Abstract
Most known archaeal DNA polymerases belong to the type B family, which also includes the DNA replication polymerases of eukaryotes, but maintain high fidelity at extreme conditions. We describe here the 2.5 Å resolution crystal structure of a DNA polymerase from the Archaea Thermococcus gorgonarius and identify structural features of the fold and the active site that are likely responsible for its thermostable function. Comparison with the mesophilic B type DNA polymerase gp43 of the bacteriophage RB69 highlights thermophilic adaptations, which include the presence of two disulfide bonds and an enhanced electrostatic complementarity at the DNA–protein interface. In contrast to gp43, several loops in the exonuclease and thumb domains are more closely packed; this apparently blocks primer binding to the exonuclease active site. A physiological role of this “closed” conformation is unknown but may represent a polymerase mode, in contrast to an editing mode with an open exonuclease site. This archaeal B DNA polymerase structure provides a starting point for structure-based design of polymerases or ligands with applications in biotechnology and the development of antiviral or anticancer agents.
Keywords: x-ray structure, disulfide bonds, replication, Archaea, exonuclease
Propagation of cells requires faithful DNA replication. This is performed in vivo by DNA polymerases (pols), which attach the appropriate dNTP to the nascent DNA primer strand to match its paired template. Different families of pols are involved in different DNA polymerization processes including not only DNA replication (1, 2) but also repair and recombination (3, 4), a heterogeneity also reflected by varying polypeptide structures and/or subunit compositions (3, 5). Some pols complement polymerase activity with 3′ → 5′ exonuclease activity (editing activity) and/or 5′ → 3′ “structure-specific endonuclease” activity, often located in separate structural domains on the same polypeptide chain (4–8).
Crystal structures are available for most known polymerase families, including the A family DNA polymerases (9–14), pol β (15–17), HIV reverse transcriptase (18–20), and recently, the B family pol gp43 from bacteriophage RB69 (21). All share a functional polymerase structure, which resembles a right hand built by the palm, fingers and thumb domains (see ref. 7 for review). Although the fingers and thumb domains are highly diverse among the different families, the palm domains, which contain the conserved catalytic aspartate residues, show a similar topology among all families except pol β. The polymerase nucleotidyl transfer was studied in detail for the A family polymerases, HIV reverse transcriptase, and pol β, and was shown to involve two metal ions (summarized in ref. 7).
Considerably less is known for the family of type B pols, which are replicative enzymes in eukaryotes and most likely also Archaea (22, 23). The structure of gp43 from bacteriophage RB69 (21) provided an excellent first insight into this family. In addition to the three polymerase domains, gp43 contains an 3′ → 5′ exonuclease domain and an N-terminal domain. The exonuclease and palm domains share the topology and active site of A family enzymes, implying similar metal-assisted mechanisms for polymerase and exonuclease activities (21). The thumb and finger domains are apparently unrelated to the other polymerase families. The function of the N-terminal domain remains unknown, but may help assemble the multicomponent replication apparatus (21).
Much is known about the replication of phages (24–26), viruses (1, 27), Prokaryota, (28) and Eukaryota (1, 3, 29, 30), which in general involves pols but also primases, helicases, RNaseH, sliding clamps, and other factors (31). Considerably less is known for archaeal replication, where mostly B type polymerases, similar to eukaryotic replication enzymes pol α and δ, have been identified (6, 22, 23, 32–34). This relative ignorance is surprising, because such crucial biotechnological applications as cloning and PCR require the thermostability and fidelity typical of archaeal polymerases (6). Thus, in addition to satisfying basic research interests, structural information could assist, for example, the engineering of variant enzymes with tailored nucleotide incorporation rates or the design of antiviral and anticancer polymerase inhibitors. For these reasons, we have determined the structure of a DNA polymerase from Thermococcus gorgonarius (Tgo), an extremely thermophilic sulfur-metabolizing archaeon isolated from a geothermal vent in New Zealand (35). This enzyme possesses pol and a 3′ → 5′ exonuclease activity, which together ensure thermostable replication with high fidelity (error rate: 3.3–2.2 × 10−6; see ref. 36). The 2.5 Å structure shows a topological similarity to gp43 and gives insight in the structural biology of archaeal DNA polymerases, including the identification of several mechanisms for thermophilic adaptation.
MATERIALS AND METHODS
Materials.
All materials were of the highest grade commercially available.
Bacterial Strains.
Escherichia coli LE392 containing pUBS520 was used as described (36). E. coli B834 (DE3) (hsd metB) was a generous gift of Nediljko Budisa (Max-Planck-Institut).
Expression Vectors.
PBTac2 was obtained from Roche Molecular Biochemicals.
Cloning, Expression, and Protein Purification.
The gene for the 89.8-kDa DNA-dependent pol (Tgo pol) was cloned from Tgo (Deutsche Sammlung von Mikroorganismen no. 8976) and expressed in E. coli LE392pUBS520 pBtac2Tgo (Deutsche Sammlung von Mikroorganismen no. 11328) as described (36). Tgo pol was purified essentially as described (36) with the substitution of the TSK butyl Toyopearl column by Blue-Trisacryl M (Serva) and with an additional concentration step on Poros 50 HQ anion exchange medium (Roche Molecular Biochemicals). Active fractions were combined, concentrated to 30 mg/ml, and transferred to 20 mM sodium phosphate, pH 8.2/10 mM 2-mercaptoethanol/500 mM NaCl.
The gene for a selenomethionine-containing variant of Tgo pol (Se-Tgo pol) was expressed in E. coli B842 (DE3) (hsd metB) using a published protocol (37). Se-Tgo pol was purified by using the wild-type protocol.
Crystallization.
Crystals of purified Tgo pol (or Se-Tgo pol) were grown by using sitting-drop vapor-diffusion technique at 4°C with high-salt conditions (2:2 μl protein:reservoir—100 mM Tris, pH 8/2.0M ammonium sulfate) and diffracted to 3.0 Å (in-house) and to 2.5 Å [beamline BW6 at Deutsches Elektronen Synchrotron (DESY), Hamburg]. Low-salt conditions (100 mM Tris, pH 7.0/200 mM ammonium sulfate/30% PEG 400) yielded only poorly diffracting crystals but allowed soaks (including heavy atoms) with some cell constant modulation (a, b, c = 63.6, 105.0, 160.5) but minimal loss of resolution.
Data Collection and Processing.
Data were collected with a MAR imaging plate or a Bruker AXS X1000 mounted on a Rigaku rotating anode source, or with a MAR imaging plate or a MAR CCD (charge-coupled device) at beamline BW6 at DESY, Hamburg. The data were processed with saint (Bruker AXS), mosflm (Mar CCD; ref 38), or denzo (MAR imaging plate; ref 39), scaled with scala (40) or scalepack (39), and reduced with truncate (40).
Structure Determination.
The structure was solved by multiple isomorphous replacement and anomalous scattering (MIRAS) by using data from crystals transferred to low-salt conditions (Table 1). Crystallographic calculations were done with programs from the CCP4 suite (40). Heavy atom positions of major sites were located in difference Patterson maps and were refined with mlphare (40) to calculate protein phase angles to 3.5 Å resolution. A partial polyalanine model was built into interpretable portions of secondary structural elements of the miras map by using main (41). The quality of the electron density was improved by phase combination of the partial model with the experimental phases by using sigmaa (40), and several cycles of solvent flattening to 3.0 Å by using solomon (40). At this stage, no interpretable density was found for a significant portion of the molecule, comprising residues 147–154, 283–306, 653–728, and 752–773.
Table 1.
Data collection and isomorphous replacement
| Derivative | Resolution limit, Å | Total observations | Unique observations | Completeness, % | Rsym, % | Riso, % | Phasing power |
|---|---|---|---|---|---|---|---|
| Native | 3.0 | 136,953 | 21,529 | 91.9 | 7.0 | ||
| U | 3.2 | 25,981 | 16,119 | 89.5 | 11.9 | 18.1 | 0.31 |
| PT1 | 3.7 | 65,464 | 11,692 | 98.2 | 12.0 | 22.6 | 1.54 |
| PT2 | 4.0 | 53,870 | 6,293 | 66.9 | 13.3 | 13.8 | 1.77 |
| PT3 | 3.4 | 85,226 | 14,155 | 92.8 | 10.5 | 17.1 | 1.16 |
| PT4 | 2.6 | 387,925 | 27,487 | 90.6 | 5.7 | 35.5 | 0.67 |
| PT5 | 2.7 | 358,695 | 24,543 | 88.7 | 6.1 | 36.2 | 0.83 |
| PTU | 3.5 | 79,629 | 11,437 | 82.4 | 9.4 | 24.7 | 1.54 |
| PB | 3.5 | 78,190 | 11,336 | 92.9 | 11.5 | 12.8 | 0.41 |
| PBPT | 3.4 | 94,557 | 14,129 | 91.6 | 8.0 | 19.6 | 1.04 |
| OS | 2.8 | 600,108 | 24,026 | 91.2 | 5.3 | 38.4 | 1.82 |
| Overall figure of merit (15.0–3.0 Å): 0.73 | |||||||
Heavy-atom derivatives were prepared by soaking the crystals in low-salt buffer containing the heavy atom as follows: U, 0.5 mM uranyl acatate for 2 h; PT1, 5 mM K2PtCl4 for 1 d, PT2, 5 mM K2PtCl6 for 2 d; PT3, saturated cis-dichlorodipyridine-Pt(II) for 2 d; PT4, 5 mM K2PtCl6 for 7 d; PT5, saturated cis-dichlorodipyridine-Pt(II) for 7 d; PTU, PT1 + U; PB, saturated dinitrophenyl-Pb(NO3)2 for 7 d; PBPT, PB + PT5; OS, saturated K2OsO4 for 7 d. NAT1, PT1, PT2, PT3, PTU, PB and PBPT were collected with a Mar imaging plate and U was collected with a Bruker AXS area detector on a Rigaku rotating anode source. All other data sets were collected with a Mar charge-coupled device at beamline BW6 at DESY, Hamburg.
Model Building and Refinement.
The partial model (R factor 35%) was used to phase the 2.5 Å resolution data of the Se-Tgo pol (high-salt conditions). The model was oriented with amore (40). The correlation coefficient of 22.0% and the R factor of 50.3% showed divergence of the high- and low-salt structures. After bulk solvent correction, anisotropic B factor correction and rigid-body minimization (treating five domains independently), the partial model was iteratively refined and extended with simulated annealing, Powell minimization, restrained individual B factor refinement with cns (42), and manual model building with main (41) by using data from 25.0–2.5 Å resolution (Table 2, Fig. 1).
Table 2.
Crystallographic refinement, high-salt form
| Space group | P212121 |
| Cell dimensions, Å | a = 58.1, b = 105.2, c = 154.2 |
| Observations, 25–2.5 Å | |
| Total | 482,448 |
| Unique | 30,451 |
| Completeness, % | |
| Total | 91.1 |
| Last shell | 86.0 |
| Rsym, % | |
| Total | 7.1 |
| Last shell | 26.2 |
| R factor (Rfree), % | 20.9 (27.1) |
| rms deviation in bond lengths, Å | 0.008 |
| rms deviation in bond angles, ° | 1.5 |
| No. of nonhydrogen atoms | |
| Protein | 6,328 |
| Water | 339 |
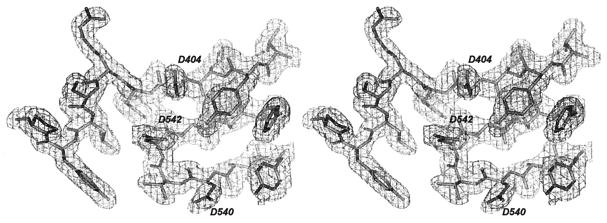
Figure 1.
Stereorepresentation of the electron-density map. The 2 (Fo − Fc) electron density contoured at 1.0 σ at the polymerase active site is well defined for the refined model (stick representation).
RESULTS AND DISCUSSION
Structure of Tgo pol.
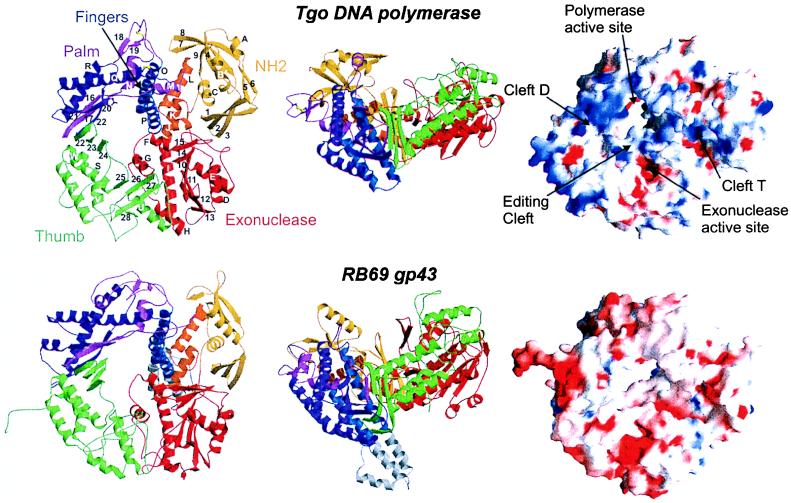
Tgo pol is a ring shaped molecule with dimensions 50 Å × 80 Å × 100 Å. The single polypeptide chain of 773 aa is folded into five distinct structural domains (Fig. 2): the N-terminal domain (residues 1–130), the 3′ → 5′ exonuclease domain (131–326), the palm (369–449 and 500–585), fingers (450–499), and thumb (586–773) domains of the polymerase unit, and a helical interdomain insertion (327–368) between the exonuclease and palm domains. The polymerase unit forms the DNA-binding crevice, reminiscent of a right hand, which is the identifying characteristic of pols. Gp43 from bacteriophage RB69 also shows this overall domain topology (21).
Figure 2.
Structure of Tgo pol and comparison with gp43 form bacteriophage RB69. (Left and middle) Ribbon representation of Tgo DNA polymerase (Upper) in two orthogonal views with labeled secondary structure elements. The molecule is composed of five domains: N-terminal domain (yellow), 3′ → 5′ exonuclease (red), palm [light and dark magenta represent the N-terminal and C-terminal segment, respectively (see text)], fingers (blue), and thumb (green), which are arranged to form a ring. An interdomain helical segment between the exonuclease domain and the palm is orange. The conserved carboxylates in the active site and the two disulfide bridges are shown as magenta and yellow ball-and-sticks, respectively. Tgo pol has the same overall architecture and domain topology than gp43 of RB69 (Lower). The 50-residue insertion in the fingers of gp43 is gray. (Right) Comparison of molecular surfaces of Tgo pol (Upper) and RB69 gp43 (Lower). Red and blue denote negative and positive electrostatic surface potentials, respectively. In contrast to gp43, Tgo pol has a strongly enhanced positive potential at the putative DNA-binding clefts.
Three clefts extend radially from the polymerase active site at the center of the ring: two of them in opposite directions, forming a large cleft across the molecule, and one perpendicular to these. Based on active-site homology to pol A family enzymes, the two opposite clefts probably bind duplex DNA (cleft D, according to ref. 21) and single-strand template DNA (cleft T), respectively. The perpendicular (editing) cleft links the polymerase active site and the exonuclease active site and binds the primer strand in editing mode (21).
The exonuclease domain is structurally equivalent to the 3′ → 5′ exonuclease domain of pol A family (43). Like gp43, however, it is bound at the opposite side of the polymerase unit by noncovalent contacts to the thumb domain at the editing cleft, on one side, and by covalent and noncovalent contacts to the N-terminal and palm domains and the 42 residue interdomain helix, on the other side. This latter segment is located at the base of cleft T, which is additionally bounded by the exonuclease, N-terminal, and palm domains.
The topology of the palm domain is conserved among polymerase families (5), with two long helices (Q and R) packed against the five-stranded antiparallel β-sheet that contains the three conserved aspartate residues involved in nucleotidyl transfer. The fingers emerge from the palm domain as an α-helix-rich insertion. Its 50 residues are folded into two antiparallel coiled α-helices of approximately equal size: helix P contains the conserved Kx3NsxYGx2G motif of B type polymerases and is related to the O helix of A type enzymes (see below). The ≈50-residue insertion between helix O and P in RB69 and T4 gp43 is missing in Tgo pol, where both helices and 4 residue linker are much shorter than their equivalents in gp43. The shorter fingers of Tgo pol presumably reflect the typical structure of the nonbacteriophage B type fingers (pol α, pol δ, and E. coli pol II). The thumb domain topology, similar to that of gp69, is unrelated to other polymerase types. However, like the thumb of A type enzymes, a bundle of α-helices at its base protrude from the active site β-sheet. Distal to the active site, the thumb contains a 75-residue subdomain (665–729), which fixes the exonuclease domain and contributes to the editing channel, explaining why mutations in the exonuclease domain of B-type polymerases affect the polymerase activity and vice versa (44; 45).
Weakly defined density across the base of the thumb domain was modeled as the C-terminal 6 residues with a polyalanine chain. The C terminus thus does not protrude from the core molecule as in the RB69 polymerase (21). Because the C terminus of the T4 pol are involved in sliding-clamp binding (46), it is likely, however, that these residues become ordered on any similar holoenzyme formation.
Sequence Alignment of Archaeal DNA Polymerases.
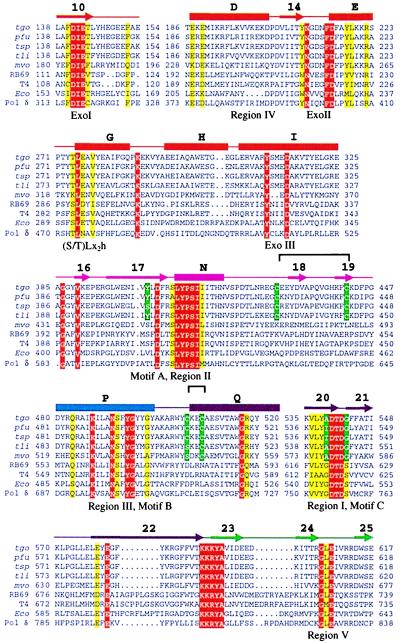
The structure of Tgo pol allows the generation of a structure based sequence alignment of the archaeal subfamily of type B DNA polymerases, the location of conserved and unique residues, and the comparison with other type B DNA polymerases (Fig. 3).
Figure 3.
Sequence alignment of B family DNA polymerases. The alignment has been adapted from ref. 21 to highlight specific residues from the class of archaeal pols. The secondary structure of Tgo pol is indicated on top of the alignment with helices (bars), strands (arrows) and loops (lines) colored according to domains with the same color code as Fig. 2B. Strictly conserved residues of type B polymerases are red, and additional conserved residues are yellow. Uniquely conserved residues of archaeal type B enzymes—as discussed in the text—are green. Disulfide bonds are shown by a bar on top of the alignment. Abbreviations: tgo, Thermococcus gorgonarius pol; pfu, Pyrococcus furiosus pol; tsp, Thermococcus sp. pol; tli, Thermococcus litoralis pol; mvo, Methanococcus voltae pol; RB69, bacteriophage RB69 pol; T4, bacteriophage T4 pol; Eco; E. coli pol II; pol δ; human pol δ.
Polymerase Active Site.
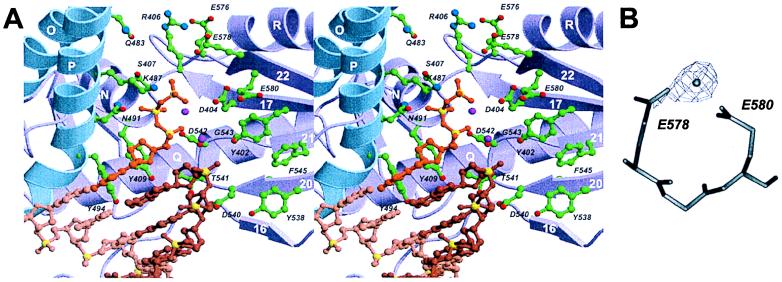
The polymerase active site is formed by the central β-sheet (strands 16, 17, 20, 21, and 22) and helix N of the palm domain and helix P located in the fingers and is highly conserved among B family polymerases (Fig. 4). Three carboxylates required for nucleotidyl transfer in B family polymerases, two of which coordinate two metal ions (14) are superimposably conserved among A family enzymes, B family enzymes, and reverse transcriptase (21). Superposition of Tgo pol and T7 replication complex (14) places the dNTP near the proposed nucleotide-binding site in helix P, the K487x3NSxYGx2G motif (Fig. 3) and suggests interactions of the carboxylates with metals and the phosphate tail of the bound dNTP (Fig. 4). Reorientation of the strictly conserved Lys-487 allows it to mimic the Lys-522–phosphate tail interaction in T7. Tyr-494 (Kx3NSxY494Gx2G) and Tyr-409 (SLY409PSII) form the bottom of the nucleotide-binding site.
Figure 4.
Polymerase active site. (A) Stereoribbon representation (using color code as in Fig. 2) with modeled DNA. Active-site residues are shown as ball-and-stick representations with carbon (green), nitrogen (blue), and oxygen (red) atoms. The DNA template (light brown), primer (light brown), and dNTP (orange) complex has been taken from the coordinates of T7 replication complex (15) by superimposing D404, D542, and adjacent residues with corresponding residues in T7 pol (D475 and D654). Phosphorus atoms are yellow. The two metals of the T7 replication complex are shown as magenta spheres. (B) Experimentally observed metal-binding site for Mn2+ (Fo − Fc density contoured at 5σ) and Zn2+ in the “low salt” crystal form. The carboxylates E578 and E580 are conserved in type B polymerases (Fig. 3).
The active site of B family polymerases contains a DTDS motif, which, however is DTDG in the archaeal subfamily. In Tgo pol, the relatively conserved Tyr-402 from the adjacent strand provides an alternate alcohol group, at a position appropriate for metal coordination or binding of the 3′ end of the primer. The orientation of Tyr-402 is stabilized by an aromatic cluster that also includes Phe-545 and Tyr-538. Archaeal Methanococcus voltae and Thermococcus sp. pol’s (see Fig. 3) have Tyr at position 545—Phe in Tgo—rather than at 402, but might also supply an alcohol group. The displacement of a functional alcohol from serine in DTDS to Tyr-402 or Tyr-545 might stabilize its orientation as an adaptation for thermostability.
The conserved cluster of acidic amino acids (E578, E580) form an unexpected metal-binding site for Mn2+ and Zn2+ (Fig. 4). Its proximity to Asp-404 and to the expected location of the dNTP γ-phosphate suggests a supporting role in nucleotide binding and/or catalysis.
3′ → 5′ Exonuclease Active Site.
Tgo pol is characterized by a strong 3′ → 5′ exonuclease activity, unlike eukaryotic B type polymerases (unpublished results). The exonuclease active site is formed at the interface between the exonuclease domain and the tip of the thumb (Fig. 5). All residues required for catalysis are located in the exonuclease domain, which, at least for T4 gp43, retains activity when dissociated from the polymerase (47). However, the thumb domain, with, for example, RB69 gp43’s Phe-123–base intercalation, partially controls the binding geometry of single strand DNA (21, 43).
Figure 5.
3′ → 5′ exonuclease active site. (A) Stereoribbon representation with modeled DNA using the color code of Fig. 4A (ball-and-stick) and Fig. 2 (ribbons). Active-site residues are shown as ball-and-stick representation. The single-stranded DNA has been taken from the coordinates of RB69–single-stranded DNA complex (21). The orientation of the DNA has been obtained by superimposing the DIE143 motif in Tgo pol with corresponding motif of RB69 gp43 (DIE116). Strand 27 and its preceding loop from the thumb (green) is apparently in collision with the modeled DNA. (B) Comparison of the exonuclease–thumb interface between Tgo pol (color code of Fig. 2B) and RB69 gp43 (gray). In Tgo pol, the lid of the editing site (red) is bent outward compared with the equivalent loop of gp43 (yellow), allowing the tip of the thumb to move several Å (⤡) closer to the exonuclease. This conformation is incompatible with formation of an editing complex [the p(dT)4 of gp43 is shown as brown space-filling model].
The exonuclease structures of Tgo and gp43 DNA polymerases are similar at the editing site but differ considerably at the exonuclease–thumb interface. Strand 10 contains the metal-binding D141IE motif and readily superimposes with the equivalent strand from gp43, allowing modeling of a single-strand DNA segment into the exonuclease site based on theRB69 gp43-p(dT)4 complexes (21). The conserved residues Asp-141 and Glu-143 in the Exo I motif, Tyr-209, Asn-210, Phe-214, and Asp-215 in Exo II, and Tyr-311 and Asp-315 in Exo III are in approximate DNA-binding conformations (Fig. 5A). However, the editing cleft is constricted by a displacement of the tip of the thumb toward the exonuclease domain to prohibit single-strand binding (Fig. 5B). This shift is correlated with a large change in the loop between strands 10 and 11. In RB69 gp43 (and likewise in T4 gp43), this loop forms a lid over the 3′ base and contains Phe-123, which intercalates between the first two bases. In Tgo pol, the loop is curved outward, away from the thumb, and Phe-152 (the equivalent of gp43’s Phe-123) attaches to Phe-214 10 Å away from intercalation site. This shift allows the tip of the thumb to move into the editing channel and to block the exonuclease site.
Are There Different Conformations in Polymerase and Editing Mode?
If a closed conformation of the exonuclease domain prohibits single strand binding, an open conformation is required for editing. The observed closed conformation may represents the enzyme in “polymerase” mode. Preliminary analysis of the crystal structure of Tgo pol in the low-salt conditions indicates a structural change at the interface of exonuclease and thumb, possibly reflecting a transition between open and closed forms. The closed conformation observed here may, however, be a nonphysiological artifact of the high ionic strength used for crystallization. Crystal structures of the enzyme in both polymerase and editing modes are required.
Adaptation to High Temperatures.
Tgo is a sulfur-metabolizing, extremely thermophilic archaeon, with a growth range between 55°C and 98°C. For accurate replication at this temperature range, the polymerase must not only be stable, but must also adequately bind substrate DNA. A comparison with gp43 from the mesophilic bacteriophage RB69 indicates several such adaptations to high temperatures. Several loops are shorter in Tgo pol than in gp43 (Fig. 2), and there is an increase in hydrogen bonded β-strand content: Tgo pol secondary structure includes 41% helix, 22% β-strands, and 19% turns (calculated according to ref. 48), whereas gp43 has 42% helix, 17% β-strands, and 19% turns.
Although rare among cytoplasmic or nuclear proteins, two disulfide bridges might be formed: cysteine pairs 428–442 and 506–509, although reduced, are poised for attachment (Fig. 2). Model refinement and electron density inspection with and without constraints for the disulfide bonds verified the reduced state (observed unrestrained SG–SG distance: 2.8 Å and 3.0 Å). This is consistent with our E. coli expression and further rules out structural perturbation by nonnative oxidation. These cysteines are located in the palm domain and are conserved among B type enzymes from hyperthermophilic sulfur-metabolizing archaeons, but not among mesophile homologs (Fig. 3). The Cys-428–Cys-442 bridge stabilizes the compact fold of the loop segment between helix N in the palm domain and helix O in the fingers and presumably also the relative orientation of these helices. In addition, the loop segment packs against helix Q in the palm domain. Helix Q, the spine of the palm domain, is further stabilized at the first helical turn by the second disulfide bridge between Cys-506 and Cys-509.
A much enhanced complementary positive potential for all three DNA-binding clefts of Tgo pol is observed relative to gp43 (Fig. 2). Thus, in addition to hydrogen bonding and specific DNA–protein interactions, binding to Tgo pol has an additional strong stabilizing electrostatic component.
An increase in the number of salt bridges is often associated with thermostability. Although Tgo pol has a greater total number of charged residues (262) than gp43 (245), both molecules have 54 salt bridges within a 3–5 Å bound. However, in the 5–7 Å range of charge distance, Tgo pol has 77 ion pairs compared with 43 for gp43. This large increase results in a more highly charged surface of Tgo pol, accompanied by a more balanced charge distribution, compared with gp43 where charges are often located in patches (Fig. 2).
Acknowledgments
We thank Dr. Uwe Jacob and Martin Augustin for stimulating discussions, Dr. Nediljko Budisa for help with expression of selenomethionine variants, and Dr. Hans Bartunik and Dr. Gleb Bourenkov at DESY, Hamburg for help with data collection.
ABBREVIATIONS
- Tgo
Thermococcus gorgonarius
- pol
DNA polymerase
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY, 11973 (PDB ID code 1TGO).
References
- 1.Waga S, Stillman B. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 2.Maga G, Hübscher U. Biochemistry. 1996;35:5764–5777. doi: 10.1021/bi952455k. [DOI] [PubMed] [Google Scholar]
- 3.Wang T S F. Annu Rev Biochem. 1991;60:513–552. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- 4.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 5.Joyce C M, Steitz T A. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 6.Perler F B, Kumar S, Kong H. Adv Protein Chem. 1996;48:377–435. doi: 10.1016/s0065-3233(08)60367-8. [DOI] [PubMed] [Google Scholar]
- 7.Brautigam C A, Steitz T A. Curr Opin Struct Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 8.Arnold E, Ding J, Hughes S H, Hostomsky Z. Curr Opin Struct Biol. 1995;5:27–38. doi: 10.1016/0959-440x(95)80006-m. [DOI] [PubMed] [Google Scholar]
- 9.Ollis D L, Brick P, Hamlin R, Xuong N G, Steitz T A. Nature (London) 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 10.Freemont P S, Friedman J M, Beese L S, Sanderson M R, Steitz T A. Proc Natl Acad Sci USA. 1988;85:8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiefer J R, Mao C, Hansen C J, Basehore S L, Hogrefe H H, Braman J C, Beese L S. Structure (London) 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 12.Kiefer J R, Mao C, Braman J C, Beese L S. Nature (London) 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y, Eom S H, Wang J, Lee D S, Suh S W, Steitz T A. Nature (London) 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 14.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Nature (London) 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 15.Davies J F, Almassy R J, Hostomska Z, Ferre R A, Hostomsky Z. Cell. 1994;76:1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- 16.Sawaya M R, Pelletier H, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 17.Pelletier H, Sawaya M R, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 18.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 19.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, et al. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jager J, Smerdon S J, Wang J, Boisvert D C, Steitz T A. Structure (London) 1994;2:869–876. doi: 10.1016/s0969-2126(94)00087-5. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Sattar A K, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 22.Edgell D R, Doolittle W F. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- 23.Cann I K O, Komori K, Toh H, Kanai S, Ishino Y. Proc Natl Acad Sci USA. 1998;95:14250–14255. doi: 10.1073/pnas.95.24.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debyser Z, Tabor S, Richardson C C. Cell. 1994;77:157–166. doi: 10.1016/0092-8674(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 25.Nossal N G. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 43–53. [Google Scholar]
- 26.Nossal N G. FASEB J. 1992;6(3):871–878. doi: 10.1096/fasebj.6.3.1310946. [DOI] [PubMed] [Google Scholar]
- 27.Fanning E, Knippers R. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 28.Kelman Z, O’Donnell M. Annu Rev Biochem. 1996;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 29.De Pamphilis M L, editor. DNA Replication in Eukaryotic Cells. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. [Google Scholar]
- 30.Waga S, Stillman B. Nature (London) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 31.Baker T A, Bell S P. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 32.Uemori T, Sato Y, Kato I, Doi H, Ishino Y. Genes Cells. 1997;2(8):499–512. doi: 10.1046/j.1365-2443.1997.1380336.x. [DOI] [PubMed] [Google Scholar]
- 33.Braithwaite D K, Ito J. Nucleic Acids Res. 1993;21(4):787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayre J D, Venter J C. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 35.Miroshnichenko M L, Gongadze G M, Rainey F A, Kostyukova A S, Lysenko A M, Chernyh N A, Bonch-Osmolovskaya E A. Int J Syst Bacteriol. 1998;48:23–29. doi: 10.1099/00207713-48-1-23. [DOI] [PubMed] [Google Scholar]
- 36.Bonch-Osmolovskaya, E., Svetlichny, V., Ankenbauer, W., Schmitz-Agheguian, G., Angerer, B., Ebenbichler, C. & Laue, F. (1996) Eur. Patent Appl. EP 0 834 570 A1.
- 37.Budisa N, Steipe B, Demange P, Eckerskorn C, Kellermann J, Huber R. Eur J Biochem. 1995;320:788–796. doi: 10.1111/j.1432-1033.1995.tb20622.x. [DOI] [PubMed] [Google Scholar]
- 38.Leslie A G W. Mosflm User Guide. Cambridge, UK: MRC Laboratory of Molecular Biology; 1994. , Version 5.1. [Google Scholar]
- 39.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project, No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 41.Turk D. Ph.D. Thesis. München: Technische Universität; 1992. [Google Scholar]
- 42.Adams P D, Pannu N S, Read R J, Brunger A T. Proc Natl Acad Sci USA. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Yu P, Lin T C, Konigsberg W H, Steitz T A. Biochemistry. 1996;35:8110–8119. doi: 10.1021/bi960178r. [DOI] [PubMed] [Google Scholar]
- 44.Sattar A K, Lin T C, Jones C, Konigsberg W H. Biochemistry. 1996;35:16621–16629. doi: 10.1021/bi961552q. [DOI] [PubMed] [Google Scholar]
- 45.Reha-Krantz L J, Nonay R L. J Biol Chem. 1994;269:5635–5643. [PubMed] [Google Scholar]
- 46.Berdis A J, Soumillion P, Benkovic S J. Proc Natl Acad Sci USA. 1996;93:12822–12827. doi: 10.1073/pnas.93.23.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin T C, Karam G, Konigsberg W H. J Biol Chem. 1994;269:19286–19394. [PubMed] [Google Scholar]
- 48.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]