Abstract
Flagellated bacteria, such as Escherichia coli, are propelled by helical flagellar filaments, each driven at its base by a reversible rotary motor, powered by a transmembrane proton flux. Torque is generated by the interaction of stator proteins, MotA and MotB, with a rotor protein FliG. The physiology of the motor has been studied extensively in the regime of relatively high load and low speed, where it appears to operate close to thermodynamic equilibrium. Here, we describe an assay that allows systematic study of the motor near zero load, where proton translocation and movement of mechanical components are rate limiting. Sixty-nanometer-diameter gold spheres were attached to hooks of cells lacking flagellar filaments, and light scattered from a sphere was monitored at the image plane of a microscope through a small pinhole. Paralyzed motors of cells carrying a motA point mutation were resurrected at 23°C by expression of wild-type MotA, and speeds jumped from zero to a maximum value (≈300 Hz) in one step. Thus, near zero load, the speed of the motor is independent of the number of torque-generating units. Evidently, the units act independently (they do not interfere with one another), and there are no intervals during which a second unit can add to the speed generated by the first (the duty ratio is close to 1).
Keywords: colloidal gold, molecular motor, motility
In Escherichia coli, a motor torque-generating unit is composed of four copies of MotA and two copies of MotB, enclosing two proton-conducting transmembrane channels (1). Protonation and subsequent de-protonation of Asp-32 at the cytoplasmic end of either channel is thought to drive conformational changes that exert forces on the periphery of the rotor, via electrostatic interactions between MotA and the rotor protein FliG (2). Estimates of the number of torque-generating units per motor range from 8 to 11 (3, 4). At high loads and low speeds, motors operate near thermodynamic equilibrium, where rates of ion movement or conformational change do not matter (5). One can heat cells up or cool them down or change the solvent from H2O to D2O without changing motor torque; however, at low loads and high speeds, rates do matter, and motors run more slowly at lower temperatures or in D2O (6, 7).
Because power output is the product of torque and speed, it is of interest to measure the torque generated at different speeds. The torque-speed relationship is useful for testing specific models of motor rotation (1, 5). In E. coli, motor torque is maximum at stall, it falls ≈10% between 0 and ≈170 Hz (at room temperature), and then it drops rapidly, reaching 0 at ≈330 Hz (6, 8). We have measured torque-speed curves for motors with different numbers of torque-generating units by linking latex spheres of different sizes to flagellar filament stubs on motA cells and then inducing expression of wild-type MotA (9). This work suggested that the family of torque-speed curves might converge to the same zero-torque value, as expected if torque generators move independently and have a high duty ratio (10); however, the smallest latex bead that we were able to use was 300 nm in diameter, preventing close approach to the zero-torque limit. Subsequently, a motor model that explains most features of the torque-speed curves predicted that, near zero torque, one torque-generating unit would spin the motor substantially faster than two or three, because the units would interfere with one another (11). To learn more about motor behavior near zero torque, we devised the assay described here, finding that one torque-generating unit is as effective as many, implying that torque generators move independently with a duty ratio close to one. The experiments were done in two ways, by inducing the expression of wild-type MotA at a high level and monitoring the speed of the motor as a function of time, or by inducing the expression of wild-type MotA at a low level or a moderate level and comparing speeds at steady state. Experiments were repeated at high load with latex spheres of diameter 1,000 nm, so that behaviors at low and high loads could be compared.
Results
Temporal Behavior.
We constructed two isogenic strains deleted for genes encoding FliC (flagellin) and CheY (the chemotaxis response regulator) and carrying a motA point mutation. These cells lacked flagellar filaments but had proximal hooks (the short flexible coupling between the end of the drive shaft and the base of the filament). Their motors were paralyzed, but when cured by expression of wild-type MotA, were destined to spin exclusively counterclockwise. The first strain (JY22) carried a plasmid expressing wild-type MotA under control of the rhamnose promoter (12), and the second strain (JY23) carried, in addition, a compatible plasmid expressing the FliC sticky (FliCst) allele (13). Hooks of cells of the first strain were labeled with 60-nm-diameter gold spheres (see Materials and Methods). Filaments of the second strain were reduced to stubs by viscous shear and were labeled with 1,000-nm-diameter latex spheres.
Cells were allowed to adhere to a glass coverslip that was mounted in a flow cell at 23°C. The expression of wild-type MotA was leaky, so some spheres rotated before the addition of inducer. When working with 60-nm gold, these spheres were ignored. We positioned the image of a stationary sphere over the pinhole, added the inducer [1 mM rhamnose in motility medium containing 10% (vol/vol) tryptone broth] and then waited to see what might happen. Fig. 1 A and B show dramatic results. In ≈1 in 10 trials, a sphere would suddenly spin at full speed, as shown in Fig. 1A. Usually, the speed would remain high thereafter, but sometimes it would toggle between zero and full speed, as shown in Fig. 1B, presumably because attachment of the first torque generator was transient. Five such spheres were found, all behaving in a similar way. To confirm that these spheres were able to rotate and did not simply break free, we compared signals due to their random motion before rotation to signals obtained with spheres that were stuck, and found that the former signals were much larger and more erratic than the latter ones (data not shown). Stuck spheres were easy to identify by eye, because they could be brought into sharp focus.
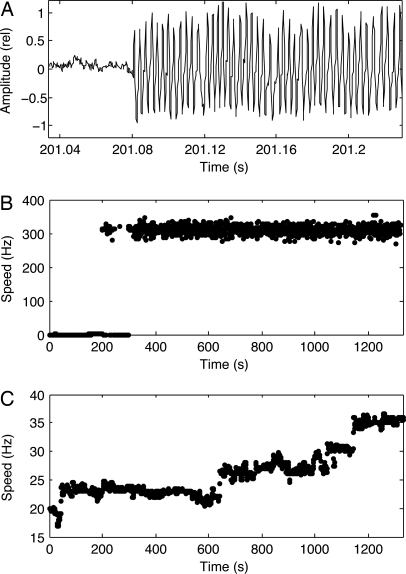
Fig. 1.
Resurrection traces. (A) Photomultiplier output (AC coupled) showing sudden onset of rotation of a 60-nm gold sphere on the hook of a paralyzed cell (strain JY22) after induction of wild-type MotA. (B) Speed as a function of time after induction for the sphere tracked in A. (C) Speed as a function of time after induction for a 1-μm latex sphere on a filament stub (strain JY23).
When working with 1,000-nm latex, we saved time by starting with a sphere that already had begun to rotate, added the inducer, and then followed the behavior of the sphere over time, as shown in Fig. 1C. Now, the motor increased its speed in a series of equally spaced increments (five in Fig. 1), indicating that several wild-type torque-generating units were able to displace defective copies over the time span of the experiment. At either low or high load, the speed increased within 10 min after addition of the inducer, and at high load, it reached its maximal value within an additional 20–30 min. It is reasonable to expect similar behavior in either load regime, because the only genetic difference between the two strains is the additional low-copy-number plasmid in strain JY23 that encodes FliCst. We conclude that the speed of the motor is independent of the number of stator units near zero external load.
Steady-State Behavior.
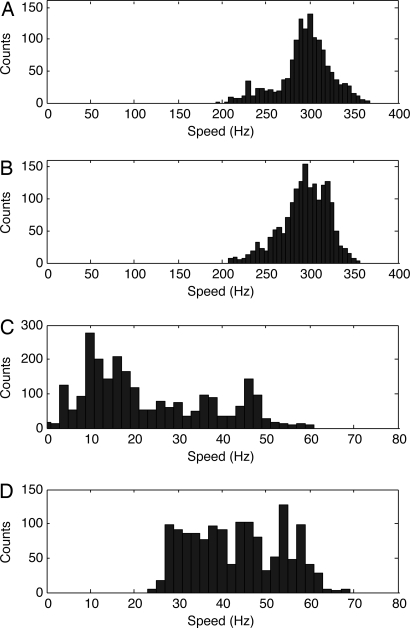
Another way to do these experiments is to monitor the steady-state speeds of populations of cells labeled with 60 nm gold or 1,000 nm latex in which expression of wild-type MotA is induced with 5 or 100 μM rhamnose. The corresponding speed histograms are shown in Fig. 2. At steady state, the number of torque generating units per motor depends on the level of induction (4). This number did not affect the speeds of motors operating near zero load (Fig. 2 A and B), but it did affect the speeds of motors operating at high load (Fig. 2 C and D). So once again, we conclude that motor speed is independent of the number of torque-generating units near zero load.
Fig. 2.
Speed histograms for steady-state induction of MotA. Each sphere was observed for 30 s with its speed computed once per second. (A) Histogram for gold spheres at low induction (5 μM rhamnose, 34 spheres on cells of strain JY22, 5-Hz bins). (B) Histogram for gold spheres at high induction (0.1 mM rhamnose, 40 spheres on cells of strain JY22, 5-Hz bins). (C) Histogram for latex spheres at low induction (5 μM rhamnose, 44 spheres on cells of strain JY23, 2 Hz bins). (D) Histogram for latex spheres at high induction (0.1 mM rhamnose, 27 spheres on cells of strain JY23, 2-Hz bins).
Discussion
One expects a single torque generating unit to drive the rotor as rapidly as many if torque generators do not interfere with one another (see below) and if they have a high duty ratio, i.e., if they remain linked to the rotor for most of their mechanochemical cycle (5, 9, 10). Zero-load speeds were all ≈300 Hz, a value inferred earlier from experiments with tethered cells spun by electro-rotation (8). Standard deviations in speed were relatively small (≈30 Hz). The smallest load on a free-running motor studied previously was that of a 300-nm-diameter latex sphere on a filament stub (9). Because the rotational viscous drag of a sphere scales with the cube of its radius, a 60-nm-diameter gold sphere on a hook reduces the lowest load ever studied by more than two orders of magnitude.
For a sphere of radius a rotating about its center in a medium of viscosity η, the rotational frictional drag coefficient is fθ = 8πηa3. If the rotation is about an axis offset by a distance l from the center, the drag coefficient is larger by an amount 6πηal2. For a 60-nm gold sphere on a hook of diameter 15-nm, assuming an additional 5-nm of macromolecules around the gold sphere and 5-nm of antibodies around the hook, the frictional drag coefficient ranges from 1.1 to 2.5 × 10−3 pN nm per rad s−1 (for a = 35 nm and l = 0 to 47.5 nm, respectively). The load might be larger if the hook is bent, but given the l2 dependence, by not more than a factor of ≈4. The rotational drag coefficient of the rotor was estimated to be 2 × 10−2 pN nm per rad s−1, modeled as a sphere of radius 20 nm in lipid of viscosity 100 cp (5). Therefore, with our method, the external load on the motor is probably smaller than the intrinsic load due to internal friction.
The hook stiffness was measured previously, kθ = 400 pN nm rad−1 (14, 15), yielding a relaxation time for the hook τ = fθ/kθ ≤ 6 μs. For a rotational frequency ≈300 Hz and 26 steps per revolution (16), the time between each step is ≈128 μs, which is much larger than the relaxation time for the hook. Therefore, for modeling purposes, the effect of the gold on motor function can be absorbed into the rotor by increasing its rotational drag coefficient and assuming that the external load is zero. A recent model of the flagellar motor (11) predicts that, at zero external load, the speed will decrease as the number of torque-generating units increases, because the stator elements interfere with one another. One way to avoid this problem is to uncouple the stator elements from one another by increasing the compliance between each unit and the rigid framework of the cell wall. We repeated the simulation of Xing et al. (11) at zero load by including the effect of stator springs, finding that speed is independent of the number of torque-generating units if the springs have a stiffness of 0.5 pN/nm or less (in a direction tangent to the edge of the rotor); see supporting information (SI) Text. Springs this compliant, if linear, would stretch a distance of order 10 nm when motors run at high load, which seems unlikely. However, the linkage to the peptidoglycan layer provided by the C-terminal end of MotB is more nearly parallel than perpendicular to the axis of rotation, and proteins are neither homogeneous nor isotropic, so the springs might be highly nonlinear.
Materials and Methods
Strains.
Strains were derived from the wild-type RP437 (17). An in-frame deletion in fliC was generated by using the λ-Red system/FLP recombinase protocol (18). Other constructs were made by using the λ-Red system to replace the chromosomal gene with a DNA fragment carrying kanR and ccdB under control of the rhamnose promoter, and then selecting for allele replacement by survival on rhamnose minimal medium (K. A. Datsenko and B. L. Wanner, personal communication). motA448 (19) was PCR-amplified from MS5037 (20), ΔcheY from pVS20 (21), and wild-type motA from RP437. The latter fragment was subcloned into pRha67 under a rhamnose-inducible promoter (12). The mutation in motA448 was found by sequencing to be G43E.
Labeling Hooks with Gold Spheres.
Colloidal gold (diameter 60 nm; British Biocell International) was conjugated with anti-rabbit IgG (R5506, Sigma) following a method adapted from Liao et al. (22): IgG was activated with succinimidyl 6-[3-(2-pyridyldithio)-propionamido]hexanoate (LC-SPDP; 21651, Pierce) following the instructions from the manufacturer; 2.5 μl of the final solution was added to 500 μl of the gold suspension, and the mixture was incubated at 23°C for 2 h; then 10 μl of 1 mM O-[2-(3-mercaptopropionylamino)ethyl]-O′-methylpolyethylene glycol 5000 (mPEG-SH 5000; 11124, Fluka) was added, and the mixture was incubated at 23°C overnight. Cells were grown in tryptone broth (at 33°C to OD600 ≈0.6) and washed and concentrated three times in motility medium (10 mM potassium phosphate, 0.1 mM EDTA, 10 mM lactate, pH 7.0); 2.5 μl of anti-FlgE antibody (1 mg/ml) was added to a 100-μl aliquot, and the mixture was incubated at 23°C for 25 min. The antibody-treated cells were washed twice with 300 μl of motility medium and gently resuspended in 40 μl of motility medium. One hundred microliters of the IgG-conjugated gold solution was centrifuged at 1,100 × g for 3 min, and the pellet was mixed with the 40-μl cell suspension. The mixture was left at 23°C for 25 min, and then the cells were centrifuged, decanted, and resuspended in 200 μl of motility medium; 30 μl of this suspension was added to a polylysine coated glass coverslip, allowed to stand for 5 min, and then assembled in a flow cell (23), which was rinsed with motility medium. The bottom window of the flow cell was quartz.
Labeling Filament Stubs with Latex Spheres.
Cells were prepared, adsorbed to polylysine-coated glass, and exposed to 1,000-nm-diameter latex spheres (Polysciences), as described in ref. 9.
Laser Scattering Microscopy.
A truncated 60° prism, made by cutting 30° sectors off of opposite ends of a 25 × 25 × 10 mm silica window (leaving one side of the window square and the other rectangular), was mounted on a Nikon Optiphot microscope above a long-working-distance phase-contrast condenser. This allowed gold spheres to be monitored by light scattering and cells and latex spheres to be observed by phase contrast. The bottom of the flow cell was mated to the top of the prism (the square side of the window) with glycerol. Temperature was controlled by a Peltier system similar to that described in ref. 24 and checked with a small thermocouple. Light from a 655-nm diode laser (DL5147-042, run at ≈8 mW with a DLC500 controller, Thorlabs), with its plane of polarization horizontal, was reflected by a mirror sitting on the stage at the side of the prism at an angle below the critical angle for total internal reflection at the quartz-water interface but above the critical angle for total internal reflection at the glass-air interface, thus allowing the light to pass through the flow cell and reflect at the coverslip. Cells were imaged from above with a Nikon Plan 40/0.65 BM objective, and light from gold or latex spheres was focused onto a 0.2-mm-diameter pinhole in front of a photomultiplier tube (R7400U-20, Hamamatsu). The output was AC-coupled to an 8-pole low-pass Bessel filter (3384, Krohn-Hite) with a cutoff frequency of 2 kHz and sampled at 3 kHz using LabVIEW. Power spectra were computed for successive 1-s blocks of data. Speed-time plots for the resurrection experiments were smoothed with a median filter of rank 4.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Sandy Parkinson, Karen Fahrner, and Veda Nathan for advice on strain construction, Carl Gunderson and Anca Segall (San Diego State University, San Diego, CA) for the gift of rhamnose-inducible expression vectors, Karen Fahrner and Tom Shimizu for helpful discussions, and David Blair, Karen Fahrner, Gabriel Hosu, Birgit Scharf, and Tom Shimizu for comments on the manuscript. This work was supported by National Institutes of Health Grant AI016478.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711539105/DC1.
References
- 1.Blair DF. Flagellar movement driven by proton translocation. FEBS Lett. 2003;545:86–95. doi: 10.1016/s0014-5793(03)00397-1. [DOI] [PubMed] [Google Scholar]
- 2.Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochemistry. 2001;40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 3.Blair DF, Berg HC. Restoration of torque in defective flagellar motors. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 4.Reid SW, Leake MC, Chandler JH, Lo C-J, Armitage JP, Berry RM. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci USA. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Berg HC. Torque-speed relationship of the flagellar rotary motor. Biophys J. 2000;78:1036–1041. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Berg HC. Solvent-isotope and pH effects on flagellar rotation in Escherichia coli. Biophys J. 2000;78:2280–2284. doi: 10.1016/S0006-3495(00)76774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg HC, Turner L. Torque generated by the flagellar motor of Escherichia coli. Biophys J. 1993;65:2201–2216. doi: 10.1016/S0006-3495(93)81278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryu WS, Berry RM, Berg HC. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature. 2000;403:444–447. doi: 10.1038/35000233. [DOI] [PubMed] [Google Scholar]
- 10.Leibler S, Huse DA. Porters versus rowers: A unified stochastic model of motor proteins. J Cell Biol. 1993;121:1357–1368. doi: 10.1083/jcb.121.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing J, Bai F, Berry R, Oster G. Torque-speed relationship of the bacterial flagellar motor. Proc Natl Acad Sci USA. 2006;103:1260–1265. doi: 10.1073/pnas.0507959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giacalone MJ, Gentile AM, Lovitt BT, Berkley NL, Gunderson CW, Surber MW. Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. BioTechniques. 2006;40:355–364. doi: 10.2144/000112112. [DOI] [PubMed] [Google Scholar]
- 13.Scharf BE, Fahrner KA, Turner L, Berg HC. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Block SM, Blair DF, Berg HC. Compliance of bacterial flagella measured with optical tweezers. Nature. 1989;338:514–517. doi: 10.1038/338514a0. [DOI] [PubMed] [Google Scholar]
- 15.Block SM, Blair DF, Berg HC. Compliance of bacterial polyhooks measured with optical tweezers. Cytometry. 1991;12:492–496. doi: 10.1002/cyto.990120605. [DOI] [PubMed] [Google Scholar]
- 16.Sowa Y, Rowe AD, Leake MC, Yakushi T, Homma M, Ishijima A, Berry RM. Direct observation of steps in rotation of the bacterial flagellar motor. Nature. 2005;437:916–919. doi: 10.1038/nature04003. [DOI] [PubMed] [Google Scholar]
- 17.Parkinson JS. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armstrong JB, Adler J. Genetics of motility in Escherichia coli: Complementation of paralyzed mutants. Genetics. 1967;56:363–373. doi: 10.1093/genetics/56.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silverman M, Simon M. Operon controlling motility and chemotaxis in E. coli. Nature. 1976;264:577–580. doi: 10.1038/264577a0. [DOI] [PubMed] [Google Scholar]
- 21.Sourjik V, Berg HC. Location of components of the chemotaxis machinery of Escherichia coli using fluorescent-protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 22.Liao H, Hafner JH. Gold nanorod bioconjugates. Chem Mater. 2005;17:4636–4641. [Google Scholar]
- 23.Berg HC, Block SM. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J Gen Microbiol. 1984;130:2915–2920. doi: 10.1099/00221287-130-11-2915. [DOI] [PubMed] [Google Scholar]
- 24.Khan S, Berg HC. Isotope and thermal effects in chemiosmotic coupling to the flagellar motor of Streptococcus. Cell. 1983;32:913–919. doi: 10.1016/0092-8674(83)90076-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.