Abstract
The heat shock protein (Hsp)70 family of molecular chaperones interacts with unfolded proteins through a C-terminal substrate-binding domain (SBD) that is controlled by nucleotide binding to the N-terminal domain. The ATPase cycle is regulated by cochaperones, including DnaJ proteins that accelerate ATP hydrolysis to stabilize the Hsp70–substrate complex. We found that R197 in hamster BiP, which resides at the surface of the nucleotide-binding domain, is critical for both association with endoplasmic reticulum DnaJ proteins and interaction with the SBD. Decreasing the positive charge at this residue enhanced basal ATPase activity, destabilized interaction with the SBD, and reduced substrate release both in vitro and in vivo. Mutation of three glutamic acids in the SBD mimicked many of these effects. Our data provide insights into communications between the two domains and suggest a mechanism by which DnaJ proteins increase ATP hydrolysis.
Keywords: ATPase activity, heat shock protein 70
The heat shock cognate (Hsc)70/heat shock protein (Hsp)70 family of molecular chaperones is highly conserved and is expressed in all cells and organelles. These chaperones regulate an extremely diverse array of functions including transcription, DNA replication, signal transduction, protein translocation into organelles, protein folding, extraction of proteins from organelles, and protein degradation. This remarkable ability to contribute to so many distinct cellular functions is achieved by their ability to bind to unfolded regions on proteins through a C-terminal substrate-binding domain (SBD). Peptide-binding studies have been performed for several Hsp70 family members (1–3), and although there are some differences in apparent specificities, it appears that sequences of ≈7–9 aa in length with alternating hydrophobic residues constitute the recognition motif on unfolded proteins. This is most consistent with Hsp70 proteins recognizing extended polypeptide chains, which would orient the hydrophobic residues in a single direction. NMR (4) and crystallographic (5) studies have further supported this possibility. An early study calculated that sequences with these characteristics would occur every 16–36 aa on the average protein (1, 3), which explains the ability of Hsp70 proteins to interact with such a vast number of proteins and thus contribute to so many distinct cellular functions.
The binding of Hsp70 proteins to substrates is regulated by nucleotide binding to their N-terminal nucleotide-binding domain (NBD) (6, 7), which is even more highly conserved than the SBD. Hsp70s vacillate between an ATP- and an ADP-bound state, with one state representing a fast substrate-binding/fast release form and the other representing a slow substrate-binding/slow release form, respectively (8). Hsp70s are regulated by DnaJ family proteins, which preferentially bind to the ATP form of their partner Hsp70 and stimulate its ATPase activity (9, 10), and by nucleotide-exchange or -releasing factors that further accelerate the cycle (11, 12). Both groups of regulatory proteins are known to interact with the ATPase domain of their designate Hsp70 protein. Hsp70s associate with unfolded proteins when they are in an “open” ATP-bound state. DnaJ proteins bind to the Hsp70, and in some cases directly to the unfolded substrate, and catalyze the rapid hydrolysis of ATP to ADP (11, 13, 14), thus “closing” Hsp70 onto the unfolded protein. The next step in the Hsp70 ATPase cycle is the exchange of ATP back into the nucleotide-binding cleft, which “reopens” Hsp70 and releases the unfolded protein, allowing it to fold. In addition to the changes in the ATPase domain that regulate the operation of the SBD, the binding of peptides to this domain is known to stimulate the ATPase activity of the NBD (15, 16). Thus, it has long been appreciated that interdomain interactions are critical to the functioning of this group of proteins, but, in the absence of a full-length structure, it has been unclear how these two domains communicate. However, two recent studies have shed light on the underlying mechanism of Hsp70 activity (17, 18).
BiP is the mammalian endoplasmic reticulum (ER) Hsp70 ortholog (19, 20). It plays a role in all known functions of the ER, including gating the translocon (21), folding nascent proteins (22, 23), targeting misfolded proteins for degradation (24, 25), regulating the unfolded protein response (26, 27), and contributing to ER calcium stores (28). Except for binding calcium, all of these functions require the ATPase activity of BiP. To date, five ER-localized DnaJ-domain-containing proteins (ERdjs) have been identified (29–33), and at least one of them can bind to substrate proteins (34). Previous studies have identified mutants in the bacterial Hsp70 protein DnaK that are unable to interact with DnaJ (35, 36). Based on these mutants, we produced a BiP R197H mutant that was unable to interact with ERdjs (37). To better understand the interactions between BiP, ERdjs, and substrates, we made additional mutations at this site and on the SBD. These mutants provide insights into the interaction between the substrate binding and NBDs of BiP and suggest possibilities on how DnaJ proteins stimulate the ATPase activity of Hsp70 proteins.
Results
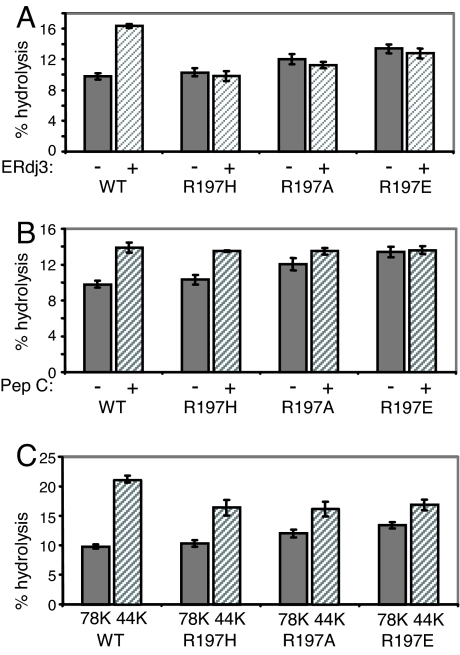
Previously, we constructed a mutation in hamster BiP, R197H (37), which corresponds to a DnaK mutant that does not interact genetically or biochemically with DnaJ (38). The arginine at this position in Hsp70 proteins associates with the highly conserved HPD motif in the J domain of DnaJ proteins (35, 36, 39). Before using this mutant for in vivo studies, we decided to make two additional mutants that could further decrease the affinity of mutant BiP for ERdjs. We reasoned that, although the shorter R group on histidine may not support the type of stable interaction our binding assay measures, it may interact transiently with J domains and interfere with a clear interpretation of in vivo studies. Thus, we also chose to substitute R197 with alanine, a neutral amino acid, and glutamic acid, a negatively charged amino acid. First, we assessed the ability of the various recombinant mutants to bind to a recombinant J domain from ERdj3. Wild-type BiP readily bound in the presence of ATP, but none of the three R197 mutants showed detectable levels of binding [supporting information (SI) Fig. 8]. Second, we measured the ATPase activity of each of the three mutants either alone or with recombinant ERdj3 (Fig. 1A). As previously reported, the basal ATPase activity of wild-type BiP was stimulated by ERdj3, whereas the R197H mutant was not stimulated, even though its basal ATPase activity was very similar to that of wild-type BiP (37). As expected, the R197A and R197E mutants were also not stimulated by ERdj3; however, we did detect a modest but statistically significant increase in basal ATPase activity as the positive charge at residue 197 decreased [R197A (P = 0.034) and R197E (P = 0.012), whereas that of R197H (P = 0.221) was not statistically different from that of wild-type BiP].
Fig. 1.
Characterization of BiP R197 mutants. (A) Recombinant protein was isolated for full-length and N-terminal nucleotide-binding fragment of wild-type BiP and three different mutants with amino acid substitutions at residue 197. Full-length recombinant BiP proteins were assayed for ATPase activity when stimulated with full-length recombinant ERdj3 (A) or peptide C (B). (C) The basal ATPase activity of the full-length and NBD of wild-type and mutant proteins was measured.
Another highly conserved but incompletely understood property of Hsp70 proteins is the ability of peptide binding to the SBD to stimulate their ATPase activity (15). When the four proteins were tested, we found that peptide addition stimulated the ATPase activity of all three mutants and wild-type BiP to the same level, even though their basal ATPase levels varied. Furthermore, the peptide-stimulated rates for all four proteins were very similar to the basal rate of ATP hydrolysis by R197E BiP (Fig. 1B). The SBD of at least two different Hsp70 proteins is known to interact with the NBD in a way that negatively regulates their ATPase activity (6, 40, 41). We produced recombinant protein corresponding to the NBD of wild-type BiP and each of the three mutants and measured the ATPase activity of each protein (Fig. 1C). Removing the SBD from wild-type BiP increased its basal ATPase activity by ≈2.0-fold, in keeping with previous results (40). In the case of the three mutants, we found that removal of their SBD resulted in proteins with rates of hydrolysis that were nearly identical to each other, even though the basal rate of hydrolysis for each of the full-length proteins varied. Together, the properties of these mutants began to suggest that differences in ATP hydrolysis were not intrinsic to the ATPase domain. Instead, they argued that by decreasing the positive charge at residue 197, we may have been altering the interaction of the SBD with the NBD and relieving the suppression that this domain normally provides (6, 40, 41).
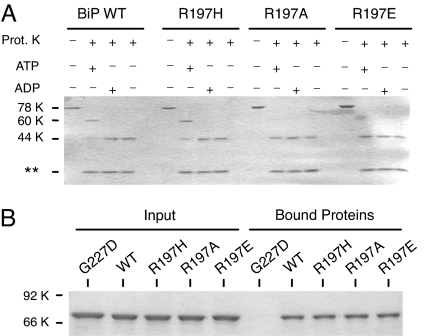
To test this possibility, the mutants were next examined for nucleotide-induced changes in their conformation, as measured by a protease sensitivity assay (Fig. 2A). Both wild-type and R197H BiP demonstrated the characteristic ATP-induced protection of an ≈60-kDa fragment after incubation with proteinase K that includes both the NBD and SBD and is considered to represent the open form of Hsp70 proteins in terms of substrate binding (6, 41, 42). Binding of ADP to these two proteins protected only the ≈44-kDa NBD, which represents the closed form for interacting with substrate. When the R197A and R197E mutants were similarly analyzed, we found that incubation with ATP did not induce the conformational change leading to protection of the ≈60-kDa fragment. Instead, these mutants remained in the closed form, which is indicative of an ADP-bound state. This was not because these mutants are unable to bind to ATP stably, because all three mutants bound ATP–agarose as well as wild-type BiP (Fig. 2B), indicating that the mutations did not affect their ability to bind stably to ATP, but instead argued that this did not lead to the characteristic conformational change in the protein. This strongly implied that as the charge at residue 197 decreased, the SBD was less able to interact with the ATPase domain in the characteristic way and raised the possibility that in the absence of this close contact with the NBD, the SBD may remain in a closed conformation, which would be less permissive for both substrate binding and substrate release.
Fig. 2.
Mutations at residue 197 do not affect nucleotide binding but do alter nucleotide-induced conformational changes. (A) Recombinant BiP proteins were incubated in the absence of proteinase K or with proteinase K, along with ATP, ADP, or nucleotide. Samples were analyzed by SDS/PAGE. Full-length proteins migrated at ≈78 kDa, whereas the normal ATP-protected fragment migrated at ≈60 kDa, and the ADP-protected fragment migrated at ≈44 kDa. Proteinase K can be observed at the bottom of the gel. (B) The three R197 mutants, as well as wild-type and G227D BiP, were incubated with ATP–agarose to assess the effects of the mutations on nucleotide binding. The left side of the gel is the input protein, and right side is the protein bound to the ATP–agarose beads. G227D BiP serves as a control for a non-ATP-binding mutant.
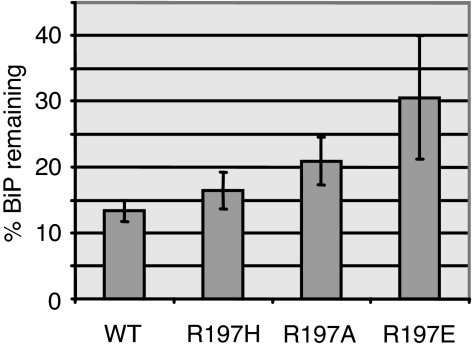
This possibility was first tested by isolating Ig heavy chain (HC) complexes with either wild-type BiP or each of the R197 mutants and testing their ability to be released in vitro with ATP. COS cells were cotransfected with cDNAs encoding Ig HC and the various BiP mutants. Cell lysates were prepared and immunoprecipitated with anti-BiP or with protein A–Sepharose to isolate HC and associated BiP protein. The protein A-precipitated material was either run directly on SDS gels or incubated with ATP to release BiP before electrophoresis. All of the R197 mutants were coprecipitated with Ig HCs, demonstrating that they were capable of binding substrate (Fig. 3). When the BiP signals remaining associated with HC after ATP addition were normalized to the amount of HC that was precipitated in three different experiments and averaged, we found that the R197H mutant was released nearly as effectively as wild-type BiP (16.5 ± 2.8% vs. 13.35 ± 1.7% remaining, respectively). On the other hand, the R197E mutant (30.6 ± 9.4%) and the R197A mutant were partially defective in ATP-mediated release (20.9 ± 3.61%), even though both proteins are able to bind and hydrolyze ATP as well as or better than wild-type BiP. This is consistent with their inability to achieve the ATP-induced conformation represented by protection of an ≈60-kDa fragment in the presence of protease. It is important to note that, in all cases, there was also endogenous wild-type BiP associated with the HC, which was released with ATP, and that the amount of mutant BiP that was expressed in the cells was likely to vary slightly in each experiment.
Fig. 3.
ATP-mediated release of R197 mutants from substrate in vitro. COS cells were cotransfected with Ig HCs and either wild-type BiP or the three R197 mutants. Cells were metabolically labeled, and lysates were divided for immunoprecipitation with anti-BiP or protein A–Sepharose to isolate Ig HCs. One set of HCs was left untreated, and one set was incubated with ATP for each group. Samples were electrophoresed on 10% SDS gels under reducing conditions. BiP signals were quantitated by phosphorimaging, normalized to HC signals, and expressed as the percentage remaining after ATP treatment. Results are from three independent experiments, and error bars represent SD.
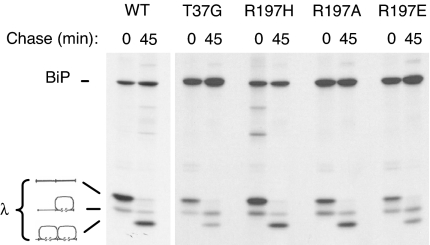
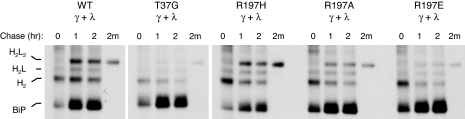
The residue 197 mutants were next examined for their ability to support light chain folding and Ig assembly in vivo. Wild-type and the T37G BiP ATPase mutant (42) were included as controls. λ light chains were coexpressed with each of the BiP constructs, and cells were pulse-labeled in the presence of DTT to accumulate nonoxidized intermediates of light chain, which are a substrate for BiP binding (Fig. 4). Cells were either lysed directly (0 min) or chased for 45 min in complete media lacking DTT before lysing and immunoprecipitating with anti-λ LC antiserum. After 45 min, most of the light chains coexpressed with either wild-type or R197H BiP had reached the fully oxidized form. However, similar to the results obtained with the T37G mutant, we reproducibly found that, in cells expressing R197E BiP, approximately half of the light chains reached the fully oxidized state, whereas the remaining half were only partially oxidized. The R197A mutant was only slightly less active than wild-type or R197H BiP in this assay.
Fig. 4.
R197 substitutions can affect Ig light chain folding. COS cells were transfected with the indicated BiP constructs along with mouse λ light chains. Cells were pulse-labeled with [35S]methionine in the presence of DTT (0) and then chased in complete media without DTT for 45 min. Cell lysates were immunoprecipitated with anti-λ antibodies, and proteins were electrophoresed under nonreducing conditions. The migration of completely reduced, partially oxidized, and completely oxidized light chains is indicated at the right.
As another test of in vivo activity, we measured the ability of the R197 mutants to support Ig assembly. COS cells were cotransfected with Ig HCs and light chains along with each of the BiP mutants. Cells were pulse-labeled and then chased for 1 and 2 h. Cell lysates were incubated with protein A–Sepharose to isolate Ig HCs, and the precipitated material was analyzed under nonreducing conditions to monitor the assembly state of the HCs. After the initial pulse, most of the HCs were dimers (H2), regardless of with which BiP construct they were coexpressed (Fig. 5). Within 1 h, completely assembled (H2L2) molecules were readily detected in the wild-type and R197H BiP-expressing cells, and, by 2 h, a significant proportion of HCs had been secreted. When R197E-expressing cells were examined, we found that the pattern of Ig assembly and secretion resembled that obtained with T37G BiP. The total amount of HCs in the cells was reduced during the chase period compared with wild-type BiP cells, and the relative amount of completely assembled and secreted Ig was also diminished. This demonstrates that the R197E mutant is also defective in this in vivo test for function. The R197A mutant was found to be slightly better than the R197E mutant in this assay but showed significantly less activity than R197H BiP. Thus, various substitutions at position 197 demonstrate the full range of activity from wild-type to ATPase mutant in in vivo assays, and the degree of activity correlates with the charge at this residue. The BiP mutant with a positive charge retained full activity, with the neutral substitution retaining partial activity, and a negative charge at this residue completely inhibited in vivo activity.
Fig. 5.
Effects of R197 substitutions on Ig assembly. COS cells were transfected with the indicated BiP constructs along with vectors encoding Ig γ HC and λ light chain. Cells were pulse-labeled (0) and chased for the indicated times. The media from the 2-h chase (2m) was saved, and both cell lysates and culture supernatants were immunoprecipitated with anti-γ HC antiserum. Samples were analyzed under nonreducing conditions, and the mobility of assembly intermediates is indicated at the left.
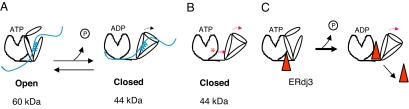
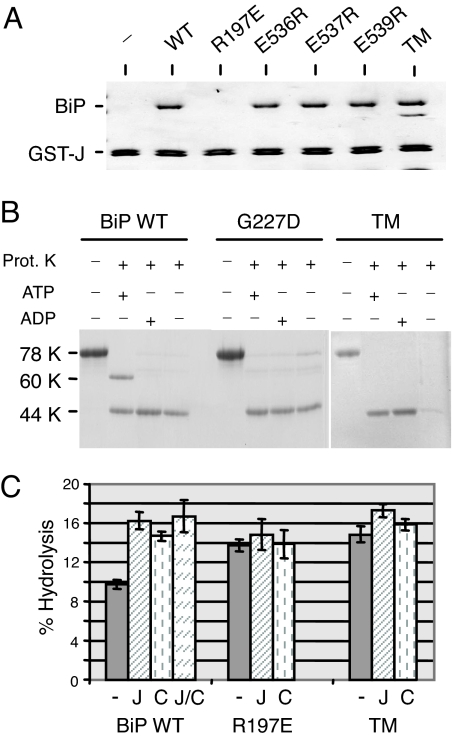
Together, these data led us to propose a model in which the SBD of Hsp70 proteins suppresses ATPase activity when they are in the ATP, or open, form. When DnaJ proteins bind to the NBD via interactions with R197, we suggest that they stimulate the ATPase activity and closing of the SBD, at least in part, by pushing the two domains away from each other. We hypothesized that, by substituting decreasingly positive charges at position 197, we mimicked this action because of steric hindrance or charge clashes with the SBD (Fig. 6). Thus, we inspected the recent full-length crystal structure of bovine Hsc70 (17, 43) and found three highly conserved glutamine acid residues on the SBD in the vicinity of R197. It is important to note that the structure is for the ADP-bound form of Hsc70, but, nonetheless, we mutated each of these residues to arginine alone and together. All four mutants were still able to bind to the J domain (Fig. 7A), and each of the single mutants adopted the 60-kDa open form with ATP (SI Fig. 9A), but the triple mutant (TM) did not (Fig. 7B). When the recombinant proteins were tested for ATPase activity, we found that the basal ATPase activity of the single mutants was similar or only slightly higher than wild-type BiP and could be stimulated with both J domain and peptide (SI Fig. 9B). Conversely, when all three negative charges in the SBD were mutated together, the basal ATPase activity the TM was significantly higher than wild-type BiP (P = 0.003) and was even slightly higher than that of R197E (Fig. 7C). Similar to R197E, the TM was not stimulated by peptide and was only very weakly stimulated by the J domain, even though it bound stably to the J domain. When the TM was tested for its ability to be released from HC in vitro with ATP, we found that it was as defective as the R197E mutant in this assay, whereas the single mutants were only slightly inhibited (SI Fig. 10). Thus, mutation of all three negatively charged amino acids on the SBD that lie near R197 to a positively charged arginine resulted in a protein that was unable to form the 60-kDa fragment with ATP, that had higher basal ATPase activity, which was not appreciably stimulated by J domain or peptide, and that was unable to release from HC with ATP.
Fig. 6.
Model for functional interactions between nucleotide and substrate-binding domains. (A) When BiP is in the ATP-bound form, which is open for substrate binding, the SBD interacts with the NBD in a way resulting in the protection of a 60-kDa fragment when the protein is treated with protease. Hydrolysis of ATP induces closing of the SBD and an alteration in its interaction of the NBD, so that only a 44-kDa fragment is now protected. (B) Mutation of R197 (red asterisk) on the NBD to a neutral or negatively charged residue mimics the ADP-bound form in that the two domains no longer interact in a way that allows protection of a 60-kDa fragment and induces closing of the SBD. (C) ERdj3 (triangle) binds to the ATP-bound form of BiP and serves to push the SBD domain away from the NBD and thus relieve the suppression to ATP hydrolysis that this domain provides and concomitantly to close the SBD.
Fig. 7.
Mutation of negative residues on the SBD of BiP can affect interdomain communication and ATP hydrolysis. (A) Recombinant proteins corresponding to wild-type BiP, three single-point mutants in the SBD, and a TM in which all three residues were combined were incubated with a fusion protein corresponding to ERdj3 J domain and GST. Bound material was subjected to SDS/PAGE analysis and detected by Coomassie blue staining. (B) The indicated recombinant BiP proteins were incubated with ATP, ADP, or no nucleotide and treated with proteinase K as described in Fig. 2. (C) Recombinant BiP proteins were analyzed for ATPase activity alone (−), with ERdj3 (J), with peptide C (C), and with the two together (J/C).
Discussion
In this study, we produced and characterized three different mutations at R197 in hamster BiP. To our surprise, the initial in vitro characterization of the corresponding recombinant proteins demonstrated that, in addition to playing a critical role in binding to J domain proteins, changes in this residue affected a number of additional BiP parameters in a charge-dependent manner. Because the charge at amino acid 197 became less positive, the mutant had higher rates of ATP hydrolysis and showed less of an ability to protect a 60-kDa fragment with ATP or to be stimulated with peptide. The increased ATP hydrolysis by the R197A and R197E mutants did not appear to be attributable to direct changes in the ability of the mutants to hydrolyze ATP because the ATPase rate for the 44-kDa domain alone was very similar for each of the mutants. Instead, the differences in ATP hydrolysis could be due to changes in the ability of the SBD to hamper ATP hydrolysis (6, 41) when these mutations are present. We interpret these data to argue that R197 interacts with the SBD when BiP is in the ATP-bound state and positions the SBD in a manner that diminishes ATP hydrolysis. This could serve to diminish nonnecessary hydrolysis in the absence of a J protein or substrate. The recent structure obtained for full-length bovine Hsc70 (17) and mutational analysis of the NBD and linker of DnaK (18) are consistent with this interpretation. Mutation of the corresponding R151 on the NBD (18) or D393 in the linker (44) of DnaK dramatically interfered with relaying conformational changes to the SBD upon ATP binding, as demonstrated by changes in tryptophan fluorescence. In the case of Hsc70, several additional charged residues exist in the vicinity of R171 (corresponds to R197 in BiP) and form ionic interactions with residues in the SBD (17). Mutation of these residues resulted in altered rates of ATP hydrolysis and affected the ability of ATP to protect an ≈60-kDa fragment. We found that, by decreasing the positive charge at R197 in BiP, the SBDs of our mutants interacted less stably or not at all with the NBD, as judged by the ability to form the 60-kDa fragment. This implied that this residue may normally interact with a negatively charged amino acid(s) on the SBD or hinge to hold the two domains together. Although there is no full-length structure of the ATP-bound form of any Hsp70 protein, there were several glutamic acid residues juxtaposed to R171 in the ADP-bound form of Hsc70 (17) that are conserved in BiP and other Hsp70 proteins. Indeed, we found that mutation of all three amino acids to arginine yielded a BiP mutant that was phenotypically very similar to the R197E mutant. Although this mutant supports our model, clearly these data must be interpreted cautiously in the absence of an ATP structure for an Hsp70. However, a recent crystallographic structure has been obtained for the ATP bound form of Ssel, a member of the high molecular weight Hsp70s that are thought to serve as nucleotide exchange factors for Hsp70s (45). The structure revealed extensive interaction between NBD and the linker, the lid of the SBD (SBD alpha), and the SBD itself (SBD beta) when Ssel is bound to ATP.
In addition, we found that the less positive the charge at residue 197 was, the more compromised the mutant was in terms of substrate release both in vivo and in vitro. This could be interpreted to suggest that interaction of the SBD with the ATPase domain holds the lid open, supported by the Ssel ATP structure (45), whereas hydrolysis to ADP loosens the interaction, allowing the lid to close. Our data would argue that R197 plays a critical role in the interaction between the domains and, therefore, the opening and closing of the SBD. We hypothesized that when mutations in the NBD effectively “pushed” the SBD away, it adopted a closed configuration. Indeed, this interpretation is supported by data on similar mutants in Hsc70 (17), and in DnaK, where auxilin and peptide release respectively was negatively affected (18). The fact that our R197E mutant was as unable as a BiP ATPase mutant to properly fold or assemble substrate proteins in vivo argues that the interactions that are affected in our in vitro analyses are at the heart of the protein's function in vivo.
The presence of an arginine at this site is highly conserved in all Hsp70 proteins and is a major site for interaction with the J domain of DnaJ proteins (35, 39, 46). Although the substitution of a histidine at this position in BiP affected its ability to interact with J domains and to gate the translocon in an earlier study (37), it did not appear to dramatically alter the interaction of the SBD and NBD, as measured by protease sensitivity or to affect in vivo protein folding and assembly of Ig proteins. It is striking that this residue should be intimately involved in both interdomain communication and in interactions with the J domain. A function of J domain proteins is to bind the ATP-bound form of Hsp70 proteins and stimulate the hydrolysis of ATP. Based on the data presented here, we suggest that part of the mechanism by which DnaJ proteins stimulate Hsp70 ATPase activity is by serving as a wedge to push the SBD away from the NBD, an idea that is supported by recent crystallographic data obtained for a portion of Hsc70 and a J domain (39). This would serve to both stimulate ATP hydrolysis and to close the lid on the SBD. In keeping with this, our SBD TM (E536R, E537R, and E539R) retained the ability to bind to a J domain, but its ATPase activity was not appreciably enhanced by this binding. This argues that binding of the J domain is not necessarily sufficient to stimulate ATP hydrolysis, but that the J domain must interact with BiP when the NBD and SBD are juxtaposed to stimulate hydrolysis. In the case of DnaK, it has been shown that full function (47) and maximal stimulation of ATPase activity by DnaJ requires that it interacts with both the arginine (R151) on the NBD and residues within the substrate-binding cleft, which could be observed even when the two signals were delivered in trans (14, 48). However, we did not observe a similar synergist stimulation of ATPase activity when J and peptide were added to BiP together (Fig. 7C), suggesting that there may be some intrinsic differences between these proteins.
In summary, we produced BiP mutants that were anticipated to be defective in the interaction with ER-localized mammalian DnaJ proteins. The substitution of neutral or negatively charged amino acids at R197 resulted in proteins that were also defective in interdomain communications between the NBD and SBD that interfered with protein folding and assembly in cells. This extends recent studies on DnaK and Hsc70 to an ER homolog and underscores the importance of this residue in interactions with J domains and with the SBD both in vivo and in vitro, thus providing additional insights into Hsp70/DnaJ/substrate biology.
Materials and Methods
Generation of BiP Mutants.
To produce BiP mutants at R197, the initial CGG codon in a hamster BiP cDNA was mutated to CAC for histidine, GCG for alanine, and GAG for glutamic acid by site-directed mutagenesis with the use of Quikchange Site-Directed Mutagenesis kit (Stratagene). Residues E536, E537, and E539 of the SBD were mutated to arginine either alone or in combination. After sequencing, each mutant was inserted into pQE vector to produce full-length recombinant protein in bacteria and into pMT vector for expression in mammalian cells. Each of the NBD mutants was also expressed in the context of the ATPase domain alone.
Protein Expression and Purification.
Recombinant BiP proteins were expressed in Escherichia coli M15 cells under nondenaturing conditions and purified on Ni–agarose beads (Qiagen). A bacterial expression vector for full-length human ERdj3 with an N-terminal hexahistidine tag was kindly provided by Yi Jin in our laboratory and one for a GST–fusion protein that encodes the J domain and a portion of the G/F linker was a generous gift from David Haslam (Washington University, St. Louis, MO) and has been described previously (31).
GST Pull-Down Assay.
GST pull-down assays were performed as described previously (31). Recombinant BiP proteins bound to ERdj3-coupled glutathione agarose beads were subjected to SDS/PAGE analysis and detected by Coomassie blue staining.
ATP-Binding and ATPase Assays.
ATP-binding assays (49) and ATPase assays were performed as described previously (32, 42). To measure ATP hydrolysis, the various recombinant BiP proteins were incubated alone at a concentration of 1 μM each, with 0.5 μM full-length ERdj3 or with 5 mM C peptide (1) at 37°C for 20 min. After chromatography, the radioactive ATP and free phosphate signals were quantified by phosphorimaging analysis (Molecular Dynamics) with the use of Image Quant software. The free phosphate signal was expressed as a percentage of the total phosphate signal. Data were deduced from three independent experiments, and the error bars represent SDs.
Antibodies.
Polyclonal anti-rodent BiP antisera has been described previously (40). Goat anti-human Ig HC and goat anti-mouse λ antibodies were purchased from Southern Biotechnology Associates.
Transient Expression and Immunoprecipitation.
COS-1 cells were transfected with the indicated vectors by using the FuGENE 6 transfection reagent (Boehringer Mannheim). For BiP release experiments, cells were metabolically labeled for 2 h with 100 μCi [35S]-Translabel (ICN). Cell lysates were prepared and divided into three aliquots. One was immunoprecipitated with BiP and two with protein A–Sepharose to isolate Ig HCs. One of the latter samples was then incubated with 1 mM ATP to release BiP, as described previously (50). Samples were analyzed on SDS gels, and signals for BiP and HC were quantified by phosphorimaging. BiP bound to HC was normalized to the HC signal, and the amount of BiP remaining after ATP addition was calculated as a percentage of the BiP bound without ATP. Three separate experiments were performed for each mutant to calculate the percentage of BiP remaining. For the light chain folding experiments, cells were labeled in the presence of 10 mM DTT for 20 min and chased in media devoid of DTT as described in ref. 22. Samples were separated on SDS gels under nonreducing conditions to visualize the light chain folding intermediates. To examine Ig assembly and secretion, transfected cells were pulse metabolically labeled for 20 min and chased for 1 and 2 h. Culture supernatants from the 2-h chase and cell lysates were immunoprecipitated with the indicated antibodies. Precipitated proteins were analyzed on SDS gels under nonreducing conditions. Signals were enhanced with Amplify (Amersham) for radiographic visualization.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Yi Jin and Melissa Mann in our laboratory for generous gifts of reagents, assistance with experiments, and scientific advice. This work was supported by National Institutes of Health Grant GM54068 (to L.M.H.), National Cancer Institute Cancer Center Core Grant CA21765, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702132105/DC1.
References
- 1.Flynn GC, Pohl J, Flocco MT, Rothman JE. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 2.Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 3.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landry SJ, Jordan R, McMacken R, Gierasch LM. Nature. 1992;355:455–457. doi: 10.1038/355455a0. [DOI] [PubMed] [Google Scholar]
- 5.Zhu X, Zhao X, Burkholder WG, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappell TG, Konforti BB, Schmid SL, Rothman JE. J Biol Chem. 1987;262:746–751. [PubMed] [Google Scholar]
- 7.Flaherty KM, DeLuca Flaherty C, McKay DB. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 8.Liberek K, Skowyra D, Zylicz M, Johnson C, Georgopoulos C. J Biol Chem. 1991;266:14491–14496. [PubMed] [Google Scholar]
- 9.Cheetham ME, Caplan AJ. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banecki B, Zylicz M. J Biol Chem. 1996;271:6137–6143. doi: 10.1074/jbc.271.11.6137. [DOI] [PubMed] [Google Scholar]
- 13.Bukau B, Horwich AL. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 14.Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Proc Natl Acad Sci USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flynn GC, Chappell TG, Rothman JE. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 16.McCarty JS, Buchberger A, Reinstein J, Bukau B. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 17.Jiang J, Prasad K, Lafer EM, Sousa R. Mol Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel M, Bukau B, Mayer MP. Mol Cell. 2006;21:359–367. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Munro S, Pelham HR. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 20.Haas IG, Meo T. Proc Natl Acad Sci USA. 1988;85:2250–2254. doi: 10.1073/pnas.85.7.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamman BD, Hendershot LM, Johnson AE. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y-K, Brewer JW, Hellman R, Hendershot LM. Mol Biol Cell. 1999;10:2209–2219. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang N, Daniels R, Hebert DN. Mol Biol Cell. 2005;16:3740–3752. doi: 10.1091/mbc.E05-05-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P. J Cell Biol. 2002;158:247–257. doi: 10.1083/jcb.200204122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 27.Shen J, Chen X, Hendershot L, Prywes R. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 28.Lievremont JP, Rizzuto R, Hendershot L, Meldolesi J. J Biol Chem. 1997;272:30873–30879. doi: 10.1074/jbc.272.49.30873. [DOI] [PubMed] [Google Scholar]
- 29.Tyedmers J, Lerner M, Bies C, Dudek J, Skowronek MH, Haas IG, Heim N, Nastainczyk W, Volkmer J, Zimmermann R. Proc Natl Acad Sci USA. 2000;97:7214–7219. doi: 10.1073/pnas.97.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer HA, Grau H, Kraft R, Kostka S, Prehn S, Kalies KU, Hartmann E. J Biol Chem. 2000;275:14550–14557. doi: 10.1074/jbc.275.19.14550. [DOI] [PubMed] [Google Scholar]
- 31.Yu M, Haslam RH, Haslam DB. J Biol Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- 32.Shen Y, Meunier L, Hendershot LM. J Biol Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- 33.Cunnea PM, Miranda-Vizuete A, Bertoli G, Simmen T, Damdimopoulos AE, Hermann S, Leinonen S, Huikko MP, Gustafsson JA, Sitia R, et al. J Biol Chem. 2003;278:1059–1066. doi: 10.1074/jbc.M206995200. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y, Hendershot LM. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Proc Natl Acad Sci USA. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gassler CS, Buchberger A, Laufen T, Mayer MP, Schroder H, Valencia A, Bukau B. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alder NN, Shen Y, Brodsky JL, Hendershot LM, Johnson AE. J Cell Biol. 2005;168:389–399. doi: 10.1083/jcb.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh WC, Lu CZ, Gross CA. J Biol Chem. 1999;274:30534–30539. doi: 10.1074/jbc.274.43.30534. [DOI] [PubMed] [Google Scholar]
- 39.Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Mol Cell. 2007;28:1–12. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendershot LM, Wei J-Y, Gaut JR, Lawson B, Freiden PJ, Murti KG. Mol Biol Cell. 1995;6:283–296. doi: 10.1091/mbc.6.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kassenbrock CK, Kelly RB. EMBO J. 1989;8:1461–1467. doi: 10.1002/j.1460-2075.1989.tb03529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei J-Y, Gaut JR, Hendershot LM. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- 43.Jiang J, Lafer EM, Sousa R. Acta Crystallogr F. 2006;62:39–43. doi: 10.1107/S1744309105040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogel M, Mayer MP, Bukau B. J Biol Chem. 2006;281:38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, Hendrickson WA. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landry SJ. Biochemistry. 2003;42:4926–4936. doi: 10.1021/bi027070y. [DOI] [PubMed] [Google Scholar]
- 47.Davis JE, Voisine C, Craig EA. Proc Natl Acad Sci USA. 1999;96:9269–9276. doi: 10.1073/pnas.96.16.9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karzai AW, McMacken R. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- 49.Milarski KL, Morimoto RI. J Cell Biol. 1989;109:1947–1962. doi: 10.1083/jcb.109.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaut JR, Hendershot LM. J Biol Chem. 1993;268:7248–7255. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.