Abstract
Myosin Va is a well known processive motor involved in transport of organelles. A tail-inhibition model is generally accepted for the regulation of myosin Va: inhibited myosin Va is in a folded conformation such that the tail domain interacts with and inhibits myosin Va motor activity. Recent studies indicate that it is the C-terminal globular tail domain (GTD) that directly inhibits the motor activity of myosin Va. In the present study, we identified a conserved acidic residue in the motor domain (Asp-136) and two conserved basic residues in the GTD (Lys-1706 and Lys-1779) as critical residues for this regulation. Alanine mutations of these conserved charged residues not only abolished the inhibition of motor activity by the GTD but also prevented myosin Va from forming a folded conformation. We propose that Asp-136 forms ionic interactions with Lys-1706 and Lys-1779. This assignment locates the GTD-binding site in a pocket of the motor domain, formed by the N-terminal domain, converter, and the calmodulin in the first IQ motif. We propose that binding of the GTD to the motor domain prevents the movement of the converter/lever arm during ATP hydrolysis cycle, thus inhibiting the chemical cycle of the motor domain.
Keywords: actin, trafficking, molecular motor, lever arm, regulation
It is generally accepted that motor protein activity must be tightly regulated in cells to transport cargo efficiently and to prevent the futile hydrolysis of ATP. One of the best characterized regulations of myosin motor proteins is the regulation of vertebrate myosin Va.
Myosin Va consists of two identical heavy chains that dimerize through the formation of a coiled-coil structure to form a homodimer. At the amino terminus is the motor domain containing ATP- and actin-binding sites. The motor domain is followed by a neck that consists of six IQ motifs with the consensus sequence IQXXXRGXXXR, which act as the binding sites for calmodulin (CaM) or CaM-like light chains. The next ≈500 aa are predicted to form a series of coiled-coils separated by several flexible regions. The last ≈400 aa form a C-terminal globular tail domain (GTD). The GTD, in conjunction with a portion of the coiled-coil region, mediates myosin Va binding to specific membrane-bounded organelles, such as melanosomes (1–4).
Significant progress has recently been achieved in understanding the mechanism of myosin Va regulation, although it was shown as early as 1993 that the actin-activated ATPase activity of myosin Va purified from chick brain is well regulated by Ca2+ (5). It was found that Ca2+-activation of the ATPase activity of myosin-Va is accompanied by a large conformational transition, from a 14S folded conformation in the absence of Ca2+ to an 11S open conformation in the presence of micromolar Ca2+ (6–8). In contrast, myosin Va heavy meromyosin (HMM), a constitutively active fragment lacking the C-terminal globular tail domain, is not well regulated by Ca2+ and does not undergo a large conformational transition like full-length myosin Va (6–8). These results suggest that the tail domain plays a key role in the Ca2+-dependent regulation of myosin Va by inhibiting the ATPase of the head. Based on these findings, a tail-inhibition model for the regulation of myosin Va has been proposed (6–8). In this model, myosin Va in the inhibited state is in a folded conformation such that the tail domain interacts with and inhibits myosin Va motor activity; high Ca2+ or cargo binding may reduce the interaction between the head and tail domains, thus activating motor activity.
Using negative staining electron microscopy, Knight and colleagues first observed a triangular shape of myosin Va in the absence of Ca2+ but not in its presence: The head domains apparently fold back to interact with the tail domain (7). Later, it was found that the GTD directly inhibits the activity of the motor domain of myosin-Va: Myosin-Va HMM is inhibited and folded by exogenous GTD (9, 10). We found that strong inhibition of motor activity by the GTD requires not only the two-headed structure, but also the presence of an intact first long coiled-coil segment of the tail (9). Based on these findings, we proposed that the GTD simultaneously binds to the motor domain and the C terminus of the first long coiled-coil segment of the tail, thus forming a triangular conformation (9). However, the precise interaction between the GTD and the motor domain and thus the mechanism of inhibition remained unclear.
Based on averaged images of inhibited myosin Va and molecular modeling, Knight and colleagues proposed that the GTD binds to a lobe of the motor domain (P117-P137) that contains several conserved acidic residues (10). Because the assigned GTD-binding site has no direct interaction with the ATP-binding site, they proposed that the GTD regulates motor activity allosterically (10). However, Taylor and colleagues assigned the GTD-binding site to loop 1, which is near the entrance of the ATP-binding pocket in the motor domain (11). They proposed that binding of the GTD to loop 1 decreases the rates of nucleotide exchange, thus inhibiting the ATPase activity of the motor domain (11). However, the recently solved crystal structure of the motor domain of chicken myosin Va (12, 13) and the GTD of Myo2p, a yeast myosin V (14), show that it is unlikely that the GTD binds to lobe P117-P137 and loop-1 simultaneously.
In this study, we identify a conserved acidic residue (D136) in lobe P117-P137 in the motor domain and two conserved basic residues (K1706 and K1779) in the GTD as critical residues for the inhibition of myosin Va. Based on these findings, we propose a model for inhibition of the motor domain by the GTD.
Results
The Conserved Residue D136 in the Motor Domain of Class V Myosin Is Responsible for the Inhibition of Myosin Va ATPase Activity by the GTD.
It was proposed that the GTD binds to lobe P117–P137 in the motor domain of mouse myosin Va to inhibit its ATPase activity (10). Among the four acidic residues in lobe P117-P137, two acidic residues, D134 and D136, are highly conserved in class V myosins but not in other classes. To investigate the role of these two acidic residues on the regulation of myosin Va, we examined the effect of D134A and/or D136A mutations on the inhibition of the actin-activated ATPase activity of myosin Va by the GTD.
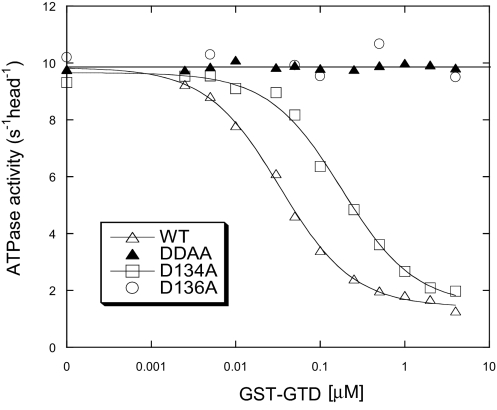
Previously, we showed that the exogenous GTD is capable of inhibiting the actin-activated ATPase activity of myosin-Va heavy meromyosin (M5HMM) in a Ca2+-dependent manner (9). As shown in Fig. 1, GST-GTD strongly inhibited the actin-activated ATPase activity of M5HMM-WT and -D134A, but it failed to inhibit these of D134A/D136A and D136A mutants. Whereas D134A mutation decreased the affinity between GST-GTD and M5HMM fivefold, the ATPase activity of the D134A mutant was still strongly inhibited by GST-GTD. These results indicate that D136 is essential for the inhibition of myosin Va motor function by the GTD.
Fig. 1.
Inhibition of the actin-activated ATPase activity of M5HMM with D134A and/or D136A mutations by GST-GTD. The dissociation constants (Kd) of GST-GTD and M5HMM, obtained by a quadratic fit, are 0.058 μM (M5HMM-WT) and 0.341 μM (M5HMM-D134A). DDAA, D134A/D136A.
The Conserved Residue D136 in the Motor Domain of Class V Myosin Is Critical for the Folded Conformation of Myosin Va.
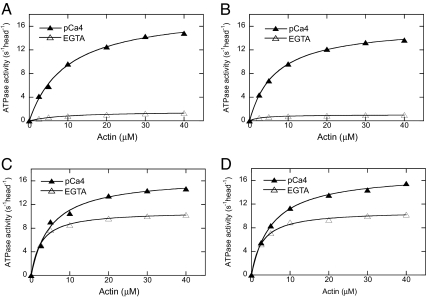
To further determine the role of D134 and D136 on the regulation of myosin Va, we introduced D134A and D136A mutations individually in full-length mouse myosin Va (M5Full). Consistent with previous reports (5–8), we found that the actin-activated ATPase activity of M5Full-WT was well regulated by Ca2+; i.e., it had a low actin-activated ATPase activity in the absence of Ca2+ (EGTA conditions) and high ATPase activity in the presence of Ca2+ [Fig. 2A and supporting information (SI) Table 1]. High Ca2+ enhanced the ATPase activity of myosin Va by ≈10-fold. Similar to that of WT, the actin-activated ATPase activity of M5Full-D134A was significantly inhibited in EGTA (Fig. 2B and SI Table 1). In contrast, we found that the actin-activated ATPase activity of M5Full-D136A in EGTA was markedly increased compared with that of WT under the same conditions, and Ca2+ only slightly enhanced its ATPase activity (Fig. 2C and SI Table 1). The ATPase activity of M5Full-D136A in the absence of Ca2+ was similar to a truncated myosin Va without the GTD (8). These results indicate that the EGTA-induced inhibition of myosin Va was strongly dampened by D136A mutation, suggesting the importance of this residues for the regulation.
Fig. 2.
Effect of D134A, D136A, and K1706A/K1779A mutations on the regulation of the actin-activated ATPase activity of M5Full. (A) WT. (B) D134A. (C) D136A. (D) K1706A/K1779A. Data were fit with a hyperbola to define Vmax (maximal ATPase activity) and Kactin (the concentration of actin that stimulates the ATPase activity to 50% of Vmax). SI Table 1 summarizes the Vmax and Km values for multiple assays.
It has been shown that the activation of myosin Va by Ca2+ is accompanied by a transition from a folded conformation (14S in the absence of Ca2+) to an open conformations (11S in the presence of Ca2+) (6–8). We took two approaches to investigate the effect of D134A and D136A mutations on the conformational changes of myosin Va.
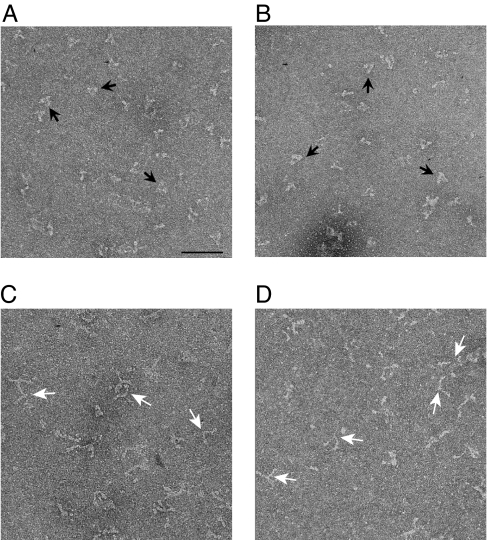
First, we used negative staining electron microscopy to visualize the conformation of M5Full. We found that the majority of M5Full-WT and -D134A molecules were in the folded conformation in EGTA (Fig. 3 A and B). In contrast, M5Full-D136A molecules were in the open conformation in the same conditions (Fig. 3C).
Fig. 3.
Effect of D134A, D136A, and K1706A/K1779A mutations on the conformation of M5Full. EM Images of negatively stained M5Full in EGTA conditions. (A) WT. (B) D134A. (C) D136A. (D) K1706A/K1779A. Black arrows indicate triangular shaped molecules, and white arrows indicate open Y-shaped molecules. (Scale bars: 100 nm.)
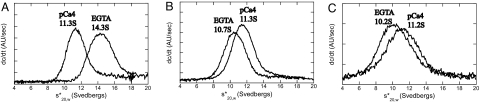
Second, we performed an analytical ultracentrifuge analysis. Similar to those of WT (6), the sedimentation coefficient (s value) of the M5Full-D134A decreased from 14.3S in EGTA conditions to 11.3S in pCa4 conditions (Fig. 4A). In contrast, M5Full-D136A underwent only a small change in the s value, from 10.7S in EGTA conditions to 11.3S in pCa4 conditions (Fig. 4B). Note that the direction of this change is opposite to those for the WT (6) and D134A. In pCa4 conditions, the s values of both mutants are essentially identical to that of the WT (6), which is in the open conformation, indicating that both mutants are in the open conformation in pCa4 conditions. The s value of M5Full-D136A in EGTA conditions is lower than in pCa4 conditions, suggesting that the conformation of the M5Full-D136A is more open in EGTA conditions than in pCa4 conditions. Taken together, these results show that D136 in the motor domain plays a critical role in the inhibition of the ATPase activity by the GTD and in the production of the folded conformation of myosin Va.
Fig. 4.
Sedimentation analysis of M5Full mutants in the absence of Ca2+ (EGTA) and it presence (pCa4). (A) D134A. (B) D136A. (C) K1706A/K1779A.
Two Conserved Basic Residues in the GTD Are Responsible for the Inhibition of Myosin Va.
It is known that that the folded conformation of myosin Va is very sensitive to ionic strength (6–8), suggesting that the head-tail interaction is mediated by electrostatic interaction. In the motor domain of myosin Va, we have identified a conserved acidic residue, D136, responsible for the inhibition of the motor function by GTD and a folded conformation. Crystal structures of the myosin Va motor domain show that the side chain of D136 protrudes outward (13). Thus, we expect that D136 interacts with a basic residue(s) in the GTD.
Knight and colleagues suggested that the acidic residues in lobe P117–P137 in the motor domain may interact with residues in the GTD within R1800–K1810, which contains a cluster of five basic residues (10). The homologous residues of yeast Myo2p are located in the distal end of the GTD, and well suited to match the morphology of the inhibited myosin Va (14). To determine the roles of the basic residues within R1800–K1810, we introduced alanine mutations of these basic residues in the GST-GTD and examined the effect of these mutations on the regulation. We found that all of the alanine mutants, including R1800A, R1805A/R1807A, R1809A, and K1810A, potently inhibited the ATPase activity of M5HMM, with apparent Kd similar to that of WT GST-GTD (SI Fig. 7). These results indicate that the basic residues in R1800–K1810 are not essential for inhibition.
Because the acidic residue D136 is a conserved residue in class V myosins, we hypothesized that the side chain of D136 in the motor domain interacts with conserved basic residue(s) in the GTD. Because the crystal structure of the GTD is known for Myo2p-GTD (the GTD of Myo2p, a yeast myosin V), we compared the sequence of mammalian myosin V (mouse myosin Va, rat myosin Vb, and human myosin Vc) with that of yeast Myo2p. Ten conserved basic residues were identified in the GTD (SI Fig. 8). The crystal structure of the Myo2p-GTD shows that 7 of the 10 are located on or close to the surface of the GTD; one (K1756) is an internal residue, and two (R1647 and K1820) are missing in the crystal structure (SI Fig. 9). We expected that only the conserved basic residues located on the surface of GTD would be capable of interacting with the acidic residue in the motor domain and therefore focused our effort on those residues.
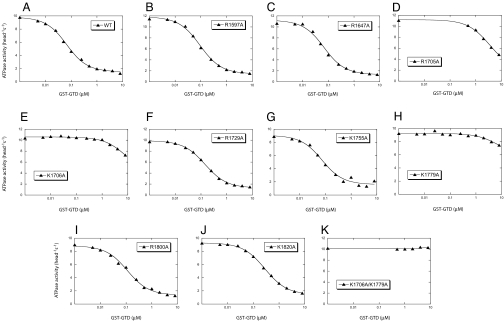
To determine the role of these residues on regulation, we mutated them into alanine and examined the inhibitory activity of the GST-GTD mutants. Among nine single alanine mutations, three, R1705A, K1706A, and K1779A, dramatically decreased the inhibitory activity of GST-GTD on the M5HMM ATPase activity (Fig. 5). The Kd of WT GST-GTD for the inhibition of M5HMM ATPase activity was 0.058 μM, whereas that of the R1705A mutant was 3.6 μM. The inhibitory activities of the K1706A and K1779A mutants were very weak, and these mutants could inhibit activity no more than 30%, even at the highest concentration tested (8 μM). In the crystal structure of Myo2p-GTD, R1705, K1706, and K1779 are located in a cluster in contrast to other conserved basic residues in the GTD (SI Fig. 9). The side chain of R1705 faces inward, but those of K1706 and K1779 protrude outwards, suggesting that both side chains of K1706 and K1779 may interact with the acidic residue of D136.
Fig. 5.
Effects of Alanine mutations of conserved basic residues in the GTD on the inhibitory activity of the GST-GTD. The actin-activated ATPase activities of M5HMM in the presence of various concentrations of GST-GTD were measured. The Kd of GST-GTD (except K1705A, K1706A, K1779A, and K1706A/K1779A) to M5HMM were obtained by a quadratic fit, whereas the Kd of GST-GTD mutants (K1705A, K1706A, and K1779A) to M5HMM were obtained by a hyperbolic fit. The calculated values are 0.058 μM (WT), 0.084 μM (R1597A), 0.058 μM (R1647A), 3.611 μM (R1705A), >8 μM (K1706A), 0.134 μM (R1729A), 0.064 μM (K1755A), >8 μM (K1779A), 0.102 μM (R1800A), and 0.271 μM (K1820A).
We thought that the weak inhibitory activity of K1706A and K1779A might come from interaction between the remaining basic residue, i.e., K1779 or K1706, respectively, in these GTD mutants and D136 in the motor domain. If so, eliminating both basic residues should completely eliminate the inhibitory activity of the GTD. Indeed, K1706A/K1779A mutations completely eliminated inhibitory activity of the GTD (Fig. 5K).
Two Conserved Basic Residues in the GTD Are Responsible for the Folded Conformation of Myosin Va.
Because K1706A/K1779A mutations abolished the inhibition of M5HMM ATPase activity by the GTD, we expected that these mutations would dampen the regulation of full-length myosin Va by Ca2+. As expected, there was significant activation of M5Full-KKAA (K1706A/K1779A) ATPase activity in EGTA (Fig. 2D and SI Table 1). This activity was a little elevated by Ca2+.
Because the inhibition of the actin-activated ATPase activity of myosin Va is correlated with the formation of a folded conformation (6–8), we expected that K1706 and K1779 would play a key role in maintaining a folded conformation of myosin Va. Negative staining electron microscopy revealed that majority of M5Full-KKAA molecules are in the open conformation even in EGTA conditions (Fig. 3D). Analytical ultracentrifuge analysis showed that M5Full-KKAA underwent only a small change from 10.2 S in EGTA conditions to 11.2 S in pCa4 conditions (Fig. 4C) similar to that observed for the D136A mutant (Fig. 4B), indicating that M5Full-KKAA molecules are in an open conformations in both conditions.
Discussion
Structure of Inhibited Myosin Va.
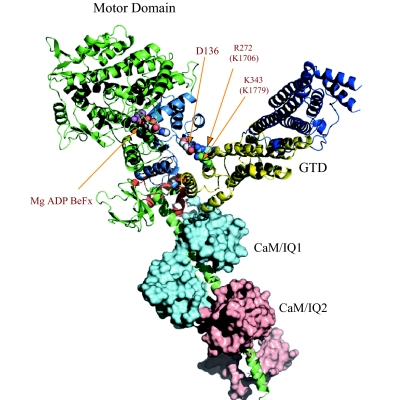
In the present study, we identified a conserved acidic residue in the motor domain (D136) and two conserved basic residues in the GTD (K1706 and K1779) as critical residues for this regulation. Alanine mutations of these conserved charged residues not only abolished the inhibition of motor activity by the GTD, but also prevented myosin Va from forming a folded conformation. Based on these findings, we propose a model for the interaction between the motor domain and the GTD (Fig. 6). In this model, an oxygen atom (Oδ) of D136 in the motor domain and a nitrogen atom (Nη) of R272 and a nitrogen atom (Nζ) of K343 in Myo2p-GTD (homolog residues of K1706 and K1779 in mouse myosin Va, respectively) are within ≈4 Å of each other, thus forming ionic bridges. This arrangement puts the GTD in a pocket of the motor domain, which is formed by the N-terminal, converter, and CaM/IQ1 domains.
Fig. 6.
Model for interaction between the GTD and the motor domain of myosin Va in the postrigor conformation. The GTD-binding pocket consists of the N-terminal domain (blue), converter (orange), and the CaM (cyan) in IQ1. Subdomain-II (yellow) of the GTD interacts with the motor domain. The distances between the oxygen atom (Oδ) of D136 in the motor domain and the nitrogen atom (Nζ) of K272 and the nitrogen atom (Nη) of R343 in Myo2p-GTD (homolog residues of K1706 and K1779 in mouse myosin Va, respectively) are ≈4 Å.
Previously, we and other groups have found that exogenous GTD potently inhibits ATPase activity of the HMM form of myosin Va but only weakly inhibits ATPase activity of the S1 form (9, 10), suggesting that either the formation of the two-headed structure or the coiled-coil domain contributes to the inhibition by the GTD. We also found that deletion of the C-terminal residues of the first long coiled-coil significantly hampers inhibition by the GTD (9). However, deletion of these residues did not influence the formation of the two-headed myosin V. Therefore, we proposed that the C-terminal region of the first coiled-coil of the myosin Va tail is critical for the formation of a folded conformation and the GTD-induced inhibition of myosin Va activity (9). We propose the following scenario for formation of the folded triangular structure. The GTD domain binds to the C-terminal residues of the first coiled-coil. The neck–neck joint is flexible enough to allow the head domain to contact the GTD residing at the C-terminal end of the first coiled-coil. The acidic side chain of D136 interacts with the basic side chains of K1706 and K1779, stabilizing the folded conformation of myosin Va.
The folded triangular conformation of inhibited myosin Va is common in various nucleotides, including ATP, ADP, and rigor (6–10). Thus, it is expected that the interactions between the motor domain and the GTD are preserved in these conditions. Indeed, molecular modeling shows that the interactions between D136 in the motor domain and K1706/K1779 in the GTD can be maintained at least in postrigor (Fig. 6) and prepower stroke conformations (SI Fig. 10).
Mechanism for Inhibition of the Motor Domain ATPase by the GTD.
The mechanism by which the GTD inhibits the ATPase activity of the motor domain is not clear. Because D136 is ≈2 nm away from the ATP-binding site and ≈7 nm away from the actin-binding site, it is less likely that interaction of the GTD basic residues with D136 directly alters ATP binding or actin binding. Knight and colleagues proposed that the GTD allosterically regulates the ATPase activity (10). They suggested two allosteric routes by which binding of the GTD could perturb ATPase activity. First, the N-terminal part of P117-P137 interacts with the C-terminal turns of helix F of the motor domain. At the N terminus of helix F is the P-loop (which is critical for ATPase activity), which could therefore be sensitive to GTD binding. Second, the residues upstream of P117, to N111, form part of the binding sites for the adenine and ribose moieties of ATP. GTD-binding could thus have a big effect on affinity for nucleotide. However, the results from this study suggest other mechanism.
A critical point is that ATP hydrolysis induces a structural change in the motor domain that is connected to the converter and lever arm (13, 15–18). The small change in the catalytic ATPase site is coupled with large conformational changes in the converter and lever arm. Most consider that the flow of structural change above is from ATP to the converter/lever arm. However, as pointed out by Fischer et al. (19), the coupling mechanism is also valid in the reverse direction, i.e., a motion in the converter (and lever arm) can lead to corresponding modifications near ATP. Consistently, a recent study showed the communication from the lever arm to the ATPase site in Dictyostelium myosin II (20). Comparison of various structures of the myosin motor has shown that the conformational changes in the motor domain (with various nucleotides bound) take place mostly by rigid-body rotations of secondary and tertiary structure elements (reviewed in ref. 21). Thus, it is likely that hindering the movements of these structural elements will slow the ATP turnover rate.
Identification of the GTD-binding site in the motor domain provides a clue on how the GTD might inhibit motor domain function. In our model, the GTD-binding site is surrounded by the N-terminal domain, converter, and CaM/IQ1 domain, which is part of the neck domain or lever arm. We thus propose a “brake mechanism” for the inhibition of myosin Va by the GTD: The binding of the GTD to the motor domain prevents movement of the converter/lever arm during the ATP hydrolysis cycle, thus inhibiting the chemical cycle of the motor domain.
In our previous kinetic study of myosin Va ATPase (22), we found that the basal ATPase of myosin Va is also regulated by Ca2+. Single turnover assay of full-length myosin Va ATPase showed that the rate constant of the predominant fraction of the basal ATPase in EGTA was 10 times slower than that in Ca2+. Thus, the basal ATPase activity is also inhibited in EGTA, supporting our model that the GTD-dependent inhibition acts allosterically on the ATPase site by constraining converter/lever arm movement.
We found that the major effect of the formation of the inhibited structure on the ATP hydrolysis cycle is strong inhibition of phosphate release (>1,000-fold) from the active site pocket (22). It is thought that Pi is released from the back door (23), and it is anticipated that a large conformational change occurs before Pi release (21). Starting from the M.ADP.Pi state, actin binds strongly to myosin, opening the backdoor, and swinging the converter/lever arm, which then allows Pi to dissociate. Thus, it is possible that binding of the GTD to the motor domain slows the movement of the converter/lever arm, thus preventing Pi release.
The proposed model for inhibited myosin Va is reminiscent of the inhibited structures of vertebrate smooth muscle myosin II (24, 25) and invertebrate striated muscle myosin II (26), in which the tip of one motor domain binds to the converter region of the other. Thus, it is plausible that the inhibition of the converter/lever-arm movement is a general strategy for myosin motor proteins to achieve the inhibited state.
Mechanism for Ca2+-Dependent Regulation of Myosin Va.
Micromolar concentration of Ca2+ (pCa6-pCa5) stimulates the ATPase activity of myosin Va and induces an open conformation (5–8). Thus, it is expected that activation is initiated by the binding of Ca2+ to CaM, which is bound to the neck domain. The proposed model for the interaction between the motor domain and the GTD immediately suggests two mechanisms for Ca2+-dependent regulation of myosin Va.
One possible mechanism is that the C-terminal domain of the CaM in IQ1 is part of or close to the GTD-binding pocket (Fig. 6), and Ca2+-induced conformational change of CaM in IQ1 may prevent the interaction between the motor domain and the GTD. This mechanism appears to be inconsistent with the report that the light chain in the IQ1 of chicken myosin Va is LC-1sa but not CaM (27). Coexpression of various light chains and CaM with a truncated chicken myosin Va containing motor domain and IQ1 showed that the relative order of affinity for the IQ1 is as follows: LC-1sa > LC-17b > CaM (27). However, the relatively low affinity between CaM and IQ1 in the truncated myosin Va does not necessarily mean a low affinity between CaM and IQ1 in the intact neck having six IQ motifs. It is known that the affinity of CaM for IQ motifs is influenced by the light chain in the neighboring IQ motif (28). The crystal structure of CaM bound to IQ1 and IQ2 of mouse myosin Va shows that there are apolar interactions between CaMs in IQ1 and IQ2 (29). Thus, it is likely that interactions between the adjacent CaMs change their affinity for IQ1 and IQ2 of myosin V heavy chain. Indeed, a recent study showed that the light chain bound to IQ1 of tissue-isolated chicken myosin Va is actually CaM (30).
Alternatively, Ca2+-induced activation is initiated from the CaM in IQ2 and transduced to the CaM in IQ1 through interaction between these CaMs as shown in the crystal structure of light chain-binding domain of myosin V (29, 31). It appears that activation of ATPase activity of myosin Va by high Ca2+ is correlated with Ca2+-induced dissociation of CaM from a single specific IQ motif (32). Several groups have identified the specific IQ motif to be IQ2 and proposed that myosin Va is regulated by Ca2+ via the CaM in IQ2 in a way similar to scallop myosin and smooth muscle myosin (30, 33–35). Currently, there is no direct evidence to distinct these two mechanisms and further experiments are required.
Materials and Methods
Proteins.
Full-length mouse melanocyte-type myosin Va (M5Full) and myosin Va HMM residues of M1-A1234 (M5HMM) were prepared as described in ref. 9. The GST-GTD fusion protein comprised the C-terminal 410 residues of myosin Va, encompassing the entire GTD, which was subcloned into pGEX-4T2 (Amersham) from pFastHTb (Invitrogen) (9) by using BamH1 and XhoI sites, was expressed and purified by standard techniques. The purified GST-GTD protein was dialyzed against 5 mM Tris·HCl (pH 7.5 at 20°C), 0.2 M NaCl, and 1 mM DTT on ice overnight. The concentration of GST-GTD was measured by absorbance at 280 nm, using a molar extinction coefficient of 71,990 liters·mol−1·cm−1.
Point mutations were introduced by Quikchange Site-directed mutagenesis with Ultra High Fidelity Pfu (Stratagene) and standard molecular biological techniques. cDNA derived from PCR were sequenced to confirm the presence of intended mutations and absence of unintended mutation. Note that for easy comparison with previous publications, we used the nucleotide number of X57377, which contains exon-B and F but lacks exon-D. The melanocyte-type myosin Va contains exon-D and -F but lacks exon-B (36). Other proteins and reagents are as described in ref. 9.
ATPase Assay.
The ATPase activity was measured in the presence of an ATP regeneration system containing pyruvate kinase and phosphoenolpyruvate (PEP). The amount of liberated pyruvate was detected by dinitrophenyl hydrazine. The ATPase activity for M5Full was measured in the presence of various concentrations of actin in a solution containing 20 mM Mops-KOH (pH 7.0), 0.1 M NaCl, 1 mM MgCl2, 1 mM DTT, 0.25 mg/ml BSA, 12 μM calmodulin, 0.5 mM ATP, 2.5 mM PEP, 20 units/ml pyruvate kinase, 10–100 nM M5Full, 1 mM EGTA, and various concentrations of actin at 25°C for EGTA conditions (in the absence of Ca2+). EGTA (1 mM) was replaced with 0.9 mM EGTA and 1 mM CaCl2 for pCa4 conditions (in the presence of Ca2+). The ATPase activity of M5HMM in the presence of GST-GTD was measured in EGTA conditions in a solution containing 20 mM Mops-KOH (pH 7.0), 0.1 M NaCl, 1 mM MgCl2, 1 mM DTT, 0.25 mg/ml BSA, 12 μM calmodulin, 0.5 mM ATP, 2.5 mM PEP, 20 units/ml pyruvate kinase, 20–50 nM M5HMM, 40 μM actin, 1 mM EGTA, and various concentrations of GST-GTD at 25°C. The reaction was stopped at various times between 4 and 60 min by adding 25 μl of reaction solution to a well of 96-well plate (Flat Bottom) containing 100 μl of 0.36 mM 2,4-dinitrophenyl hydrazine (Sigma–Aldrich) and 0.4 M HCl. After incubation at 37°C for 15 min, 50 μl of 2.5M NaOH and 0.1 M EDTA were added to each well, and the absorptions at 450 nm were recorded in a microplate reader.
Calculation of Dissociation Constant (Kd) of GST-GTD and M5HMM.
Other Assays.
Negative staining electron microscopy was performed as described in ref. 9. The sedimentation coefficients of M5Full were measured by analytical ultracentrifugation as described with minor modification (6). The purified M5Full was dialyzed against 10 mM Mops-KOH (pH 7.0), 0.2 M NaCl, 1 mM DTT, and 1 mM EGTA on ice overnight. Just before running, MgCl2 was adjusted to 1 mM for EGTA conditions; MgCl2 and CaCl2 were adjusted to 1 mM and 1.1 mM, respectively, for the pCa4 conditions.
Model Building.
The model of the head domain and the binding of the GTD to the head domain were created by using Swiss PDB viewer software. The model of the motor domain and the first two IQ motifs domain of myosin Va was created from the motor domain from chicken myosin Va ADP.BeFx (1w7j.pdb) (13) and the first two IQ motifs domain from mouse myosin Va (2ix7.pdb) (29) by superposing the backbone of R767-I773. The model of the interaction between the head domain and the GTD was created from the head domain of myosin Va (above) and the Myo2p-GTD, the GTD of a yeast myosin V (2f6h.pdb) (14), by positioning the oxygen atom (Oδ) of D136 in myosin Va and nitrogen atom (Nη) of R272 and nitrogen atom (Nζ) of K343 in Myo2p-GTD (homologous to K1706 and K1779 of mouse myosin Va, respectively) within a distance of ≈4 Å and avoiding collision with the N-terminal domain and converter domain.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Kimberley Crowley (University of Massachusetts Medical School) for technical assistance in analytical ultracentrifugation. This work was supported by an American Heart Association Scientist Development Grant (to X.-d.L.) and National Institutes of Health Grants AR41653, AR048526, and DC006103 (to M.I.) and AR34711 (to R.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709741105/DC1.
References
- 1.Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA., III Nat Cell Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- 2.Wu X, Wang F, Rao K, Sellers JR, Hammer JA., III Mol Biol Cell. 2002;13:1735–1749. doi: 10.1091/mbc.01-12-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukuda M, Kuroda TS. J Cell Sci. 2004;117:583–591. doi: 10.1242/jcs.00891. [DOI] [PubMed] [Google Scholar]
- 4.Li XD, Ikebe R, Ikebe M. J Biol Chem. 2005;280:17815–17822. doi: 10.1074/jbc.M413295200. [DOI] [PubMed] [Google Scholar]
- 5.Cheney RE, O'Shea MK, Heuser JE, Coelho MV, Wolenski JS, Espreafico EM, Forscher P, Larson RE, Mooseker MS. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 6.Li XD, Mabuchi K, Ikebe R, Ikebe M. Biochem Biophys Res Commun. 2004;315:538–545. doi: 10.1016/j.bbrc.2004.01.084. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Thirumurugan K, Stafford WF, Hammer JA, III, Knight PJ, Sellers JR. J Biol Chem. 2004;279:2333–2336. doi: 10.1074/jbc.C300488200. [DOI] [PubMed] [Google Scholar]
- 8.Krementsov DN, Krementsova EB, Trybus KM. J Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li XD, Jung HS, Mabuchi K, Craig R, Ikebe M. J Biol Chem. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- 10.Thirumurugan K, Sakamoto T, Hammer JA, III, Sellers JR, Knight PJ. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 12.Coureux PD, Wells AL, Menetrey J, Yengo CM, Morris CA, Sweeney HL, Houdusse A. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- 13.Coureux PD, Sweeney HL, Houdusse A. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pashkova N, Jin Y, Ramaswamy S, Weisman LS. EMBO J. 2006;25:693–700. doi: 10.1038/sj.emboj.7600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 16.Houdusse A, Szent-Gyorgyi AG, Cohen C. Proc Natl Acad Sci USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CA, Rayment I. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 18.Fisher AJ, Smith CA, Thoden JB, Smith R, Sutoh K, Holden HM, Rayment I. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 19.Fischer S, Windshugel B, Horak D, Holmes KC, Smith JC. Proc Natl Acad Sci USA. 2005;102:6873–6878. doi: 10.1073/pnas.0408784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malnasi-Csizmadia A, Toth J, Pearson DS, Hetenyi C, Nyitray L, Geeves MA, Bagshaw CR, Kovacs M. J Biol Chem. 2007;282:17658–17664. doi: 10.1074/jbc.M701447200. [DOI] [PubMed] [Google Scholar]
- 21.Geeves MA, Holmes KC. Annu Rev Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- 22.Sato O, Li XD, Ikebe M. J Biol Chem. 2007;282:13228–13239. doi: 10.1074/jbc.M610766200. [DOI] [PubMed] [Google Scholar]
- 23.Yount RG, Lawson D, Rayment I. Biophys J. 1995;68:44S–47S. discussion 47S–49S. [PMC free article] [PubMed] [Google Scholar]
- 24.Wendt T, Taylor D, Trybus KM, Taylor K. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. J Mol Biol. 2007;312:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padron R. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 27.De La Cruz EM, Wells AL, Sweeney HL, Ostap EM. Biochemistry. 2000;39:14196–14202. doi: 10.1021/bi001701b. [DOI] [PubMed] [Google Scholar]
- 28.Martin SR, Bayley PM. Protein Sci. 2002;11:2909–2923. doi: 10.1110/ps.0210402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houdusse A, Gaucher JF, Krementsova E, Mui S, Trybus KM, Cohen C. Proc Natl Acad Sci USA. 2006;103:19326–19331. doi: 10.1073/pnas.0609436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koide H, Kinoshita T, Tanaka Y, Tanaka S, Nagura N, Meyer zu Horste G, Miyagi A, Ando T. Biochemistry. 2006;45:11598–11604. doi: 10.1021/bi0613877. [DOI] [PubMed] [Google Scholar]
- 31.Terrak M, Rebowski G, Lu RC, Grabarek Z, Dominguez R. Proc Natl Acad Sci USA. 2005;102:12718–12723. doi: 10.1073/pnas.0503899102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nascimento AA, Cheney RE, Tauhata SB, Larson RE, Mooseker MS. J Biol Chem. 1996;271:17561–17569. doi: 10.1074/jbc.271.29.17561. [DOI] [PubMed] [Google Scholar]
- 33.Homma K, Saito J, Ikebe R, Ikebe M. J Biol Chem. 2000;275:34766–34771. doi: 10.1074/jbc.M003132200. [DOI] [PubMed] [Google Scholar]
- 34.Trybus KM, Krementsova E, Freyzon Y. J Biol Chem. 1999;274:27448–27456. doi: 10.1074/jbc.274.39.27448. [DOI] [PubMed] [Google Scholar]
- 35.Trybus KM, Gushchin MI, Lui H, Hazelwood L, Krementsova EB, Volkmann N, Hanein D. J Biol Chem. 2007;282:23316–23325. doi: 10.1074/jbc.M701636200. [DOI] [PubMed] [Google Scholar]
- 36.Seperack PK, Mercer JA, Strobel MC, Copeland NG, Jenkins NA. EMBO J. 1995;14:2326–2332. doi: 10.1002/j.1460-2075.1995.tb07227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.