspergillus, Penicillium and Paecilomyces assays have been previously reported.7,9 Corresponding values for the other assays used for the first time in this study ranged from 1.81 to 2.00 for the amplification factors and from less than one to approximately four conidia per sample for the extrapolated mean sensitivities, based upon a normalized G. candidum reference assay CT value of 17.7 for 2 × 106 conidia of this organism per sample (results not shown).
To monitor the precision of the QPCR measurements, a total of 29 positive control samples, containing ~103 or 104 conidia of each of the strains listed in Table 1 were extracted and subjected to analyses by the same panel of assays over the same time period as the dust samples. Using the pooled variance among all assay CT values for these control samples and the overall correlation between target assay and reference assay CT values as previously described,9 the variance of ΔCT was determined to be 2.49. This corresponded to a 95% occurrence range about the mean of approximately 40 to 250% for individual measurements. Analyses of 15 sets of no DNA template, negative control samples over the same time period, using the same panel of assays, consistently produced no signals (CT = 40).
QPCR analyses, using the G. candidum reference assay, gave a mean CT value of 20.16, SD = 1.11, for the 37 dust sample extracts, compared with a mean value of 18.90, SD = 1.05, for the 29 control sample extracts. Three of the dust sample reference assay CT values were three standard deviations higher than the mean of the control sample results. However, further analyses of these samples provided no indications of matrix related PCR inhibition.9 Based upon these results, the mean recovery of fungal DNA from the dust samples in the extraction process was 43% of the control samples and 20% of the normalized standard curve samples. This would indicate, on average, a five-fold reduction in the reported assays’ sensitivities caused by the dust matrices. No significant difference was seen in the reference assay CT values of the MH and RH samples (P = 0.69).
Differences between MH and RH in QPCR results
The average total concentrations of mold spores or conidia in MH and RH, as determined from the combined QPCR assay results, were 44 300 and 54 300 spores per 5 mg dust, respectively, and these concentrations were not significantly different (P = 0.43). There were also no significant differences in the concentration of any individual species or assay groups between MH and RH, nor was there a significant difference (P = 0.57) between the average number of different species found in MH (24.5) and in RH (25.5). There were 57 species or assay groups that were only rarely encountered (less than 2 spores per 5 mg dust on average) and these were eliminated from further analysis. Of the remaining species or groups, 6 had GM ratios (MH : RH) > 1 and 19 had GM ratios (MH : RH) < 1 (Table 2).
Table 2.
Geometric Mean (GM) of concentrations of measured mold spores in moldy homes (MH) and reference homes (RH) and the ratios (MH : RH) of the GMs. Species with at least 2 cells on average in both MH and RH are indicated in bold. Group I (MH : RH > 1) are underlined
| Mold species and assay groups | MHa #/5 mg | RHa #/5 mg | Ratio MH : RH |
|---|---|---|---|
| Aspergillus caespitosus | 0.33 | 0.32 | |
| Aspergillus carbonarius | 0 | 0 | |
| Aspergillus candidus | 0.78 | 1.68 | |
| Aspergillus cervinus | 0 | 0 | |
| Aspergillus clavatus grp.b | 0 | 0 | |
| Aspergillus flavipes | 0 | 0 | |
| Aspergillus flavus grp.c | 0.94 | 1.89 | |
| Aspergillus fumigatus grp.d | 3.4 | 1.02 | |
| Aspergillus niger grp.e | 8.67 | 10.43 | 0.83 |
| Aspergillus niveus | 0 | 0 | |
| Aspergilus ochraceus grp.f | 7.69 | 4.12 | 1.87 |
| Aspergillus paradoxus | 0 | 0 | |
| Aspergillus parasiticus | 0 | 0 | |
| Aspergillus penicillioides | 280 | 209 | 1.34 |
| Aspergillus puniceus | 0 | 0.29 | |
| Aspergillus restrictus grp.g | 2.05 | 1.93 | |
| Aspergillus sclerotiorum | 1.77 | 1.68 | |
| Aspergillus sydowii | 0.26 | 0.95 | |
| Aspergillus tamari | 0.30 | 1.29 | |
| Aspergillus terreus | 1.22 | 0.36 | |
| Aspergillus unguis | 21.6 | 12.4 | 1.74 |
| Aspergillus ustus | 0.77 | 1.64 | |
| Aspergillus versicolor | 7.25 | 2.9 | 2.50 |
| Aspergillus wentii | 0 | 0 | |
| Emericella nidulans grp.h | 0.74 | 1.46 | |
| Emericalla variecolor | 0 | 0 | |
| Eurotium grp.i | 273 | 145 | 1.89 |
| Penicillium atramentosum | 3.50 | 1.32 | |
| Penicillium brevicompactum | 6.70 | 16.9 | 0.40 |
| Penicillium canescens | 0 | 0 | |
| Penicillium chrysogenum svar.2j | 10 | 17.3 | 0.60 |
| Penicillium citreonigrum | 0 | 0 | |
| Penicillium citrinum grp.k | 0 | 0 | |
| Penicillium digitatum | 0.71 | 0.34 | |
| Penicillium grp.1l | 0.3 | 0 | |
| Penicillium grp. 2m | 0.5 | 1.6 | |
| Penicillium coprophilum | 0 | 0 | |
| Penicillium corylophilum | 0.53 | 198 | |
| Penicillium decumbens | 0 | 0 | |
| Penicillium expansum | 0 | 0 | |
| Penicillium fellutanum grpn | 0 | 0 | |
| Penicillium glandicola | 0 | 0 | |
| Penicillium griseofulvum | 0.22 | 0 | |
| Penicillium implicatum | 0 | 0.2 | |
| Penicillium islandicum | 0.38 | 0.37 | |
| Penicillium italicum | 0 | 0 | |
| Penicillium melinii | 0 | 0 | |
| Penicillium miczynskii | 0 | 0 | |
| Penicillium olsonii | 0.79 | 0.23 | |
| Penicillium oxalicum | 7.8 | 88.4 | 0.09 |
| Penicillium purpurogenum | 3.0 | 0.79 | |
| Penicillium raistrickii | 0 | 0 | |
| Penicillium restrictum | 0.47 | 0 | |
| Penicillium roquefortii | 1.8 | 0.23 | |
| Penicillium sclerotiorum | 1.7 | 2.9 | |
| Penicillium simplicissimum grp.o | 0.11 | 0 | |
| Penicillium spinulosum grp.p | 10.60 | 11.6 | 0.91 |
| Penicillium variabile | 11.80 | 15.9 | 0.74 |
| Paecilomyces variotii | 3.30 | 9.0 | 0.37 |
| Aureobasidium pullulans | 4685 | 6332 | 0.74 |
| Acremonium strictum | 12.8 | 25.2 | 0.51 |
| Alternaria alternata | 168 | 303 | 0.56 |
| Chaetomium globosum | 2.3 | 3.48 | 0.66 |
| Cladosporium cladosporioides-svar.1 | 4804 | 7335 | 0.65 |
| Cladosporium cladosporioides-svar. 2 | 62.20 | 149 | 0.42 |
| Cladosporium herbarum | 107 | 211 | 0.57 |
| Cladosporium sphaerospermum | 123 | 93 | 1.32 |
| Epicoccum nigrum | 6047 | 11147 | 0.54 |
| Memnoniella echinata | 0 | 0 | |
| Mucor and Rhizopus grpq | 107 | 111 | 0.97 |
| Rhizopus stolonifer | 1.75 | 2.03 | |
| Scopulariopsis brevicaulis | 2.70 | 4.7 | 0.57 |
| Scopulariopsis chartarum | 0.50 | 1.3 | |
| Stachybotrys chartarum | 1.6 | 1.09 | |
| Trichoderma asperellum grp.r | 0 | 0.2 | |
| Trichoderma harzianum | 0.83 | 0.33 | |
| Trichoderma longibrachiatum grp.s | 0.65 | 0.9 | |
| Trichoderma viride grp.t | 3.4 | 6.4 | 0.53 |
| Ulocladium atrum | 0.15 | 0.19 | |
| Ulocladium chartarum | 0.15 | 0 | |
| Ulocladium botrytis | 1.46 | 0.9 | |
| Wallemia sebi | 127 | 129 | 0.99 |
Geometric mean of measured numbers per 5 mg dust.
Includes A. clavatus and A. giganteus.
Includes A. flavus and A. oryzae.
Includes A. fumigatus and Neosartorya fischeri.
Includes A. niger, A. foetidus and A. pheonicis.
Includes A. ochraceus and A. ostianus.
Includes A. restrictus, A. caesillus and A. conicus.
Includes E. nidulans, E. quadrilineata and E. rugulosa.
Includes E. amstelodami, E. chevalieri, E. herbariorum, E. rubrum and E. repens.
Includes dominant subgroup of species.
Includes P. citrinum, P. sartoryi and P. westlingi.
Includes P. aurantiogriseum, P. freii, P. hirsutum, P. polonicum, P. tricolor, P. verrucosum and P. viridicatum.
Includes P. crustosum, P. camembertii, P. commune, P. echinulatum and P. solitum.
Includes P. fellutanum and P. charlesii.
Includes P. simplicissimum and P. ochrocloron.
Includes P. spinulosum, P. glabrum, P. lividum, P. pupurescens and P. thomii.
Includes M. amphibiorum, M. circinelloides, M. hiemalis, M. indicus, M. mucedo, M. racemosus, M. ramosissimus, R. azygosporus, R. homothalicus, R. microsporus, R. oligosporus and R. oryzae.
Includes T. asperellum and T. hamatum.
Includes T. longibrachiatum and T. citrinoviride.
Includes T. viride, T. atroviride and T. koningii.
The sum of the logs of the concentrations of molds in the > 1 category (Group I) was significantly higher in MH compared to RH (P < 0.04). Those molds with a GM ratio of < 1 (Group II) were significantly lower in MH compared to RH (P < 0.02). The logistic regression analysis of the sum of the logs of the concentration of the 6 species in Group I gave a 95% probability of the house being categorized as MH, if the sum of the logs of the concentrations in Group I was found to be > 19.4 (95% fiducial limits: 14.0 to 280.4).
Comparison of QPCR and culture based analyses
The concentrations of Aspergillus fumigatus, A. niger, A. ochraceus and A. versicolor that were calculated from standard culture-based methods (101 to 102 CFU g−1) were three orders of magnitude lower than the concentrations of these same species as measured by QPCR analysis (103 to 105 cells g−1) of the same dust samples (Fig. 1). When the QPCR and cultivation results were compared in the different samples, no significant correlation was found between these two techniques.
Fig. 1.
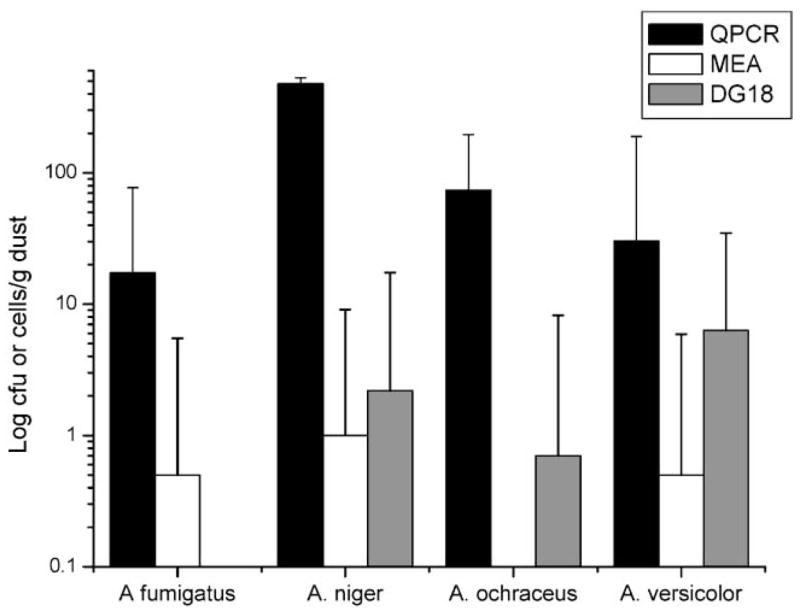
Comparison of QPCR measurements and culture-based measurements on 2% malt extract agar (MEA) and dichloran-18-glycerol agar (DG18) of four Aspergillus species in dust samples from homes in Cincinnati, OH. Concentrations are presented as geometric means and geometric standard deviations.
Discussion
QPCR offers a standardized method for the identification and enumeration of molds. It is rapid and easy to perform. The QPCR assays employed in this study are, for the most part, species specific, however, in some cases several species with identical or nearly identical rDNA sequences are simultaneously measured (Table 2). If it becomes necessary to discriminate between these species, other genes will need to be targeted.12,13 In a few cases, e.g. P. chrysogenum or C. cladosporioides, there are multiple rDNA sequevars that might eventually be separated as new species. The assay(s) used in this study detect the sequevar(s) most commonly found indoors.
Virtually all buildings contain molds that are normally introduced primarily from the outside environment.14 The average total concentration of all molds measured by QPCR in this study was essentially the same in all house dust samples examined and no individual mold was statistically significantly different in MH and RH. Therefore, an empirical process was developed to categorize the molds relevant to distinguishing MH from RH.
The various mold species or assay groups were divided into categories based upon occurrence. Most of the species or groups (n = 57) were not common enough to be evaluated in this manner and were removed from the analysis. By taking the sum of the logs of the concentrations of the 6 species in Group I, abnormal mold conditions in a home, defined in this study as visible mold damage, can be distinguished from a home with no visible mold damage. If the sum of the logs of the concentrations of species in Group I is greater than 19.4, there is at least a 95% probability that it is a MH. The use of easily collected building dust samples in conjunction with this rapid form of analysis offers great potential advantages over other sampling and analysis procedures for identifying mold problems in buildings. For example, this type of analysis could allow for the relatively effortless and inexpensive presumptive identification of mold incursions in hidden building areas such as wall cavities without destructive surface sampling or long term air sampling.
While a large number of assays were employed in this exploratory investigation, the results suggest that analyses of only a few species or groups of species may be all that is necessary to establish that there is an abnormal mold condition. More comprehensive and extensive studies will be required to determine whether this kind of sampling, limited analysis and data handling can be used to describe the mold condition of homes or other buildings in other geographic regions and under all circumstances.
As presently practiced in the industry, the cultivation of the four Aspergillus species examined in this study did not accurately represent the concentrations of these molds in the dust samples. The viable spore concentrations, as measured by cultivation, were much lower than the total spore concentrations, as measured by QPCR. This trend has been previously reported when comparing results from cultivation analysis to those from total microscopic counting.15 Whether viable or not, mold spores are still potentially allergenic and toxigenic.16 If the relationship between mold exposure and health is going to be understood, the species composition and concentrations of these species, particularly in the indoor environment, must be accurately measured. The observation that culturing underestimated the concentrations of these four representative Aspergillus species suggests that accurate risk assessments can’t be accurately made based on culture data.
Acknowledgments
This work was supported by funding from the US EPA’s National Center for Environmental Assessment’s ‘‘Children at Risk Program’’ and in part by an appointment to the Postgraduate Research Participation Program administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US DOE and the US EPA. The collection of dust samples in Cincinnati was supported by the National Institute of Health Sciences (Grant No. RO1 ES11170) as part of the study ‘‘Diesel, Aeroallergens, and Gene Interaction and Child Atopy’’. Mr. Melvin Sparks is gratefully acknowledged for his assistance with the cultivation media.
Footnotes
The US Environmental Protection Agency (EPA,) through its Office of Research and Development, partially funded and collaborated in the research described here. It has been subjected to the Agency’s peer review and has been approved as an EPA publication. Mention of trade names or commercial products does not constitute endorsement or recommendation by the EPA for use.
References
- 1.Dillon HK, Heinsohn PA, Miller D, editors. Field guide for the determination of biological contaminants in environmental samples. American Industrial Hygiene Association; Fairfax, Virginia: 1996. [Google Scholar]
- 2.Macher J, editor. Bioaerosols: Assessment and Control. American Conference of Governmental Industrial Hygienists; Cinncinnati, OH, USA: 1999. [Google Scholar]
- 3.Chew GL, Rogers C, Burge HA, Muilenberg ML, Gold DR. Allergy. 2003;58:13. doi: 10.1034/j.1398-9995.2003.00013.x. [DOI] [PubMed] [Google Scholar]
- 4.Chao HJ, Milton DK, Schwartz J, Burge HA. Mycopathologia. 2001;154:93. doi: 10.1023/a:1015592224368. [DOI] [PubMed] [Google Scholar]
- 5.Haugland RA, Vesper SJ. Identification and Quantification of Specific Fungi and Bacteria: US Patent 6 387 652. US Patent and Trademark Office; Washington, DC: 2002. [Google Scholar]
- 6.Brinkman NE, Haugland RA, Wymer LJ, Byappanahalli M, Whitman RL, Vesper SJ. Appl Environ Microbiol. 2003;69:1775. doi: 10.1128/AEM.69.3.1775-1782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haugland RA, Vesper SJ, Wymer LJ. Mol Cell Probes. 1999;13:329. doi: 10.1006/mcpr.1999.0258. [DOI] [PubMed] [Google Scholar]
- 8.Haugland RA, Brinkman NE, Vesper SJ. J Microbiol Methods. 2002;50:319. doi: 10.1016/s0167-7012(02)00037-4. [DOI] [PubMed] [Google Scholar]
- 9.Haugland RA, Varma M, Wymer LJ, Vesper SJ. Syst Appl Microbiol. 2004;27:198. doi: 10.1078/072320204322881826. [DOI] [PubMed] [Google Scholar]
- 10.Roe J, Haugland RA, Vesper SJ, Wymer LJ. J Exposure Anal Environ Epidemiol. 2001;11:12. doi: 10.1038/sj.jea.7500147. [DOI] [PubMed] [Google Scholar]
- 11.Samson RA, Flannigan B, Flannigan ME, Verhoeff AP, Adan OCG, Hoekstra EH, editors. Health Implications of Fungi in Indoor Environments. Vol. 2. Elsevier; Amsterdam: 1994. Air Quality Monographs. ch 7. [Google Scholar]
- 12.Seifert KA, Louis-Seize G. In: Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Samson RA, Pitt JI, editors. Harwood Academic Publishers; Amsterdam: 2000. pp. 163–178. [Google Scholar]
- 13.Cruse M, Telerant R, Gallagher T, Lee T, Taylor J. Mycologia. 2002;94:814. [PubMed] [Google Scholar]
- 14.Samson RA, Flannigan B, Flannigan ME, Verhoeff AP, Adan OCG, Hoekstra EH, editors. Health Implications of Fungi in Indoor Environments. Vol. 2. Elsevier; Amsterdam: 1994. Air Quality Monographs. ch. 3. [Google Scholar]
- 15.Toivola M, Alm S, Reponen T, Kolari S, Nevalainen A. J Environ Monit. 2002;4:166. doi: 10.1039/b108682k. [DOI] [PubMed] [Google Scholar]
- 16.Kozak PP, Jr, Gallup J, Cummins LH, Gillman SA. Ann Allergy. 1980;45:167. [PubMed] [Google Scholar]


